Evaluating Genome Assemblies for Optimized Completeness and Accuracy of Reference Gene Sequences in Wheat, Rye, and Triticale
Abstract
1. Introduction
2. Results
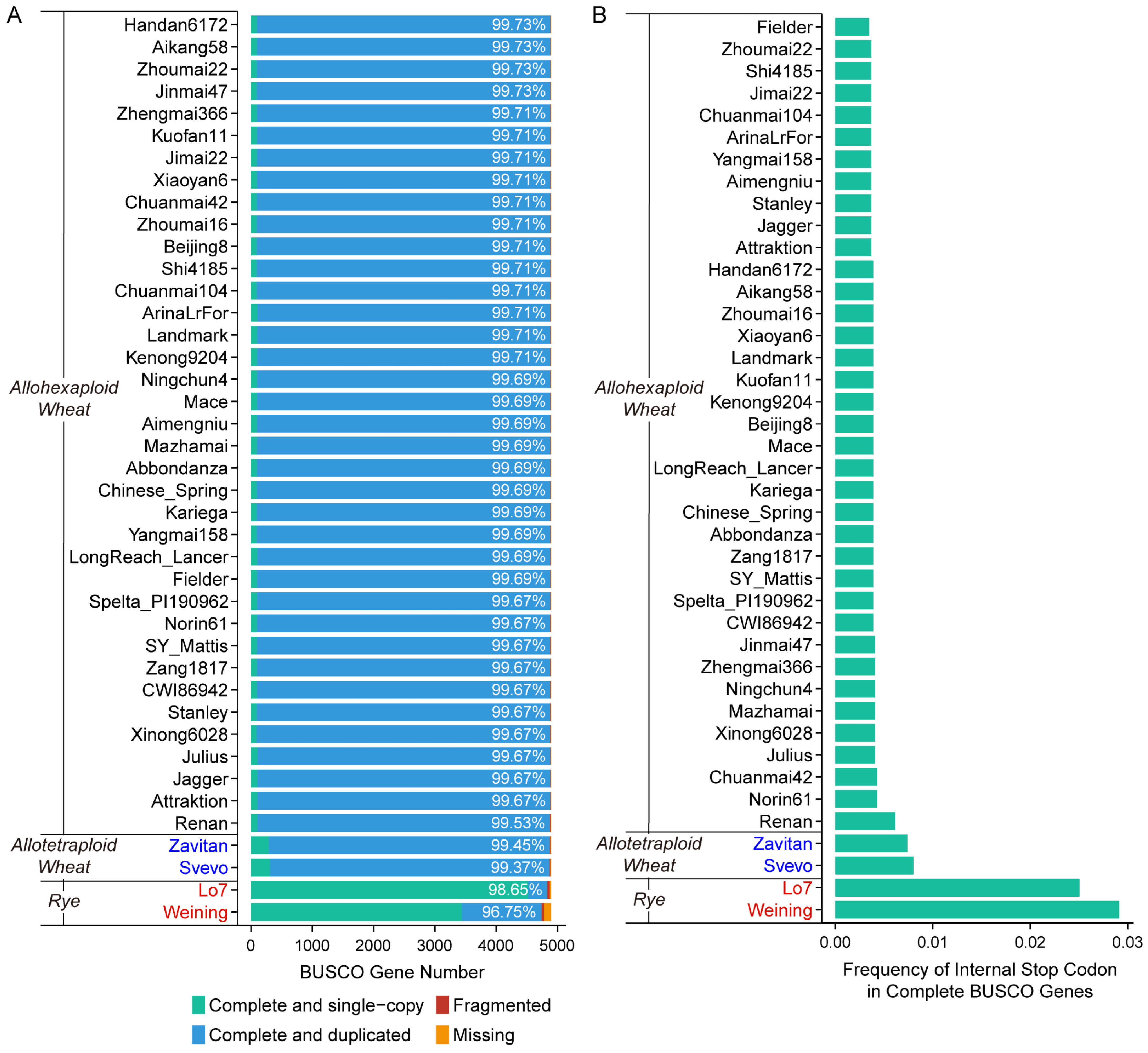
2.1. Differences in the Key Parameters of Triticeae Genome Assemblies
| Species | Genotype | Genome Type | Assembly Length (Gb) | Predicted Gene Number * | Number of Contigs | Number of Scaffolds | N50 of Contigs | N50 of Scaffolds | Percent of Gaps | Assembly Version | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Triticum aestivum | Chinese Spring | AABBDD | 14.58 | 106,914 | 306,707 | 22 | 341 Kb | 713 Mb | 1.78% | v2.1 | [32] |
| Zang1817 | AABBDD | 14.71 | 107,336 | 822,893 | 383,261 | 66 Kb | 704 Mb | 1.05% | v1 | [33] | |
| ArinaLrFor | AABBDD | 14.66 | 117,569 | 501,458 | 22 | 80 Kb | 697 Mb | 0.82% | v3.0 | [3] | |
| Jagger | AABBDD | 14.55 | 115,873 | 705,558 | 22 | 85 Kb | 705 Mb | 1.67% | v1.1 | ||
| Julius | AABBDD | 14.39 | 115,552 | 620,780 | 22 | 77 Kb | 708 Mb | 1.21% | v1.0 | ||
| Landmark | AABBDD | 14.44 | 115,515 | 662,766 | 22 | 81 Kb | 705 Mb | 1.92% | v1.0 | ||
| LongReach Lancer | AABBDD | 14.29 | 116,473 | 508,146 | 22 | 75 Kb | 688 Mb | 0.81% | v1.0 | ||
| Mace | AABBDD | 14.36 | 116,367 | 533,095 | 22 | 71 Kb | 690 Mb | 0.87% | v1.0 | ||
| Norin61 | AABBDD | 14.91 | 116,106 | 462,063 | 22 | 66 Kb | 712 Mb | 0.78% | v1.1 | ||
| spelta_PI190962 | AABBDD | 14.45 | 116,395 | 487,212 | 22 | 83 Kb | 707 Mb | 0.76% | v1.0 | ||
| Stanley | AABBDD | 14.47 | 115,781 | 603,408 | 22 | 71 Kb | 713 Mb | 1.68% | v1.2 | ||
| SY Mattis | AABBDD | 14.96 | 117,276 | 1,063,653 | 22 | 76 Kb | 698 Mb | 1.03% | v1.0 | ||
| Fielder | AABBDD | 14.70 | 116,480 | 5202 | 3795 | 20 Mb | 717 Mb | 0.00% | v1 | [34] | |
| Kariega | AABBDD | 14.68 | 116,838 | 7395 | 22 | 26 Mb | 717 Mb | 0.13% | v1 | [35] | |
| Renan | AABBDD | 14.26 | 109,653 | 12,779 | 359 | 2 Mb | 703 Mb | 1.79% | v2.1 | [36] | |
| Attraktion | AABBDD | 14.24 | NA | 1553 | 21 | 17 Mb | 703 Mb | 0.00% | NA | [37] | |
| Kenong9204 | AABBDD | 14.47 | 105,692 | 102,806 | 22 | 373 Kb | 714 Mb | 0.74% | NA | [38] | |
| Aikang58 | AABBDD | 14.76 | 119,350 | 279,897 | 158,176 | 237 Kb | 717 Mb | 0.63% | NA | [39] | |
| Chuanmai104 | AABBDD | 14.78 | 122,554 | 6065 | 5398 | 58 Mb | 727 Mb | 0.00% | v2.0 | [40] | |
| CWI86942 | AABBDD | 14.57 | 147,646 | 4780 | 22 | 44 Mb | 726 Mb | 0.00% | v1 | [41] | |
| Abbondanza | AABBDD | 14.70 | 134,090 | 7905 | 6567 | 25 Mb | 719 Mb | 0.00% | NA | [42] | |
| Aimengniu | AABBDD | 15.01 | 135,639 | 19,536 | 18,163 | 24 Mb | 727 Mb | 0.00% | NA | ||
| Beijing8 | AABBDD | 14.83 | 136,169 | 5995 | 3708 | 19 Mb | 718 Mb | 0.00% | NA | ||
| Chuanmai42 | AABBDD | 14.66 | 133,391 | 4876 | 3906 | 41 Mb | 722 Mb | 0.00% | NA | ||
| Handan6172 | AABBDD | 15.10 | 138,709 | 11,413 | 8862 | 26 Mb | 735 Mb | 0.00% | NA | ||
| Jimai22 | AABBDD | 14.62 | 134,424 | 5417 | 4268 | 33 Mb | 726 Mb | 0.00% | NA | ||
| Jinmai47 | AABBDD | 15.05 | 136,282 | 8175 | 2790 | 21 Mb | 744 Mb | 0.00% | NA | ||
| Kuofan11 | AABBDD | 14.86 | 133,789 | 9446 | 8185 | 30 Mb | 726 Mb | 0.00% | NA | ||
| Mazhamai | AABBDD | 14.67 | 135,896 | 6584 | 4985 | 32 Mb | 721 Mb | 0.00% | NA | ||
| Ningchun4 | AABBDD | 14.58 | 133,773 | 4996 | 3649 | 31 Mb | 714 Mb | 0.00% | NA | ||
| Shi4185 | AABBDD | 14.99 | 134,130 | 11,963 | 9171 | 26 Mb | 738 Mb | 0.00% | NA | ||
| Xiaoyan6 | AABBDD | 14.87 | 138,457 | 14,981 | 13,674 | 25 Mb | 720 Mb | 0.00% | NA | ||
| Xinong6028 | AABBDD | 14.69 | 136,471 | 4827 | 3522 | 31 Mb | 723 Mb | 0.00% | NA | ||
| Yangmai158 | AABBDD | 14.70 | 135,207 | 6028 | 4033 | 25 Mb | 714 Mb | 0.00% | NA | ||
| Zhengmai366 | AABBDD | 14.83 | 135,296 | 11,915 | 8003 | 26 Mb | 735 Mb | 0.00% | NA | ||
| Zhoumai16 | AABBDD | 15.06 | 136,271 | 6413 | 3366 | 33 Mb | 742 Mb | 0.00% | NA | ||
| Zhoumai22 | AABBDD | 15.04 | 135,054 | 13,808 | 10,954 | 26 Mb | 729 Mb | 0.00% | NA | ||
| Triticum turgidum | Svevo | AABB | 10.46 | 66,559 | 476,825 | 15 | 56 Kb | 722 Mb | 1.55% | v1 | [4] |
| Zavitan | AABB | 10.68 | 88,002 | 492,885 | 148,296 | 57 Kb | 747 Mb | 3.31% | v2.1 | [43] | |
| Secale cereale | Lo7 | RR | 6.74 | 34,441 | 314,632 | 8 | 72 Kb | 900 Mb | 2.31% | v1p1p1 | [44] |
| Weining | RR | 7.74 | 43,928 | 71,705 | 8 | 441 Kb | 1024 Mb | 0.10% | NA | [8] |
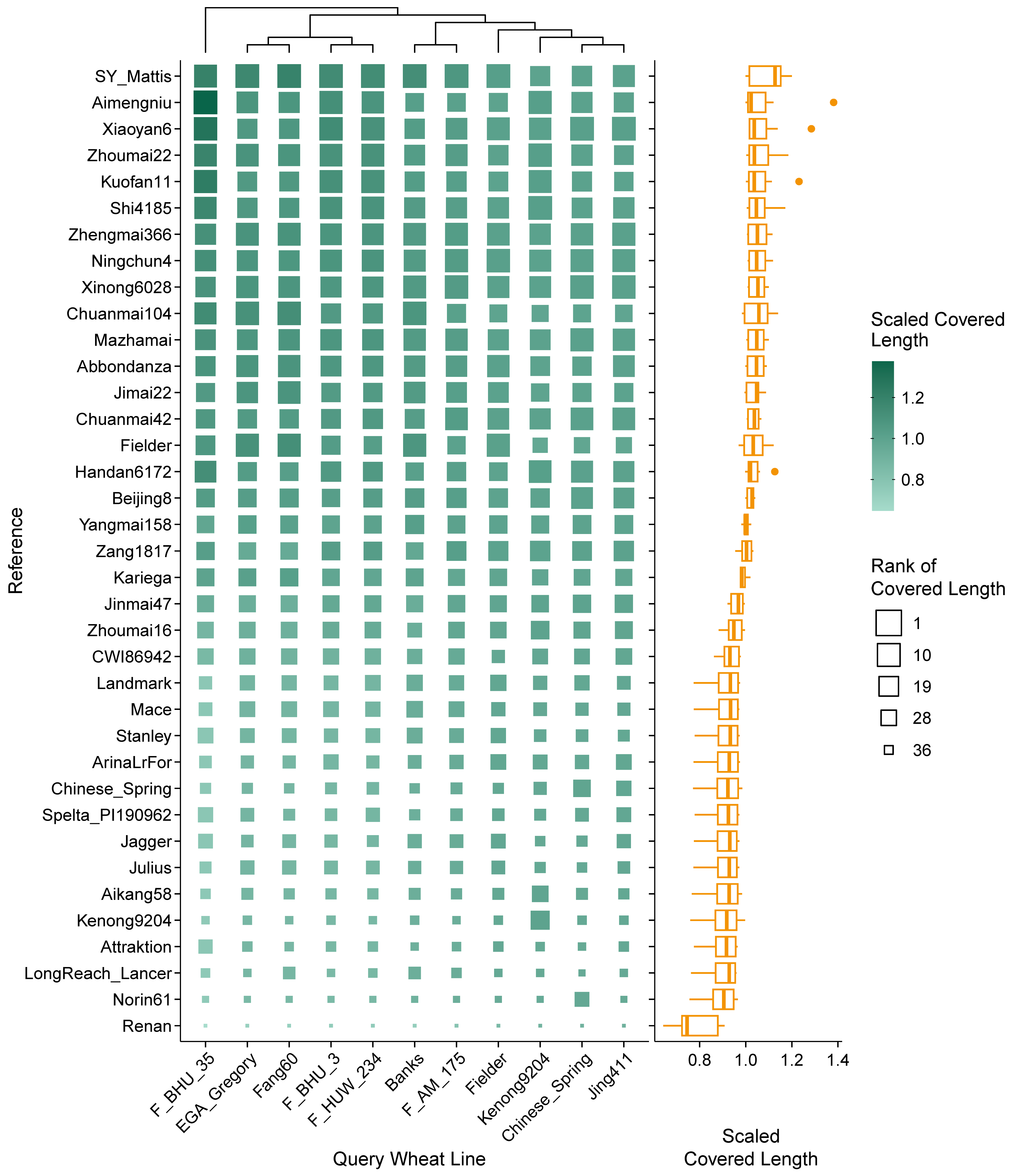
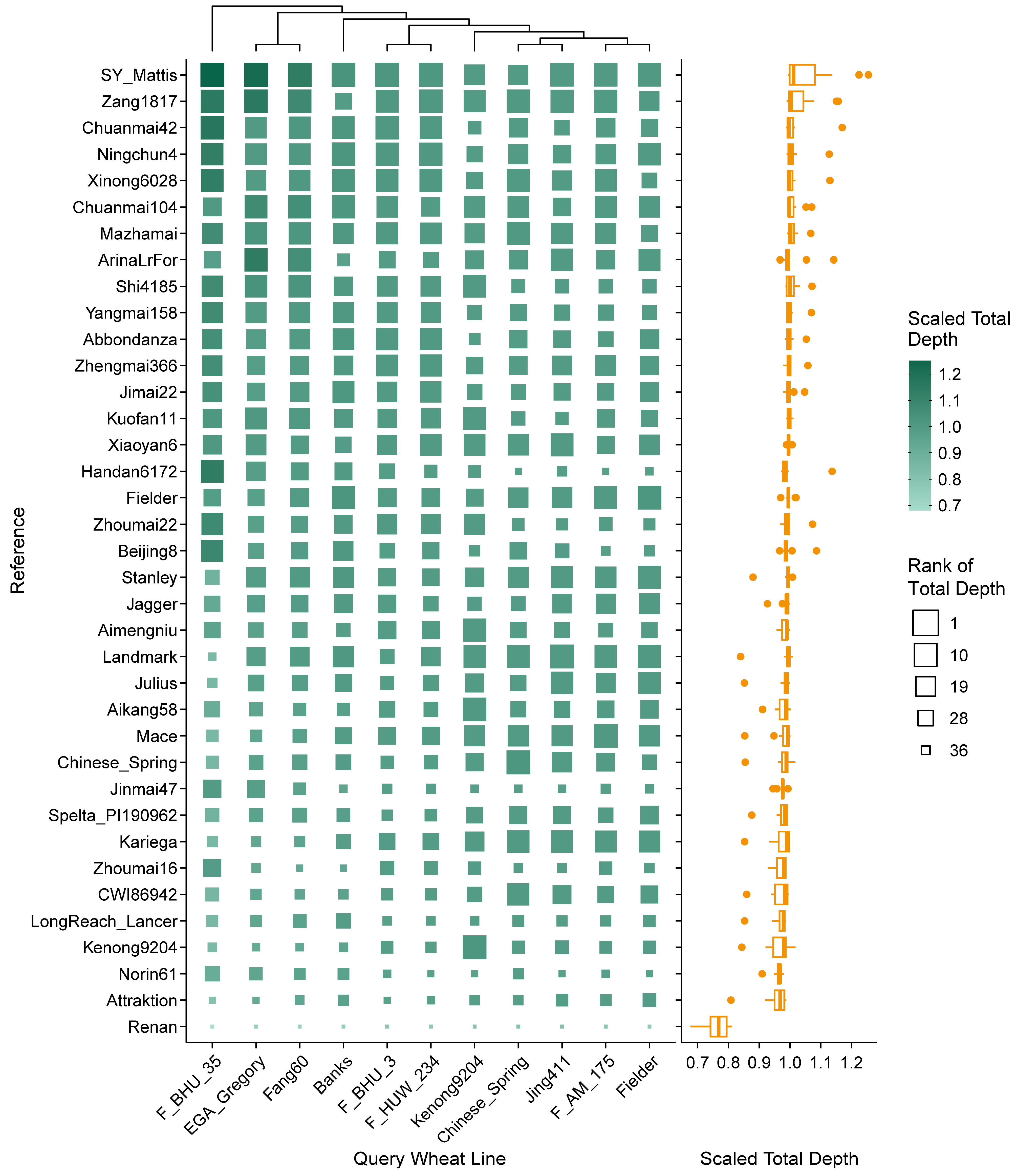
2.2. Efficiency Differences in Reference-Based Transcript Mapping in Wheat
2.3. Efficiency Differences in Reference-Based Transcript Mapping in Rye
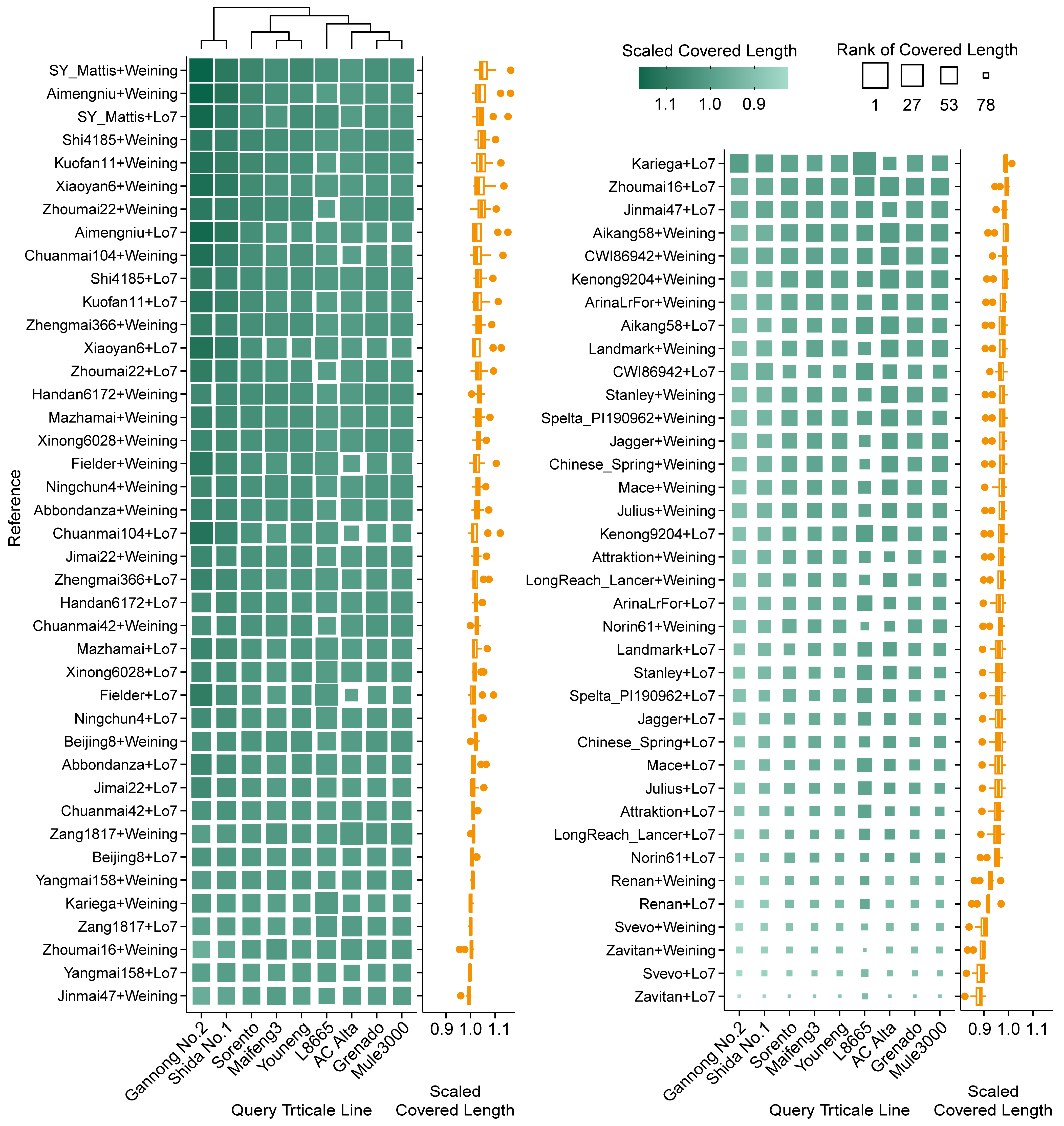
2.4. Efficiency Differences in Reference-Based Transcript Mapping in Triticale
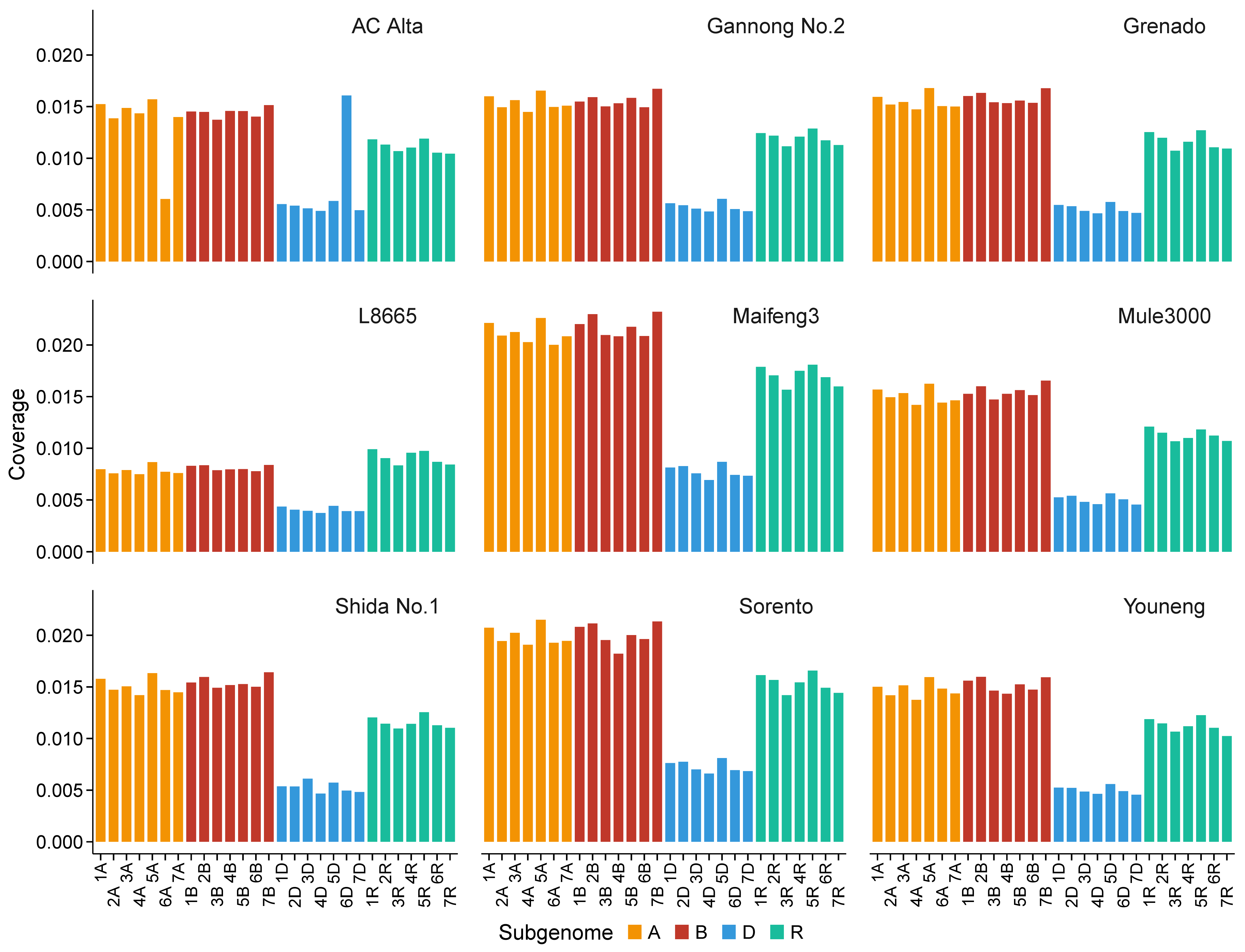
2.5. Karyotype Analysis Using RNA-Seq-Based Read Mapping in Triticale
3. Discussion
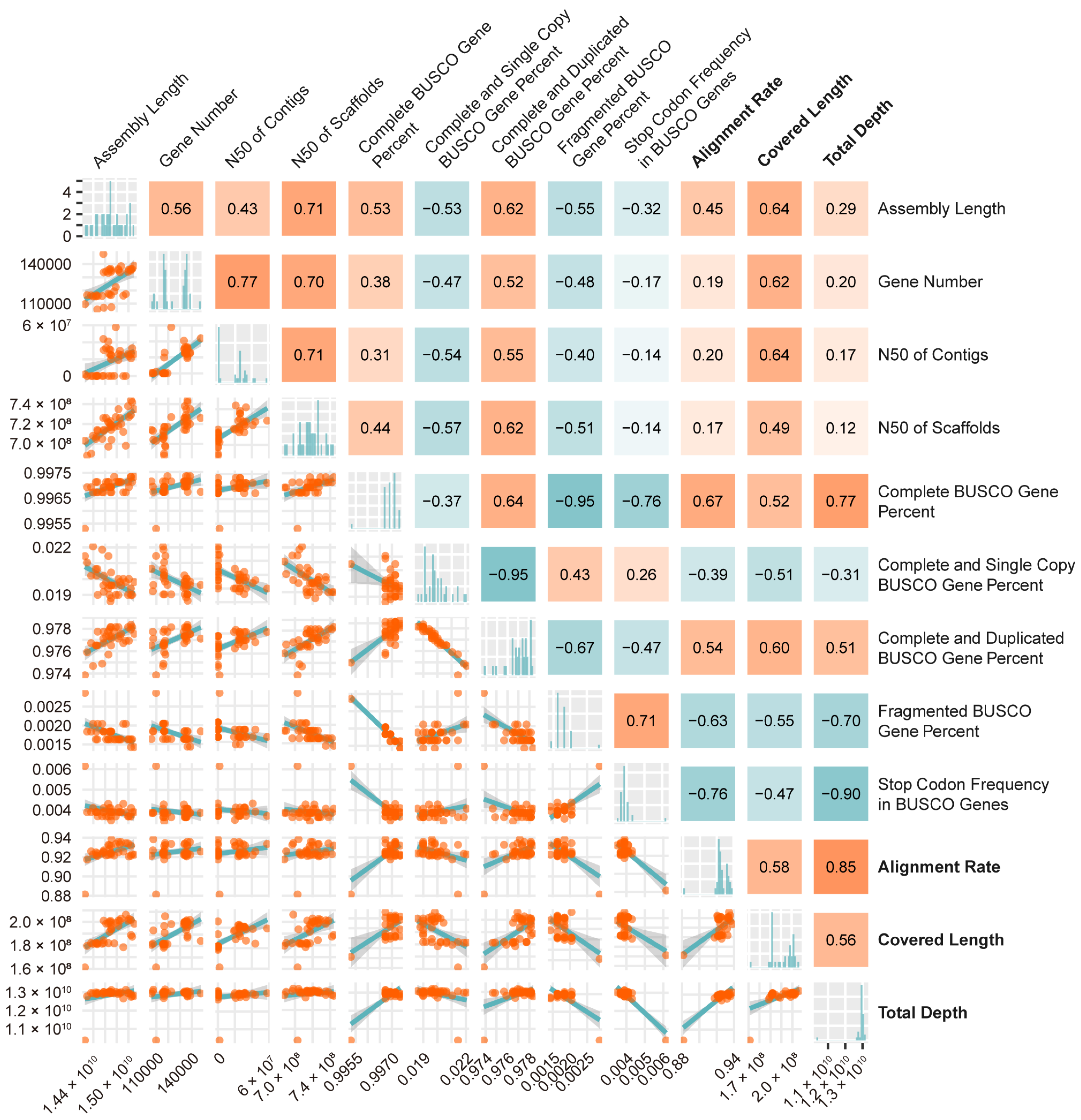
3.1. Correctness Rather than Number of Complete BUSCO Genes Is the Priority to Characterize Sequence Accuracy of Wheat Assemblies
3.2. Efficient References to Study Gene Structure and Expression in Wheat, Rye, and Triticale
4. Materials and Methods
4.1. Data Source
4.2. BUSCO Assessing of Genome Assemblies
4.3. Plant Materials and Growth Conditions
4.4. RNA Extraction, RNA-Seq Library Construction and Transcriptome Sequencing
4.5. RNA-Seq Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayalew, H.; Anderson, J.D.; Krom, N.; Tang, Y.; Butler, T.J.; Rawat, N.; Tiwari, V.; Ma, X.-F. Genotyping-by-sequencing and genomic selection applications in hexaploid triticale. G3 Genes|Genomes|Genet. 2022, 12, jkab413. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, M.; Özkan, H.; Komatsuda, T.; Mascher, M. Evolution and domestication of rye. In The Rye Genome; Rabanus-Wallace, M.T., Stein, N., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 85–100. [Google Scholar]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef]
- Randhawa, H.S.; Bona, L.; Graf, R.J. Triticale breeding—Progress and prospect. In Triticale; Eudes, F., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 15–32. [Google Scholar]
- Kuleung, C.; Baenziger, P.S.; Dweikat, I. Transferability of SSR markers among wheat, rye, and triticale. Theor. Appl. Genet. 2004, 108, 1147–1150. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Bian, J.; Lu, F.; Jiao, Y.; He, H. Triticeae crop genome biology: An endless frontier. Front. Plant Sci. 2023, 14, 1222681. [Google Scholar]
- Li, G.; Wang, L.; Yang, J.; He, H.; Jin, H.; Li, X.; Ren, T.; Ren, Z.; Li, F.; Han, X.; et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 2021, 53, 574–584. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, B.; Yao, Y.; Guo, Z.; Jia, H.; Kong, L.; Zhang, A.; Ma, W.; Ni, Z.; Xu, S.; et al. Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 2022, 65, 1718–1775. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [PubMed]
- The International Wheat Genome Sequencing Consortium; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Denton, J.F.; Lugo-Martinez, J.; Tucker, A.E.; Schrider, D.R.; Warren, W.C.; Hahn, M.W. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput. Biol. 2014, 10, e1003998. [Google Scholar] [CrossRef]
- Vuruputoor, V.S.; Monyak, D.; Fetter, K.C.; Webster, C.; Bhattarai, A.; Shrestha, B.; Zaman, S.; Bennett, J.; McEvoy, S.L.; Caballero, M.; et al. Welcome to the big leaves: Best practices for improving genome annotation in non-model plant genomes. Appl. Plant Sci. 2023, 11, e11533. [Google Scholar] [CrossRef]
- Coombes, B.; Lux, T.; Akhunov, E.; Hall, A. Introgressions lead to reference bias in wheat RNA-seq analysis. BMC Biol. 2024, 22, 56. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, X.; Wu, L.; Zhang, L.; Liu, G.; Xia, C.; Liu, X.; Kong, X. Transcriptome profiling of developing leaf and shoot apices to reveal the molecular mechanism and co-expression genes responsible for the wheat heading date. BMC Genom. 2021, 22, 468. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Yang, W.; Li, T.; Shoaib, M.; Sun, J.; Liu, D.; Li, X.; Nie, Y.; Tian, X.; Zhang, A. Combined transcriptome and proteome analysis of anthers of AL-type cytoplasmic male sterile line and its maintainer line reveals new insights into mechanism of male sterility in common wheat. Front. Genet. 2021, 12, 762332. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.-p.; Wang, Y.; Zhou, T.; Huang, J.-y.; Yue, C.-p. Combined morpho-physiological, ionomic and transcriptomic analyses reveal adaptive responses of allohexaploid wheat (Triticum aestivum L.) to iron deficiency. BMC Plant Biol. 2022, 22, 234. [Google Scholar] [CrossRef]
- Cao, D.; Wang, D.; Li, S.; Li, Y.; Hao, M.; Liu, B. Genotyping-by-sequencing and genome-wide association study reveal genetic diversity and loci controlling agronomic traits in triticale. Theor. Appl. Genet. 2022, 135, 1705–1715. [Google Scholar] [CrossRef]
- Bimpong, D.; Zhao, L.; Ran, M.; Zhao, X.; Wu, C.; Li, Z.; Wang, X.; Cheng, L.; Fang, Z.; Hu, Z.; et al. Transcriptomic analysis reveals the regulatory mechanisms of messenger RNA (mRNA) and long non-coding RNA (lncRNA) in response to waterlogging stress in rye (Secale cereale L.). BMC Plant Biol. 2024, 24, 534. [Google Scholar] [CrossRef]
- Tsers, I.; Meshcherov, A.; Gogoleva, O.; Petrova, O.; Gogoleva, N.; Ponomareva, M.; Gogolev, Y.; Korzun, V.; Gorshkov, V. Alterations in the transcriptome of rye plants following the Microdochium nivale infection: Identification of resistance/susceptibility-related reactions based on RNA-Seq analysis. Plants 2021, 10, 2723. [Google Scholar] [CrossRef]
- Krępski, T.; Piasecka, A.; Święcicka, M.; Kańczurzewska, M.; Sawikowska, A.; Dmochowska-Boguta, M.; Rakoczy-Trojanowska, M.; Matuszkiewicz, M. Leaf rust (Puccinia recondita f. sp. secalis) triggers substantial changes in rye (Secale cereale L.) at the transcriptome and metabolome levels. BMC Plant Biol. 2024, 24, 107. [Google Scholar] [CrossRef]
- Wang, P.; Wang, F. A proposed metric set for evaluation of genome assembly quality. Trends Genet. 2023, 39, 175–186. [Google Scholar] [CrossRef]
- Parra, G.; Bradnam, K.; Ning, Z.; Keane, T.; Korf, I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009, 37, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Rhie, A.; McCarthy, S.A.; Fedrigo, O.; Damas, J.; Formenti, G.; Koren, S.; Uliano-Silva, M.; Chow, W.; Fungtammasan, A.; Kim, J.; et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 2021, 592, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing genomic data quality and beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef] [PubMed]
- Earth Biogenome Project’s Scientific Subcommittee for Sequencing and Assembly. Report on Assembly Standards. Available online: https://www.earthbiogenome.org/report-on-assembly-standards (accessed on 21 January 2025).
- Whibley, A.; Kelley, J.L.; Narum, S.R. The changing face of genome assemblies: Guidance on achieving high-quality reference genomes. Mol. Ecol. Resour. 2021, 21, 641–652. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Cai, G.; Li, H.; Lu, Y.; Huang, X.; Lee, J.; Müller, P.; Ji, Y.; Liang, S. Accuracy of RNA-Seq and its dependence on sequencing depth. BMC Bioinform. 2012, 13 (Suppl. S13), S5. [Google Scholar] [CrossRef]
- Daley, T.; Smith, A.D. Predicting the molecular complexity of sequencing libraries. Nat. Methods 2013, 10, 325–327. [Google Scholar] [CrossRef]
- Cai, G.; Liang, S.; Zheng, X.; Xiao, F. Local sequence and sequencing depth dependent accuracy of RNA-seq reads. BMC Bioinform. 2017, 18, 364. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]
- Guo, W.; Xin, M.; Wang, Z.; Yao, Y.; Hu, Z.; Song, W.; Yu, K.; Chen, Y.; Wang, X.; Guan, P.; et al. Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat. Commun. 2020, 11, 5085. [Google Scholar] [CrossRef]
- Sato, K.; Abe, F.; Mascher, M.; Haberer, G.; Gundlach, H.; Spannagl, M.; Shirasawa, K.; Isobe, S. Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder’. DNA Res. 2021, 28, dsab008. [Google Scholar] [CrossRef] [PubMed]
- Athiyannan, N.; Abrouk, M.; Boshoff, W.H.P.; Cauet, S.; Rodde, N.; Kudrna, D.; Mohammed, N.; Bettgenhaeuser, J.; Botha, K.S.; Derman, S.S.; et al. Long-read genome sequencing of bread wheat facilitates disease resistance gene cloning. Nat. Genet. 2022, 54, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Aury, J.-M.; Engelen, S.; Istace, B.; Monat, C.; Lasserre-Zuber, P.; Belser, C.; Cruaud, C.; Rimbert, H.; Leroy, P.; Arribat, S.; et al. Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding. GigaScience 2022, 11, giac034. [Google Scholar] [CrossRef]
- Kale, S.M.; Schulthess, A.W.; Padmarasu, S.; Boeven, P.H.G.; Schacht, J.; Himmelbach, A.; Steuernagel, B.; Wulff, B.B.H.; Reif, J.C.; Stein, N.; et al. A catalogue of resistance gene homologs and a chromosome-scale reference sequence support resistance gene mapping in winter wheat. Plant Biotechnol. J. 2022, 20, 1730–1742. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Cui, F.; Han, X.; He, Y.; Zhao, L.; Zhang, N.; Zhang, H.; Zhu, H.; Liu, Z.; Ma, B.; et al. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204. Mol. Plant 2022, 15, 1440–1456. [Google Scholar] [CrossRef]
- Jia, J.; Zhao, G.; Li, D.; Wang, K.; Kong, C.; Deng, P.; Yan, X.; Zhang, X.; Lu, Z.; Xu, S.; et al. Genome resources for the elite bread wheat cultivar Aikang 58 and mining of elite homeologous haplotypes for accelerating wheat improvement. Mol. Plant 2023, 16, 1893–1910. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, F.; Deng, C.; Wan, H.; Tang, H.; Feng, J.; Wang, Q.; Yang, N.; Li, J.; Yang, W. Chromosome-level assembly of the synthetic hexaploid wheat-derived cultivar Chuanmai 104. Sci. Data 2024, 11, 670. [Google Scholar] [CrossRef]
- Cavalet-Giorsa, E.; González-Muñoz, A.; Athiyannan, N.; Holden, S.; Salhi, A.; Gardener, C.; Quiroz-Chávez, J.; Rustamova, S.M.; Elkot, A.F.; Patpour, M.; et al. Origin and evolution of the bread wheat D genome. Nature 2024, 633, 848–855. [Google Scholar] [CrossRef]
- Jiao, C.; Xie, X.; Hao, C.; Chen, L.; Xie, Y.; Garg, V.; Zhao, L.; Wang, Z.; Zhang, Y.; Li, T.; et al. Pan-genome bridges wheat structural variations with habitat and breeding. Nature 2024, 637, 384–393. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rodriguez, J.C.; Deal, K.R.; Avni, R.; Distelfeld, A.; McGuire, P.E.; Dvorak, J.; Luo, M.-C. Improved genome sequence of wild emmer wheat Zavitan with the aid of optical maps. G3 Genes|Genomes|Genet. 2019, 9, 619–624. [Google Scholar] [CrossRef]
- Rabanus-Wallace, M.T.; Hackauf, B.; Mascher, M.; Lux, T.; Wicker, T.; Gundlach, H.; Baez, M.; Houben, A.; Mayer, K.F.X.; Guo, L.; et al. Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 2021, 53, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, K.; Orabi, J.; Kristensen, P.S.; Sarup, P.; Jørgensen, L.N.; Jahoor, A. De novo transcriptome assembly, functional annotation, and expression profiling of rye (Secale cereale L.) hybrids inoculated with ergot (Claviceps purpurea). Sci. Rep. 2020, 10, 13475. [Google Scholar] [CrossRef]
- Abbas, Q.; Wilhelm, M.; Kuster, B.; Poppenberger, B.; Frishman, D. Exploring crop genomes: Assembly features, gene prediction accuracy, and implications for proteomics studies. BMC Genom. 2024, 25, 619. [Google Scholar] [CrossRef]
- Jauhal, A.A.; Newcomb, R.D. Assessing genome assembly quality prior to downstream analysis: N50 versus BUSCO. Mol. Ecol. Resour. 2021, 21, 1416–1421. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Keilwagen, J.; Lehnert, H.; Berner, T.; Badaeva, E.; Himmelbach, A.; Börner, A.; Kilian, B. Detecting major introgressions in wheat and their putative origins using coverage analysis. Sci. Rep. 2022, 12, 1908. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Shen, T.; Wang, X.; Yi, S.; Meng, T.; Sun, J.; Wang, X.; Qu, X.; Chen, S.; et al. A near-complete genome sequence of einkorn wheat provides insight into the evolution of wheat A subgenomes. Plant Commun. 2024, 5, 100768. [Google Scholar] [CrossRef] [PubMed]
- Rabanus-Wallace, M.T.; Wang, D.; Yang, J.; Li, G.; Stein, N. Assembling the rye genome. In The Rye Genome; Rabanus-Wallace, M.T., Stein, N., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 101–116. [Google Scholar]
- Martis, M.M.; Zhou, R.; Haseneyer, G.; Schmutzer, T.; Vrána, J.; Kubaláková, M.; König, S.; Kugler, K.G.; Scholz, U.; Hackauf, B.; et al. Reticulate evolution of the rye genome. Plant Cell 2013, 25, 3685–3698. [Google Scholar] [CrossRef]
- Kwiatek, M.T.; Nawracała, J. Chromosome manipulations for progress of triticale (×Triticosecale) breeding. Plant Breed. 2018, 137, 823–831. [Google Scholar] [CrossRef]
- Polkhovskaya, E.; Bolotina, A.; Merkulov, P.; Dudnikov, M.; Soloviev, A.; Kirov, I. Long-read cDNA sequencing revealed novel expressed genes and dynamic transcriptome landscape of triticale (x Triticosecale Wittmack) seed at different developing stages. Agronomy 2023, 13, 292. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, M.; Yang, G.; Yang, D.; Zhang, X.; Wang, Q.; Gao, J.; Mei, C. Evaluating Genome Assemblies for Optimized Completeness and Accuracy of Reference Gene Sequences in Wheat, Rye, and Triticale. Plants 2025, 14, 1140. https://doi.org/10.3390/plants14071140
Yan M, Yang G, Yang D, Zhang X, Wang Q, Gao J, Mei C. Evaluating Genome Assemblies for Optimized Completeness and Accuracy of Reference Gene Sequences in Wheat, Rye, and Triticale. Plants. 2025; 14(7):1140. https://doi.org/10.3390/plants14071140
Chicago/Turabian StyleYan, Mingke, Guodong Yang, Dongming Yang, Xin Zhang, Quanzhen Wang, Jinghui Gao, and Chugang Mei. 2025. "Evaluating Genome Assemblies for Optimized Completeness and Accuracy of Reference Gene Sequences in Wheat, Rye, and Triticale" Plants 14, no. 7: 1140. https://doi.org/10.3390/plants14071140
APA StyleYan, M., Yang, G., Yang, D., Zhang, X., Wang, Q., Gao, J., & Mei, C. (2025). Evaluating Genome Assemblies for Optimized Completeness and Accuracy of Reference Gene Sequences in Wheat, Rye, and Triticale. Plants, 14(7), 1140. https://doi.org/10.3390/plants14071140





