Plant Signaling Hormones and Transcription Factors: Key Regulators of Plant Responses to Growth, Development, and Stress
Abstract
1. Introduction
2. Plant Signaling Hormones
2.1. Auxin
2.2. Gibberellic Acid
2.3. Cytokinin
2.4. Abscisic Acid
2.5. Ethylene
2.6. Brassinosteroid
2.7. Jasmonic Acid
2.8. Salicylic Acid
2.9. Strigolactone
3. Transcription Factors
3.1. NAC
3.2. HSF
3.3. WRKY
3.4. AP2/ERF
3.5. bZIP
3.6. MADS-Box
3.7. MYB Family
| TF | Plant | Gene/Genotype/TF | Function | References |
|---|---|---|---|---|
| NAC | A. thaliana | ATAF1 overexpression | Decrease resistance while loss-of-function mutants increase resistance to B. cinerea, Pst DC3000, and Alternaria brassicicola | [534] |
| ATAF2 overexpression | Decrease resistance to Fusarium oxysporum | [535] | ||
| ANAC062/NTL6 overexpression | Increased resistance to Pst DC3000 | [536] | ||
| ANAC055 overexpression | Drought, high salinity, ABA signaling | [537] | ||
| CBNAC/NTL9 | Stomatal immunity to Pst DC3000 | [538,539] | ||
| GmNAC20 overexpression | Salt and freezing tolerance | [540] | ||
| NAC4 overexpression | Increase Pst DC3000-induced cell death | [541] | ||
| RD26 overexpression | Drought, salt, ABA signaling | [542] | ||
| Rice | OsNAC4 overexpression | Drought, salt, cold tolerance | [543] | |
| OsNAC6 overexpression | Drought and salt tolerance | [544] | ||
| ONAC10 overexpression | Drought, high salinity, low-temperature tolerance | [545] | ||
| SNAC2 overexpression | Salt, drought, salinity, cold, and wounding | [546] | ||
| Tomato | SlSRN1 | Silencing attenuated resistance to B. cinerea and Pst DC3000 | [547] | |
| JA2 | JA2-suppressed plants increased susceptibility to Pst DC3118 | [548] | ||
| Wheat | TaNAC21/TaNAC22 | Silencing increased resistance to Puccinia striiformis f. sp. tritici | [549,550] | |
| TaNAC6s overexpression | Enhance resistance to Blumeria graminis f. sp. tritici | [300] | ||
| HSF | A. thaliana | Hsf30 (TF) | Help to maintain protein stability and cellular function under high temperature | [551] |
| Hsp70 (HSP) | Prevent protein aggregation, particularly under stress conditions like heat, drought, and salinity | [552] | ||
| PtHSP17.8 overexpression | Increases survival rate and root length under heat and salt stresses | [553] | ||
| MsHSP16.9 overexpression | Improves the tolerance of a plant to heat by alleviating the damages of ROS and regulating the expression levels of stress-related genes | [554] | ||
| Tobacco | RcHSP17.8 overexpression | Resistant to high temperatures and osmotic stresses | [555] | |
| Tomato | AtHsfA1b overexpression | Improve resistance to chilling in transgenic tomato | [556] | |
| Rice | HsfA4a overexpression | Enhance Cd tolerance | [544] | |
| Wheat | ||||
| WRKY | A. thaliana | AtWRKY57 | Enhance drought tolerance | [557] |
| AtbHLH17 | Upregulate under drought and oxidative stress | [558] | ||
| AtWRKY28 | ||||
| AtWRKY23 | Participate in root development by controlling auxin distribution | [559] | ||
| VvWRKY11 overexpression | Higher tolerance to water stress induced by mannitol and response to dehydration stress | [560] | ||
| Rice | OsWRKY45 overexpression | Enhance salt and drought tolerance and increase disease resistance | [385] | |
| OsWRKY53 | Involved in pathogen defense pathways | [385] | ||
| WRKY72 overexpression | Increase sensitivity to sugar starvation stress | [561] | ||
| OsWRKY11 overexpression | Decrease plant height, tolerance to drought stress | [562] | ||
| Soybean | GmWRKY20 | Induced by ABA, salt, cold, and drought, and involved in several abiotic stress-related responses | [385] | |
| GmWRKY13 | Sensitive to salt and mannitol, negative regulator in ABA signaling | [563] | ||
| GmWRKY54 | Tolerance to salt and drought | [563] | ||
| B. campestris | BcWRKY46 | Tolerance to salt and drought | [564] | |
| Zea mays | ZmWRKY17 | Reduce salt tolerance | [565] | |
| Malus domestica | MdWRKY30 | Tolerance to salt and osmotic stress | [566] | |
| MdWRKY100 | Sensitive to salt | [567] | ||
| Ap2/ERF | A. thaliana | AtERF7 overexpression | Reduce the sensitivity of defense cells to ABA and increase water loss | [568] |
| AtERF9 AtPDF1.2 | Enhance resistance to Botrytis cinerea | [439] | ||
| AtC4H At4CL1 | Key players in phenylpropanoid metabolism and cell wall formation | [569] | ||
| ZmEREB20 | Enhance ABA sensitivity and cause delayed seed germination under salt stress by regulating abscisic acid and gibberellin-related genes | [441] | ||
| Rice | OsERF71 overexpression | Reduce water loss, resulting in increased tolerance to drought stress | [570] | |
| OsDREB2B overexpression | Improve drought tolerance | [571] | ||
| OsERF19 overexpression | Increase the tolerance of plants to salt stress | [572] | ||
| OsDREB2A OsDREB1F | Water scarcity and high salt stress tolerance | [573] | ||
| Soybean | GmERF113 | Enhance resistance to infection of Phytophthora sojae | [437] | |
| Apple | MdERF11 | Positively regulate defense responses against Botryosphaeria dothidea by promoting SA biosynthesis | [440] | |
| bZIP | A. thaliana | AtbZIP15 AtbZIP35 AtbZIP36 AtbZIP37 AtbZIP38 | Abscisic acid biosynthesis and stress signaling | [574,575] |
| AtbZIP17 AtbZIP28 | Regulate the unfolded protein response pathway | [576,577] | ||
| AtbZIP10 | Pathogen defense | [578] | ||
| Barley | HvbZIP56 | Regulate zinc deficiency | [579] | |
| HvbZIP62 | ||||
| Rice | OsbZIP23 overexpression | Improve drought resistance and salt tolerance | [580] | |
| OsbZIP46 | Mediate in ABA signaling and drought resistance | [581] | ||
| OsbZIP72 overexpression | ABA sensitivity and drought resistance | [581] | ||
| Soybean | GmbZIP14 | Salt, drought, and low-temperature stress responses | [582] | |
| GmbZIP146 | Flowering, salt and drought stress responses | [583,584,585] | ||
| GmbZIP53 | ABA signaling, salt, and low-temperature stress responses | [586] | ||
| GmbZIP45 | Pathogen response | [587] | ||
| MADS-box | A. thaliana | CmANR1 overexpression | Lateral root formation and growth, auxin biosynthesis, auxin polar transport, calcium ion signaling, ethylene biosynthesis, and cell cycle-related processes | [588] |
| AtAGL11 | Regulate fruit and transmitting tract development | [589] | ||
| B. napus | BnaAGL11 | Cause smaller, curly leaves and accelerated leaf senescence | [590] | |
| Bna.C09.AGL11 overexpression | ||||
| Bna.A04.ABI5 Bna.A05.ABI5, Bna.C04.ABI5-1 | Leaf senescence | [590] | ||
| Rice | OsMADS14, OsMADS15, | Floral meristem identity | [591] | |
| OsMADS18 | Germination, tillering, and inflorescence architecture | [592] | ||
| Sweet potato | IbMADS1 | Early-developing tuberous roots | [593] | |
| SRD1 | Metaxylem and cambium cell formation | [594] | ||
| MYB family | A. thaliana | AtMYB96 | Drought resistance by modulating cuticular wax biosynthesis | [506,507,595] |
| AtMYB41 overexpression | Synthesis and deposition of suberin-type aliphatic monomers in leaves rather than the usual cutin-type, which may result in discontinuities in the leaf cuticle and increase desiccation sensitivity in transgenic plants | [596,597] | ||
| AtMYB16 AtMYB106 | Cuticle biosynthesis | [505] | ||
| AtMYB2 AtMYB20 AtMYB73 AtMYB74 | Regulate salt tolerance by modulating gene expression under salt stress | [598] | ||
| Rice | OsMYB2 overexpression | Enhance cold resistance in transgenic rice | [599] | |
| OsMYB102 | Represses the ABA | |||
| Apple | MdMYB108L | Improve cold tolerance | [600] | |
| Tomato | SlMYB49 | Manage stress responses | [601] | |
| Pear | PbrMYB169 | Enhance lignification in the cell matrix of fruit | [602] |
4. Other Molecules Participate in Signaling and Stress Response
5. Future Prospects
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bhar, A.; Chakraborty, A.; Roy, A. Plant Responses to Biotic Stress: Old Memories Matter. Plants 2021, 11, 84. [Google Scholar] [CrossRef] [PubMed]
- Anzano, A.; Bonanomi, G.; Mazzoleni, S.; Lanzotti, V. Plant Metabolomics in Biotic and Abiotic Stress: A Critical Overview. Phytochem. Rev. 2022, 21, 5716. [Google Scholar] [CrossRef]
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Manivannan, P.; Panneerselvam, R.; Shao, M.A. Understanding Water Deficit Stress-Induced Changes in the Basic Metabolism of Higher Plants—Biotechnologically and Sustainably Improving Agriculture and the Ecoenvironment in Arid Regions of the Globe. Crit. Rev. Biotechnol. 2009, 29, 131–151. [Google Scholar] [CrossRef]
- Ahuja, I.; de Vos, R.C.; Bones, A.M.; Hall, R.D. Plant Molecular Stress Responses Face Climate Change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Pérez-Clemente, R.M.; Vives, V.; Zandalinas, S.I.; López-Climent, M.F.; Muñoz, V.; Gómez-Cadenas, A. Biotechnological Approaches to Study Plant Responses to Stress. Biomed Res. Int. 2013, 2013, 654120. [Google Scholar] [CrossRef]
- Grierson, C.; Du, J.-S.; Zabala, M.D.T.; Beggs, K.; Smith, C.; Holdsworth, M.; Bevan, M.W. Separate Cis Sequences and Trans Factors Direct Metabolic and Developmental Regulation of a Potato Tuber Storage Protein Gene. Plant J. 1994, 5, 815–826. [Google Scholar] [CrossRef]
- Sun, C.; Palmqvist, S.; Olsson, H.; Borén, M.; Ahlandsberg, S.; Jansson, C. A Novel WRKY Transcription Factor, SUSIBA2, Participates in Sugar Signaling in Barley by Binding to the Sugar-Responsive Elements of the Iso1 Promoter. Plant Cell 2003, 15, 2076–2092. [Google Scholar] [CrossRef]
- Klee, H.J. Ethylene Signal Transduction. Moving Beyond Arabidopsis. Plant Physiol. 2004, 135, 660–667. [Google Scholar] [CrossRef]
- Ghorbel, M.; Brini, F. Hormone Mediated Cell Signaling in Plants under Changing Environmental Stress. Plant Gene 2021, 28, 100335. [Google Scholar] [CrossRef]
- Raza, A.; Salehi, H.; Rahman, M.A.; Zahid, Z.; Madadkar Haghjou, M.; Najafi-Kakavand, S.; Charagh, S.; Osman, H.S.; Albaqami, M.; Zhuang, Y.; et al. Plant Hormones and Neurotransmitter Interactions Mediate Antioxidant Defenses under Induced Oxidative Stress in Plants. Front. Plant Sci. 2022, 13, 961872. [Google Scholar] [CrossRef]
- Boller, T.; He, S.Y. Innate Immunity in Plants: An Arms Race between Pattern Recognition Receptors in Plants and Effectors in Microbial Pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Dong, X. How Do Plants Achieve Immunity? Defence without Specialized Immune Cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-Triggered Immunity: From Pathogen Perception to Robust Defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Buscaill, P.; Rivas, S. Transcriptional Control of Plant Defence Responses. Curr. Opin. Plant Biol. 2014, 20, 35–46. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Liu, S.; Somssich, I.E. Transcriptional Events Defining Plant Immune Responses. Curr. Opin. Plant Biol. 2017, 38, 1–9. [Google Scholar] [CrossRef]
- Tanveer, M.; Yousaf, U. Plant Single-Cell Biology and Abiotic Stress Tolerance. In Plant Life Under Changing Environment; Tripathi, D.K., Singh, V.P., Chauhan, D.K., Sharma, S., Prasad, S.M., Dubey, N.K., Eds.; Academic Press: Amsterdam, The Netherlands, 2020; pp. 611–626. [Google Scholar] [CrossRef]
- Fahad, S.; Sonmez, O.; Saud, S.; Wang, D.; Wu, C.; Adnan, M.; Turan, V. (Eds.) Developing Climate-Resilient Crops: Improving Global Food Security and Safety; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Farooq, M.S.; Uzair, M.; Raza, A.; Habib, M.; Xu, Y.; Yousuf, M.; Yang, S.H.; Ramzan Khan, M. Uncovering the Research Gaps to Alleviate the Negative Impacts of Climate Change on Food Security: A Review. Front. Plant Sci. 2022, 13, 927535. [Google Scholar] [CrossRef]
- Vanwallendael, A.; Soltani, A.; Emery, N.C.; Peixoto, M.M.; Olsen, J.; Lowry, D.B. A Molecular View of Plant Local Adaptation: Incorporating Stress-Response Networks. Annu. Rev. Plant Biol. 2019, 70, 559–583. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic Stress, the Field Environment and Stress Combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and Biotic Stress Combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and Defense Responses in Plant Secondary Metabolites Production. Biol. Res. 2019, 52, 28. [Google Scholar] [CrossRef]
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, B.; Achard, P. Long-Distance Transport of Phytohormones through the Plant Vascular System. Curr. Opin. Plant Biol. 2016, 34, 42–47. [Google Scholar] [CrossRef]
- Kamiya, Y. Plant Hormones: Versatile Regulators of Plant Growth and Development. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef]
- Boursiac, Y.; Léran, S.; Corratgé-Faillie, C.; Gojon, A.; Krouk, G.; Lacombe, B. ABA Transport and Transporters. Trends Plant Sci. 2013, 18, 325–333. [Google Scholar] [CrossRef]
- Santner, A.; Estelle, M. Recent Advances and Emerging Trends in Plant Hormone Signaling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef]
- Park, J.; Lee, Y.; Martinoia, E.; Geisler, M. Plant Hormone Transporters: What We Know and What We Would Like to Know. BMC Biol. 2017, 15, 93. [Google Scholar] [CrossRef]
- Casanova-Sáez, R.; Voß, U. Auxin Metabolism Controls Developmental Decisions in Land Plants. Trends Plant Sci. 2019, 24, 741–754. [Google Scholar] [CrossRef]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and Its Role in Plant Development: Structure, Signaling, Regulation and Response Mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef]
- Aloni, R.; Schwalm, K.; Langhans, M.; Ullrich, C.I. Gradual Shifts in Sites of Free-Auxin Production During Leaf-Primordium Development and Their Role in Vascular Differentiation and Leaf Morphogenesis in Arabidopsis. Planta 2003, 216, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Mitina, I.; Quach, H.L.; Theologis, A. AUXIN RESPONSE FACTOR 2 (ARF2): A Pleiotropic Developmental Regulator. Plant J. 2005, 43, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Negi, N.P.; Pareek, S.; Mudgal, G.; Kumar, D. Auxin Response Factors in Plant Adaptation to Drought and Salinity Stress. Physiol. Plant. 2022, 174, e13714. [Google Scholar] [CrossRef]
- Singh, H.; Bhat, J.A.; Singh, V.P.; Corpas, F.J.; Yadav, S.R. Auxin Metabolic Network Regulates the Plant Response to Metalloids Stress. J. Hazard. Mater. 2021, 405, 124250. [Google Scholar] [CrossRef]
- Benjamins, R.; Scheres, B. Auxin: The Looping Star in Plant Development. Annu. Rev. Plant Biol. 2008, 59, 443–465. [Google Scholar] [CrossRef]
- Brumos, J.; Alonso, J.M.; Stepanova, A.N. Genetic Aspects of Auxin Biosynthesis and Its Regulation. Physiol. Plant. 2014, 151, 3–12. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin Biosynthesis and Storage Forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Ljung, K. Auxin Metabolism and Homeostasis During Plant Development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef]
- Mroue, S.; Simeunovic, A.; Robert, H.S. Auxin Production as an Integrator of Environmental Cues for Developmental Growth Regulation. J. Exp. Bot. 2018, 69, 201–212. [Google Scholar] [CrossRef]
- Mathur, P.; Tripathi, D.K.; Baluska, F.; Mukherjee, S. Auxin-Mediated Molecular Mechanisms of Heavy Metal and Metalloid Stress Regulation in Plants. Environ. Exp. Bot. 2022, 196, 10–16. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 Flavin Monooxygenase Functions in the Indole-3-Pyruvic Acid Branch of Auxin Biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Dai, X.; Mashiguchi, K.; Chen, Q.; Kasahara, H.; Kamiya, Y.; Ojha, S.; DuBois, J.; Ballou, D.; Zhao, Y. The Biochemical Mechanism of Auxin Biosynthesis by an Arabidopsis YUCCA Flavin-Containing Monooxygenase. J. Biol. Chem. 2013, 288, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Essential Roles of Local Auxin Biosynthesis in Plant Development and in Adaptation to Environmental Changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.D.; Sampson, M.B.; Neuffer, M.G.; Michalczuk, L.; Slovin, J.P.; Cohen, J.D. Indole-3-Acetic Acid Biosynthesis in the Mutant Maize Orange Pericarp, a Tryptophan Auxotroph. Science 1991, 254, 998–1000. [Google Scholar] [CrossRef]
- Wang, B.; Chu, J.; Yu, T.; Xu, Q.; Sun, X.; Yuan, J.; Xiong, G.; Wang, G.; Wang, Y.; Li, J. Tryptophan-Independent Auxin Biosynthesis Contributes to Early Embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 4821–4826. [Google Scholar] [CrossRef]
- Bhosale, R.; Giri, J.; Pandey, B.K.; Giehl, R.F.; Hartmann, A.; Traini, R.; Truskina, J.; Leftley, N.; Hanlon, M.; Swarup, K.; et al. A Mechanistic Framework for Auxin-Dependent Arabidopsis Root Hair Elongation to Low External Phosphate. Nat. Commun. 2018, 9, 1409. [Google Scholar] [CrossRef]
- Giri, J.; Bhosale, R.; Huang, G.; Pandey, B.K.; Parker, H.; Zappala, S.; Yang, J.; Dievart, A.; Bureau, C.; Ljung, K.; et al. Rice Auxin Influx Carrier OsAUX1 Facilitates Root Hair Elongation in Response to Low External Phosphate. Nat. Commun. 2018, 9, 1408. [Google Scholar] [CrossRef]
- Li, Q.; Yin, M.; Li, Y.; Fan, C.; Yang, Q.; Wu, J.; Zhang, C.; Wang, H.; Zhou, Y. Expression of Brassica napus TTG2, a Regulator of Trichome Development, Increases Plant Sensitivity to Salt Stress by Suppressing the Expression of Auxin Biosynthesis Genes. J. Exp. Bot. 2015, 66, 5821–5836. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin Biosynthesis by the YUCCA Genes in Rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell Physiol. 2020, 61, 1832–1849. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, D. Molecular Basis and Evolutionary Pattern of GA–GID1–DELLA Regulatory Module. Mol. Genet. Genom. 2014, 289, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sponsel, V.M. Signal Achievements in Gibberellin Research: The Second Half-Century. In Annual Plant Reviews; Wiley: Hoboken, NJ, USA, 2016; Volume 49, pp. 1–36. [Google Scholar] [CrossRef]
- Khush, G.S. Green Revolution: The Way Forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), “Green Revolution” Rice, Contains a Defective Gibberellin 20-Oxidase Gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin Biosynthesis and Its Regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin Signaling: Biosynthesis, Catabolism, and Response Pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef]
- Achard, P.; Gusti, A.; Cheminant, S.; Alioua, M.; Dhondt, S.; Coppens, F.; Beemster, G.T.S.; Genschik, P. Gibberellin Signaling Controls Cell Proliferation Rate in Arabidopsis. Curr. Biol. 2009, 19, 1188–1193. [Google Scholar] [CrossRef]
- Band, L.R.; Úbeda-Tomás, S.; Dyson, R.J.; Middleton, A.M.; Hodgman, T.C.; Owen, M.R.; Jensen, O.E.; Bennett, M.J.; King, J.R. Growth-Induced Hormone Dilution Can Explain the Dynamics of Plant Root Cell Elongation. Proc. Natl. Acad. Sci. USA 2012, 109, 7577–7582. [Google Scholar] [CrossRef]
- Pullen, N.; Zhang, N.; Alonso, A.D.; Penfield, S. Growth Rate Regulation Is Associated with Developmental Modification of Source Efficiency. Nat. Plants 2019, 5, 148–152. [Google Scholar] [CrossRef]
- Ogawa, M.; Hanada, A.; Yamauchi, Y.; Kuwahara, A.; Kamiya, Y.; Yamaguchi, S. Gibberellin Biosynthesis and Response During Arabidopsis Seed Germination. Plant Cell 2003, 15, 1591–1604. [Google Scholar] [CrossRef]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.P. DELLA Proteins and Gibberellin-Regulated Seed Germination and Floral Development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]
- Alabadí, D.; Gil, J.; Blázquez, M.A.; García-Martínez, J.L. Gibberellins Repress Photomorphogenesis in Darkness. Plant Physiol. 2004, 134, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, W.; Yang, W.; Kong, Z.; Xue, Y. Genome-Wide Gene Expression Profiling Reveals Conserved and Novel Molecular Functions of the Stigma in Rice. Plant Physiol. 2007, 144, 1797–1812. [Google Scholar] [CrossRef] [PubMed]
- Anfang, M.; Shani, E. Transport Mechanisms of Plant Hormones. Curr. Opin. Plant Biol. 2021, 63, 102055. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin Metabolism and Its Regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An Overview of Gibberellin Metabolism Enzyme Genes and Their Related Mutants in Rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef]
- Regnault, T.; Davière, J.M.; Heintz, D.; Lange, T.; Achard, P. The Gibberellin Biosynthetic Genes AtKAO1 and AtKAO2 Have Overlapping Roles Throughout Arabidopsis Development. Plant J. 2014, 80, 462–474. [Google Scholar] [CrossRef]
- Teng, F.; Zhai, L.; Liu, R.; Bai, W.; Wang, L.; Huo, D.; Tao, Y.; Zheng, Y.; Zhang, Z. ZmGA3ox2, a Candidate Gene for a Major QTL, qPH3.1, for Plant Height in Maize. Plant J. 2013, 73, 405–416. [Google Scholar] [CrossRef]
- Chen, Y.; Hou, M.; Liu, L.; Wu, S.; Shen, Y.; Ishiyama, K.; Kobayashi, M.; McCarty, D.R.; Tan, B.C. The Maize DWARF1 Encodes a Gibberellin 3-Oxidase and Is Dual Localized to the Nucleus and Cytosol. Plant Physiol. 2014, 166, 2028–2039. [Google Scholar] [CrossRef]
- Castro-Camba, R.; Sánchez, C.; Vidal, N.; Vielba, J. Interactions of Gibberellins with Phytohormones and Their Role in Stress Responses. Horticulturae 2022, 8, 241. [Google Scholar] [CrossRef]
- Barker, R.; Fernandez Garcia, M.N.; Powers, S.J.; Vaughan, S.; Bennett, M.J.; Phillips, A.L.; Thomas, S.G.; Hedden, P. Mapping Sites of Gibberellin Biosynthesis in the Arabidopsis Root Tip. New Phytol. 2021, 229, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.M.; Achard, P. Gibberellin Signaling in Plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.Y.; Hsing, Y.I.; Kitano, H.; Yamaguchi, I.; et al. GIBBERELLIN INSENSITIVE DWARF1 Encodes a Soluble Receptor for Gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Hauvermale, A.L.; Ariizumi, T.; Steber, C.M. Gibberellin Signaling: A Theme and Variations on DELLA Repression. Plant Physiol. 2012, 160, 83–92. [Google Scholar] [CrossRef]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Erik Olsen, C.; Weinstain, R.; et al. The Arabidopsis NPF3 Protein Is a GA Transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef]
- David, L.C.; Berquin, P.; Kanno, Y.; Seo, M.; Daniel-Vedele, F.; Ferrario-Méry, S.N. Availability Modulates the Role of NPF3.1, a Gibberellin Transporter, in GA-Mediated Phenotypes in Arabidopsis. Planta 2016, 244, 1315–1328. [Google Scholar] [CrossRef]
- Wulff, N.; Ernst, H.A.; Jørgensen, M.E.; Lambertz, S.; Maierhofer, T.; Belew, Z.M.; Crocoll, C.; Motawia, M.S.; Geiger, D.; Jørgensen, F.S.; et al. An Optimized Screen Reduces the Number of GA Transporters and Provides Insights into Nitrate Transporter 1/Peptide Transporter Family Substrate Determinants. Front. Plant Sci. 2019, 10, 1106. [Google Scholar] [CrossRef]
- Sirlin-Josserand, M.; Sakvarelidze-Achard, L.; Pflieger, D.; Davière, J.M.; Achard, P. NPF4.1 Imports Embryo-Derived GA₄ to the Endosperm to Promote Seed Germination. bioRxiv 2024, 2024.10.07.617022. [Google Scholar] [CrossRef]
- Morii, M.; Sugihara, A.; Takehara, S.; Kanno, Y.; Kawai, K.; Hobo, T.; Hattori, M.; Yoshimura, H.; Seo, M.; Ueguchi-Tanaka, M. The Dual Function of OsSWEET3a as a Gibberellin and Glucose Transporter Is Important for Young Shoot Development in Rice. Plant Cell Physiol. 2020, 61, 1935–1945. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Jung, H.-S.; Dill, A.; Kawaide, H.; Kamiya, Y.; Sun, T.-P. Repressing a Repressor: Gibberellin-Induced Rapid Reduction of the RGA Protein in Arabidopsis. Plant Cell 2001, 13, 1555–1566. [Google Scholar] [CrossRef]
- Reid, J.B. Phytohormone Mutants in Plant Research. J. Plant Growth Regul. 1990, 9, 97–111. [Google Scholar] [CrossRef]
- Zanewich, K.P.; Rood, S.B.; Williams, P.H. Growth and Development of Brassica Genotypes Differing in Endogenous Gibberellin Content. I. Leaf and Reproductive Development. Physiol. Plant. 1990, 79, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin Action in Plant Development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones Enhanced Drought Tolerance in Plants: A Coping Strategy. Environ. Sci. Pollut. Res. Int. 2018, 25, 33103–33118. [Google Scholar] [CrossRef]
- Mok, D.W.; Mok, M.C. Cytokinin Metabolism and Action. Annu. Rev. Plant Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef]
- Kisiala, A.; Kambhampati, S.; Stock, N.L.; Aoki, M.; Emery, R.N. Quantification of Cytokinins Using High-Resolution Accurate-Mass Orbitrap Mass Spectrometry and Parallel Reaction Monitoring (PRM). Anal. Chem. 2019, 91, 15049–15056. [Google Scholar] [CrossRef]
- Durán-Medina, Y.; Díaz-Ramírez, D.; Marsch-Martínez, N. Cytokinins on the Move. Front. Plant Sci. 2017, 8, 146. [Google Scholar] [CrossRef]
- Nguyen, H.N.; Lai, N.; Kisiala, A.B.; Emery, R.N. Isopentenyltransferases as Master Regulators of Crop Performance: Their Function, Manipulation, and Genetic Potential for Stress Adaptation and Yield Improvement. Plant Biotechnol. J. 2021, 19, 1297–1313. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, Biosynthesis, and Translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef]
- Svolacchia, N.; Sabatini, S. Cytokinins. Curr. Biol. 2023, 33, R10–R13. [Google Scholar] [CrossRef]
- Imamura, A.; Hanaki, N.; Umeda, H.; Nakamura, A.; Suzuki, T.; Ueguchi, C.; Mizuno, T. Response Regulators Implicated in His-to-Asp Phosphotransfer Signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 2691–2696. [Google Scholar] [CrossRef] [PubMed]
- Urao, T.; Yakubov, B.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Stress-Responsive Expression of Genes for Two-Component Response Regulator-Like Proteins in Arabidopsis thaliana. FEBS Lett. 1998, 427, 175–178. [Google Scholar] [CrossRef] [PubMed]
- To, J.P.; Haberer, G.; Ferreira, F.J.; Deruere, J.; Mason, M.G.; Schaller, G.E.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J. Type-A Arabidopsis Response Regulators Are Partially Redundant Negative Regulators of Cytokinin Signaling. Plant Cell 2004, 16, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, N.; Ju, M.; Fan, B.; Zhang, Y.; Zhu, E.; Zhang, M.; Zhang, K. ABC Transporter OsABCG18 Controls the Shootward Transport of Cytokinins and Grain Yield in Rice. J. Exp. Bot. 2019, 70, 6277–6291. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, J.; Yu, G.; Lu, X.; Mei, W.; Deng, H.; Zhang, G.; Chen, G.; Chu, C.; Tong, H.; et al. Endoplasmic Reticulum-Localized PURINE PERMEASE1 Regulates Plant Height and Grain Weight by Modulating Cytokinin Distribution in Rice. Front. Plant Sci. 2020, 11, 618560. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, D.; Zhang, G.; Gao, S.; Liu, L.; Xu, F.; Che, R.; Wang, Y.; Tong, H.; Chu, C. Big Grain3, Encoding a Purine Permease, Regulates Grain Size via Modulating Cytokinin Transport in Rice. J. Integr. Plant Biol. 2019, 61, 581–597. [Google Scholar] [CrossRef]
- Kant, S.; Burch, D.; Badenhorst, P.; Palanisamy, R.; Mason, J.; Spangenberg, G. Regulated Expression of a Cytokinin Biosynthesis Gene IPT Delays Leaf Senescence and Improves Yield under Rainfed and Irrigated Conditions in Canola (Brassica napus L.). PLoS ONE 2015, 10, e0116349. [Google Scholar] [CrossRef]
- Kuromori, T.; Sugimoto, E.; Shinozaki, K. Intertissue Signal Transfer of Abscisic Acid from Vascular Cells to Guard Cells. Plant Physiol. 2014, 164, 1587–1592. [Google Scholar] [CrossRef]
- Chen, T.T.; Liu, F.F.; Xiao, D.W.; Jiang, X.Y.; Li, P.; Zhao, S.M.; Hou, B.K.; Li, Y.J. The Arabidopsis UDP-Glycosyltransferase75B1 Conjugates Abscisic Acid and Affects Plant Response to Abiotic Stresses. Plant Mol. Biol. 2020, 102, 389–401. [Google Scholar] [CrossRef]
- Wang, C.; Yang, A.; Yin, H.; Zhang, J. Influence of Water Stress on Endogenous Hormone Contents and Cell Damage of Maize Seedlings. J. Int. Plant Biol. 2008, 50, 427–434. [Google Scholar] [CrossRef]
- Yeung, E.; van Veen, H.; Vashisht, D.; Sobral Paiva, A.L.; Hummel, M.; Rankenberg, T.; Steffens, B.; Steffen-Heins, A.; Sauter, M.; de Vries, M.; et al. A Stress Recovery Signaling Network for Enhanced Flooding Tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E6085–E6094. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.J.; Kim, W.Y. Plant Hormone-Mediated Regulation of Heat Tolerance in Response to Global Climate Change. Front. Plant Sci. 2021, 11, 627969. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.S.; Poole, R.J.; Dhindsa, R.S. Abscisic Acid-Regulated Gene Expression in Relation to Freezing Tolerance in Alfalfa. Plant Physiol. 1988, 87, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Rosales, M.; Maurel, C.; Nacry, P. Abscisic Acid Coordinates Dose-Dependent Developmental and Hydraulic Responses of Roots to Water Deficit. Plant Physiol. 2019, 180, 2198–2211. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, J.; He, J.; Qin, Y.; Hua, D.; Duan, Y.; Chen, Z.; Gong, Z. ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling PLETHORA Expression in Arabidopsis. PLoS Genet. 2014, 10, e1004791. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.; Testerink, C. Roots Withstanding Their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef]
- Seo, M.; Koshiba, T. Complex Regulation of ABA Biosynthesis in Plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Thompson, A.J.; Jackson, A.C.; Symonds, R.C.; Mulholland, B.J.; Dadswell, A.R.; Blake, P.S.; Burbidge, A.; Taylor, I.B. Ectopic Expression of a Tomato 9-Cis-Epoxycarotenoid Dioxygenase Gene Causes Overproduction of Abscisic Acid. Plant J. 2000, 23, 363–374. [Google Scholar] [CrossRef]
- Kumar, S.; Shah, S.H.; Vimala, Y.; Jatav, H.S.; Ahmad, P.; Chen, Y.; Siddique, K.H. Abscisic Acid: Metabolism, Transport, Crosstalk with Other Plant Growth Regulators, and Its Role in Heavy Metal Stress Mitigation. Front. Plant Sci. 2022, 13, 972856. [Google Scholar] [CrossRef]
- Kang, J.; Hwang, J.U.; Lee, M.; Kim, Y.Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of Arabidopsis thaliana NRT1/PTR family (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, H.; Pan, Y.; Yu, Y.; Luan, S.; Li, L. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 2014, 7, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- Léran, S.; Noguero, M.; Corratgé-Faillie, C.; Boursiac, Y.; Brachet, C.; Lacombe, B. Functional characterization of the Arabidopsis abscisic acid transporters NPF4.5 and NPF4.6 in Xenopus oocytes. Front. Plant Sci. 2020, 11, 144. [Google Scholar] [CrossRef]
- Pawela, A.; Banasiak, J.; Biała, W.; Martinoia, E.; Jasinski, M. MtABCG20 is an ABA exporter influencing root morphology and seed germination of Medicago truncatula. Plant J. 2019, 98, 511–523. [Google Scholar] [CrossRef]
- Chernys, J.T.; Zeevaart, J.A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–353. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef]
- Qin, X.; Zeevaart, J.A. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 1999, 96, 15354–15361. [Google Scholar] [CrossRef]
- Yoshiba, Y.; Aoki, C.; Iuchi, S.; Nanjo, T.; Seki, M.; Sekiguchi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of four extensin genes in Arabidopsis thaliana by differential gene expression under stress and non-stress conditions. DNA Res. Int. J. Rapid Publ. Rep. Genes Genom. 2001, 8, 115–122. [Google Scholar] [CrossRef][Green Version]
- Burg, S.P. Ethylene in Plant Growth. Proc. Natl. Acad. Sci. USA 1973, 70, 591–597. [Google Scholar] [CrossRef]
- Shin, K.; Lee, S.; Song, W.Y.; Lee, R.A.; Lee, I.; Ha, K.; Koo, J.C.; Park, S.K.; Nam, H.G.; Lee, Y.; et al. Genetic Identification of ACC-RESISTANT2 Reveals Involvement of LYSINE HISTIDINE TRANSPORTER1 in the Uptake of 1-Aminocyclopropane-1-carboxylic Acid in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Abeles, F.; Morgan, P.; Saltveit, M.J. Ethylene in Plant Biology, 2nd ed.; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- Mattoo, A.K.; Suttle, J.C. The Plant Hormone Ethylene; CRC Press: Boca Raton, FL, USA, 1991; p. 337. [Google Scholar]
- Xu, Z.H.; Xue, H.W. Plant Hormones: Function and Molecular Mechanism; Shanghai Scientific and Technical Publishers: Shanghai, China, 2012. [Google Scholar]
- Mattoo, A.K.; White, W.B. Regulation of Ethylene Biosynthesis. In The Plant Hormone Ethylene; CRC Press: Boca Raton, FL, USA, 2018; pp. 21–42. [Google Scholar]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Kende, H. Enzymes of ethylene biosynthesis. Plant Physiol. 1989, 91, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Qu, L.H.; Li, N. Tyr152 plays a central role in the catalysis of 1-aminocyclopropane-1-carboxylate synthase. J. Exp. Bot. 2005, 56, 2203–2210. [Google Scholar] [CrossRef]
- Lieberman, M. Oglobulins of Mouse. Annu. Rev. Plant Physiol. 1979, 30, 533. [Google Scholar] [CrossRef]
- Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 2014, 5, 685. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef]
- Ozga, J.A.; Reinecke, D.M. Hormonal interactions in fruit development. J. Plant Growth Regul. 2003, 22, 73–81. [Google Scholar] [CrossRef]
- Ayele, B.T.; Ozga, J.A.; Kurepin, L.V.; Reinecke, D.M. Developmental and embryo axis regulation of gibberellin biosynthesis during germination and young seedling growth of pea (Pisum sativum L.). Plant Physiol. 2006, 142, 1267–1281. [Google Scholar] [CrossRef]
- Nadeau, C.D.; Ozga, J.A.; Kurepin, L.V.; Jin, A.; Pharis, R.P.; Reinecke, D.M. Tissue-specific regulation of gibberellin biosynthesis in developing pea (Pisum sativum L.) seeds. Plant Physiol. 2011, 156, 897–910. [Google Scholar] [CrossRef]
- Van de Poel, B.; Bulens, I.; Markoula, A.; Hertog, M.L.; Dreesen, R.; Wirtz, M.; Vandoninck, S.; Oppermann, Y.; Keulemans, J.; Hell, R.; et al. Targeted Systems Biology Profiling of Tomato Fruit Reveals Coordination of the Yang Cycle and a Distinct Regulation of Ethylene Biosynthesis During Postclimacteric Ripening. Plant Physiol. 2012, 160, 1498–1514. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Lorbiecke, R.; OuYang, B.; Pochapsky, T.C.; Rzewuski, G. The Immediate-Early Ethylene Response Gene OsARD1 Encodes an Acireductone Dioxygenase Involved in Recycling of the Ethylene Precursor S-Adenosylmethionine. Plant J. 2005, 44, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Rzewuski, G.; Cornell, K.A.; Rooney, L.; Bürstenbinder, K.; Wirtz, M.; Hell, R.; Sauter, M. OsMTN Encodes a 5′-Methylthioadenosine Nucleosidase That Is Up-Regulated During Submergence-Induced Ethylene Synthesis in Rice (Oryza sativa L.). J. Exp. Bot. 2007, 58, 1505–1514. [Google Scholar] [CrossRef]
- Liang, S.; Xiong, W.; Yin, C.; Xie, X.; Jin, Y.; Zhang, S.; Yang, B.; Ye, G.; Chen, S.; Luan, W. Overexpression of OsARD1 Improves Submergence, Drought, and Salt Tolerances of Seedling Through the Enhancement of Ethylene Synthesis in Rice. Front. Plant Sci. 2019, 10, 19. [Google Scholar] [CrossRef]
- Johnstone, M.M.G.; Reinecke, D.M.; Ozga, J.A. The Auxins IAA and 4-Cl-IAA Differentially Modify Gibberellin Action via Ethylene Response in Developing Pea Fruit. J. Plant Growth Regul. 2005, 24, 214–222. [Google Scholar] [CrossRef]
- Johnson-Flanagan, A.M.; Spencer, M.S. Ethylene Production During Development of Mustard (Brassica juncea) and Canola (Brassica napus) Seed. Plant Physiol. 1994, 106, 601–606. [Google Scholar] [CrossRef]
- Washington, E.J.; Mukhtar, M.S.; Finkel, O.M.; Wan, L.; Banfield, M.J.; Kieber, J.J.; Dangl, J.L. Pseudomonas syringae Type III Effector HopAF1 Suppresses Plant Immunity by Targeting Methionine Recycling to Block Ethylene Induction. Proc. Natl. Acad. Sci. USA 2016, 113, 3577–3586. [Google Scholar] [CrossRef]
- Clouse, S.D.; Sasse, J.M. Brassinosteroids: Essential Regulators of Plant Growth and Development. Annu. Rev. Plant Biol. 1998, 49, 427–451. [Google Scholar] [CrossRef]
- Peres, A.L.; Soares, J.S.; Tavares, R.G.; Righetto, G.; Zullo, M.A.; Mandava, N.B.; Menossi, M. Brassinosteroids, the Sixth Class of Phytohormones: A Molecular View from the Discovery to Hormonal Interactions in Plant Development Stress Adaptation. Int. J. Mol. Sci. 2019, 20, 331. [Google Scholar] [CrossRef]
- Grove, M.D.; Spencer, G.F.; Rohwedder, W.K.; Mandava, N.; Cook, J.L.C. Brassinolide, a Plant Growth-Promoting Steroid Isolated from Brassica napus Pollen. Nature 1979, 281, 216–217. [Google Scholar] [CrossRef]
- Li, T.; Lei, W.; He, R.; Tang, X.; Zhang, D. Brassinosteroids Regulate Root Meristem Development by Mediating BIN2-UPB1 Module in Arabidopsis. PLoS Genet. 2020, 16, e1008883. [Google Scholar] [CrossRef]
- Bajguz, A.; Tretyn, A. The Chemical Characteristic and Distribution of Brassinosteroids in Plants. Phytochemistry 2003, 62, 1027–1046. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Ohnishi, T.; Shibata, K.; Asahina, M.; Nomura, T.; Fujita, T.; Ishizaki, K.; Kohchi, T. Occurrence of Brassinosteroids in Non-Flowering Land Plants, Liverwort, Moss, Lycophyte and Fern. Phytochemistry 2017, 136, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Zullo, M.A.T.; Bajguz, A. Brassinosteroids: Plant Growth and Development. In The Plant Hormone Brassinosteroids: Recent Advances and Future Directions; Hayat, S., Yusuf, M., Bhardwaj, R., Bajguz, A., Eds.; Springer: Singapore, 2019; pp. 1–44. [Google Scholar] [CrossRef]
- Hayat, S.; Ahmad, A.; Fariduddin, Q. Brassinosteroids: A Regulator of 21st Century; Springer: Berlin/Heidelberg, Germany, 2003; pp. 231–246. [Google Scholar]
- Bajguz, A.; Hayat, S. Effects of Brassinosteroids on the Plant Responses to Environmental Stresses. Plant Physiol. Biochem. 2009, 47, 56–62. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N.; et al. Phytohormones and Plant Responses to Salinity Stress: A Review. Plant Growth Regul. 2014, 75, 391–404. [Google Scholar] [CrossRef]
- Bajguz, A.; Chmur, M.; Gruszka, D. Comprehensive Overview of the Brassinosteroid Biosynthesis Pathways: Substrates, Products, Inhibitors, Connections. Front. Plant Sci. 2020, 11, 1034. [Google Scholar] [CrossRef]
- Choe, S.; Dilkes, B.P.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Feldmann, K.A. The DWF4 Gene of Arabidopsis Encodes a Cytochrome P450 That Mediates Multiple 22 Alpha-Hydroxylation Steps in Brassinosteroid Biosynthesis. Plant Cell 1998, 10, 231–243. [Google Scholar] [CrossRef]
- Joo, S.H.; Kim, T.W.; Son, S.H.; Lee, W.S.; Yokota, T.; Kim, S.K. Biosynthesis of a Cholesterol-Derived Brassinosteroid, 28-Norcastasterone, in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1823–1833. [Google Scholar] [CrossRef]
- Zhao, B.L.; Li, J. Regulation of Brassinosteroid Biosynthesis and Inactivation. J. Integr. Plant Biol. 2012, 54, 746–759. [Google Scholar] [CrossRef]
- Liu, L.; Jia, C.; Zhang, M.; Chen, D.; Chen, S.; Guo, R.; Guo, D.; Wang, Q. Ectopic Expression of a BZR1-1D Transcription Factor in Brassinosteroid Signalling Enhances Carotenoid Accumulation and Fruit Quality Attributes in Tomato. Plant Biotechnol. J. 2014, 12, 105–115. [Google Scholar] [CrossRef]
- Choe, S.; Fujioka, S.; Noguchi, T.; Takatsuto, S.; Yoshida, S.; Feldmann, K.A. Overexpression of DWARF4 in the Brassinosteroid Biosynthetic Pathway Results in Increased Vegetative Growth and Seed Yield in Arabidopsis. Plant J. 2001, 26, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Zolkiewicz, K.; Ahmar, S.; Gruszka, D. Genetic Manipulations of Brassinosteroid-Related Genes Improve Various Agronomic Traits and Yield in Cereals Enabling New Biotechnological Revolution: Achievements and Perspectives. Biotechnol. Adv. 2025, 108556. [Google Scholar] [CrossRef]
- Wu, C.Y.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S.; et al. Brassinosteroids Regulate Grain Filling in Rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Watanabe, B.; Sakata, K.; Mizutani, M. CYP724B2 and CYP90B3 Function in the Early C-22 Hydroxylation Steps of the Brassinosteroid Biosynthetic Pathway in Tomato. Biosci. Biotechnol. Biochem. 2006, 70, 2071–2080. [Google Scholar] [CrossRef][Green Version]
- Schröder, F.; Lisso, J.; Obata, T.; Erban, A.; Maximova, E.; Giavalisco, P.; Kopka, J.; Fernie, A.R.; Willmitzer, L.; Müssig, C. Consequences of Induced Brassinosteroid Deficiency in Arabidopsis Leaves. BMC Plant Biol. 2014, 14, 309. [Google Scholar] [CrossRef]
- Clouse, S.D. Molecular Genetics of Brassinosteroid Action. In Brassinosteroids: Steroidal Plant Hormones; Springer: Berlin/Heidelberg, Germany, 1999; pp. 163–190. [Google Scholar]
- Clouse, S.D.; Zurek, D.M.; McMorris, T.C.; Baker, M.E. Effect of Brassinolide on Gene Expression in Elongating Soybean Epicotyls. Plant Physiol. 1992, 100, 1377–1383. [Google Scholar] [CrossRef]
- McKay, M.J.; Ross, J.J.; Lawrence, N.L.; Cramp, R.E.; Beveridge, C.A.; Reid, J.B. Control of Internode Length in Pisum sativum. Plant Physiol. 1994, 106, 1521–1526. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Wang, M.; Xu, Y.Y.; Luo, W.; Liu, Y.J.; Xu, Z.H.; Li, J.; Chong, K. Engineering OsBAK1 Gene as a Molecular Tool to Improve Rice Architecture for High Yield. Plant Biotechnol. J. 2009, 7, 791–806. [Google Scholar] [CrossRef]
- Zhang, C.; Bai, M.Y.; Chong, K. Brassinosteroid-Mediated Regulation of Agronomic Traits in Rice. Plant Cell Rep. 2014, 33, 683–696. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Y.Y.; Li, J.; Powell, R.A.; Xu, Z.H.; Chong, K. Transgenic Rice Plants Ectopically Expressing AtBAK1 Are Semi-Dwarfed and Hypersensitive to 24-Epibrassinolide. J. Plant Physiol. 2007, 164, 655–664. [Google Scholar] [CrossRef]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid Confers Tolerance in Arabidopsis thaliana and Brassica napus to a Range of Abiotic Stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.; Prasad, B.D.; Liu, Q.; Grbic, V.; Sharpe, A.; Singh, S.P.; Krishna, P. Overexpression of the Brassinosteroid Biosynthetic Gene DWF4 in Brassica napus Simultaneously Increases Seed Yield and Stress Tolerance. Sci. Rep. 2016, 6, 28298. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Li, X.; Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Chen, Z.; Yu, J.-Q. H2O2 Mediates the Crosstalk of Brassinosteroid and Abscisic Acid in Tomato Responses to Heat and Oxidative Stresses. J. Exp. Bot. 2014, 65, 4371–4383. [Google Scholar] [CrossRef]
- Hussain, M.A.; Fahad, S.; Sharif, R.; Jan, M.F.; Mujtaba, M.; Ali, Q.; Ahmad, A.; Ahmad, H.; Amin, N.; Ajayo, B.S. Multifunctional Role of Brassinosteroid and Its Analogues in Plants. Plant Growth Regul. 2020, 92, 141–156. [Google Scholar] [CrossRef]
- Nakashita, H.; Yasuda, M.; Nitta, T.; Asami, T.; Fujioka, S.; Arai, Y.; Sekimata, K.; Takatsuto, S.; Yamaguchi, I.; Yoshida, S. Brassinosteroid Functions in a Broad Range of Disease Resistance in Tobacco and Rice. Plant J. 2003, 33, 887–898. [Google Scholar] [CrossRef]
- Ruan, J.J.; Zhou, Y.X.; Zhou, M.L.; Yan, J.; Khurshid, M.; Weng, W.F.; Cheng, J.P.; Zhang, K.X. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Llanes, A.; Andrade, A.; Alenabo, S.; Luna, V. Alterations of Endogenous Hormonal Levels in Plants under Drought and Salinity. Am. J. Plant Sci. 2016, 7, 1357–1371. [Google Scholar] [CrossRef]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inze, D.; Goossens, A. Mapping Methyl Jasmonate-Mediated Transcriptional Reprogramming of Metabolism and Cell Cycle Progression in Cultured Arabidopsis Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef]
- Gomi, K. Jasmonic Acid: An Essential Plant Hormone. Int. J. Mol. Sci. 2020, 21, 1261. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, Perception, Signal Transduction, and Action in Plant Stress Response, Growth, and Development. An Update to the 2007 Review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.F.; Yadav, V.; Tholl, D.; Chételat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C.; et al. A Regulatory Network for Coordinated Flower Maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates and Octadecanoids: Signals in Plant Stress Responses and Development. Prog. Nucleic Acid Res. Mol. Biol. 2002, 72, 165–221. [Google Scholar] [PubMed]
- Zhang, Y.; Turner, J.G. Wound-Induced Endogenous Jasmonates Stunt Plant Growth by Inhibiting Mitosis. PLoS ONE 2008, 3, e3699. [Google Scholar] [CrossRef]
- Farhangi-Abriz, S.; Alaee, T.; Tavasolee, A. Salicylic Acid but Not Jasmonic Acid Improved Canola Root Response to Salinity Stress. Rhizosphere 2019, 9, 69–71. [Google Scholar] [CrossRef]
- Alisof, S.; Einali, A.; Sangtarash, M.H. Jasmonic Acid-Induced Metabolic Responses in Bitter Melon (Momordica charantia) Seedlings under Salt Stress. J. Hortic. Sci. Biotechnol. 2020, 95, 247–259. [Google Scholar] [CrossRef]
- Wang, F.; Yu, G.; Liu, P. Transporter-Mediated Subcellular Distribution in the Metabolism and Signaling of Jasmonates. Front. Plant Sci. 2019, 10, 390. [Google Scholar] [CrossRef]
- Vick, B.A.; Zimmerman, D.C. The Biosynthesis of Jasmonic Acid: A Physiological Role for Plant Lipoxygenase. Biochem. Biophys. Res. Commun. 1983, 111, 470–477. [Google Scholar] [CrossRef]
- Theodoulou, F.L.; Job, K.; Slocombe, S.P.; Footitt, S.; Holdsworth, M.; Baker, A.; Larson, T.R.; Graham, I.A. Jasmonic Acid Levels Are Reduced in COMATOSE ATP-Binding Cassette Transporter Mutants. Implications for Transport of Jasmonate Precursors into Peroxisomes. Plant Physiol. 2005, 137, 835–840. [Google Scholar] [CrossRef]
- Campos, M.L.; Kang, J.H.; Howe, G.A. Jasmonate-Triggered Plant Immunity. J. Chem. Ecol. 2014, 40, 657–675. [Google Scholar] [CrossRef]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Desaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 Catalyze Two Successive Oxidation Steps of Plant Hormone Jasmonoyl-Isoleucine for Catabolic Turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [PubMed]
- Brash, A.R.; Baertschi, S.W.; Ingram, C.D.; Harris, T.M. Isolation and Characterization of Natural Allene Oxides: Unstable Intermediates in the Metabolism of Lipid Hydroperoxides. Proc. Natl. Acad. Sci. USA 1988, 85, 3382–3386. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, H.; Chen, Y.; Zhu, S.; Chen, M.; Lan, X.; Chen, G.; Liao, Z. Cold Stress Improves the Production of Artemisinin Depending on the Increase in Endogenous Jasmonate. Biotechnol. Appl. Biochem. 2017, 64, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Habibi, F.; Ramezanian, A.; Rahemi, M.; Eshghi, S.; Guillén, F.; Serrano, M.; Valero, D. Postharvest Treatments with γ-Aminobutyric Acid, Methyl Jasmonate, or Methyl Salicylate Enhance Chilling Tolerance of Blood Orange Fruit at Prolonged Cold Storage. J. Sci. Food Agric. 2019, 99, 6408–6417. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Xie, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Abscisic Acid and Jasmonic Acid Are Involved in Drought Priming-Induced Tolerance to Drought in Wheat. Crop J. 2021, 9, 120–132. [Google Scholar] [CrossRef]
- Kim, J.-M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S. Acetate-Mediated Novel Survival Strategy Against Drought in Plants. Nat. Plants 2017, 3, 17097. [Google Scholar] [CrossRef]
- Li, M.; Wang, F.; Li, S.; Yu, G.; Wang, L.; Li, Q.; Zhu, X.; Li, Z.; Yuan, L.; Liu, P. Importers Drive Leaf-to-Leaf Jasmonic Acid Transmission in Wound-Induced Systemic Immunity. Mol. Plant 2020, 13, 1485–1498. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Oikawa, T.; Suzuki, T.; Takeishi, S.; Matsuura, H.; Takahashi, K.; Hamamoto, S.; Uozumi, N.; Shimizu, T.; Seo, M.; et al. GTR1 is a Jasmonic Acid and Jasmonoyl-L-Isoleucine Transporter in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2017, 81, 249–255. [Google Scholar] [CrossRef]
- Wallis, C.M.; Galarneau, E.R.A. Phenolic Compound Induction in Plant-Microbe and Plant-Insect Interactions: A Meta-Analysis. Front. Plant Sci. 2020, 11, 580753. [Google Scholar] [CrossRef]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic Acid Biosynthesis and Metabolism. Arab. Book 2011, 9, e0156. [Google Scholar] [CrossRef]
- Raskin, I. Role of Salicylic Acid in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 439–463. [Google Scholar] [CrossRef]
- Seyfferth, C.; Tsuda, K. Salicylic Acid Signal Transduction: The Initiation of Biosynthesis, Perception, and Transcriptional Reprogramming. Front. Plant Sci. 2014, 5, 697. [Google Scholar] [CrossRef]
- Ogawa, D.; Nakajima, N.; Seo, S.; Mitsuhara, I.; Kamada, H.; Ohashi, Y. The Phenylalanine Pathway Is the Main Route of Salicylic Acid Biosynthesis in Tobacco Mosaic Virus-Infected Tobacco Leaves. Plant Biotechnol. 2006, 23, 395–398. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. An Important Role of the Pepper Phenylalanine Ammonia-Lyase Gene (PAL1) in Salicylic Acid-Dependent Signalling of the Defence Response to Microbial Pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, S.; Wang, S.; Shan, W.; Wang, X.; Lin, Y.; Su, F.; Yang, Z.; Yu, X. Defensive Responses of Tea Plants (Camellia sinensis) Against Tea Green Leafhopper Attack: A Multi-Omics Study. Front. Plant Sci. 2020, 10, 1705. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic Acid-Induced Abiotic Stress Tolerance and Underlying Mechanisms in Plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef]
- Kachroo, P.; Liu, H.; Kachroo, A. Salicylic Acid: Transport and Long-Distance Immune Signaling. Curr. Opin. Virol. 2020, 42, 53–57. [Google Scholar] [CrossRef]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic Acid Beyond Defence: Its Role in Plant Growth and Development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 Complete Salicylic Acid Biosynthesis from Isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-Derived Biosynthesis of the Plant Stress Hormone Salicylic Acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of Salicylic Acid in Plants. Plant Signal. Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Yokoo, S.; Inoue, S.; Suzuki, N.; Amakawa, N.; Matsui, H.; Nakagami, H.; Takahashi, A.; Arai, R.; Katou, S. Comparative Analysis of Plant Isochorismate Synthases Reveals Structural Mechanisms Underlying Their Distinct Biochemical Properties. Biosci. Rep. 2018, 38, BSR20171457. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defense Hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Dong, C.J.; Shang, Q.M. Genome-Wide Characterization of Phenylalanine Ammonia-Lyase Gene Family in Watermelon (Citrullus lanatus). Planta 2013, 238, 35–49. [Google Scholar] [CrossRef]
- Rao, M.V.; Paliyath, G.; Ormrod, D.P.; Murr, D.P.; Watkins, C.B. Influence of Salicylic Acid on H2O2 Production, Oxidative Stress, and H₂O₂-Metabolizing Enzymes. Plant Physiol. 1997, 115, 137–149. [Google Scholar] [CrossRef]
- Lopez-Delgado, H.; Dat, J.F.; Foyer, C.H.; Scott, I.M. Induction of Thermotolerance in Potato Microplants by Acetylsalicylic Acid and H2O2. J. Exp. Bot. 1998, 49, 713–730. [Google Scholar] [CrossRef]
- Miura, K.; Lee, J.; Miura, T.; Hasegawa, P.M. SIZ1 Controls Cell Growth and Plant Development in Arabidopsis through Salicylic Acid. Plant Cell Physiol. 2010, 51, 103–113. [Google Scholar] [CrossRef]
- Rate, D.; Cuenca, J.; Bowman, G.; Guttman, D.; Greenberg, J. The Gain-of-Function Arabidopsis acd6 Mutant Reveals Novel Regulation and Function of the Salicylic Acid Signaling Pathway in Controlling Cell Death, Defenses, and Cell Growth. Plant Cell 1999, 11, 1695–1708. [Google Scholar] [CrossRef]
- Abreu, M.E.; Munné-Bosch, S. Salicylic Acid Deficiency in NahG Transgenic Lines and sid2 Mutants Increases Seed Yield in the Annual Plant Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 1261–1271. [Google Scholar] [CrossRef]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat Stress Phenotypes of Arabidopsis Mutants Implicate Multiple Signaling Pathways in the Acquisition of Thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Knight, M.R. Protection Against Heat Stress-Induced Oxidative Damage in Arabidopsis Involves Calcium, Abscisic Acid, Ethylene, and Salicylic Acid. Plant Physiol. 2002, 128, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.M.; Mur, L.A.; Wood, J.E.; Scott, I.M. Salicylic Acid-Dependent Signaling Promotes Basal Thermotolerance but Is Not Essential for Acquired Thermotolerance in Arabidopsis thaliana. Plant J. 2004, 38, 432–447. [Google Scholar] [CrossRef] [PubMed]
- Cronjé, M.J.; Bornman, L. Salicylic Acid Influences Hsp70/Hsc70 Expression in Lycopersicon esculentum: Dose-and Time-Dependent Induction or Potentiation. Biochem. Biophys. Res. Commun. 1999, 265, 422–427. [Google Scholar] [CrossRef]
- Snyman, M.; Cronjé, M. Modulation of Heat Shock Factors Accompanies Salicylic Acid-Mediated Potentiation of Hsp70 in Tomato Seedlings. J. Exp. Bot. 2008, 59, 2125–2132. [Google Scholar] [CrossRef]
- Chang, P.-F.L.; Jinn, T.-L.; Huang, W.-K.; Chen, Y.; Chang, H.-M.; Wang, C.-W. Induction of a cDNA Clone from Rice Encoding a Class II Small Heat Shock Protein by Heat Stress, Mechanical Injury, and Salicylic Acid. Plant Sci. 2007, 172, 64–75. [Google Scholar] [CrossRef]
- Bandurska, H. The Effect of Salicylic Acid on Barley Response to Water Deficit. Acta Physiol. Plant. 2005, 27, 379–386. [Google Scholar] [CrossRef]
- Munne-Bosch, S.; Penuelas, J. Photo-and Antioxidative Protection, and a Role for Salicylic Acid During Drought and Recovery in Field-Grown Phillyrea angustifolia Plants. Planta 2003, 217, 758–766. [Google Scholar] [CrossRef]
- Abreu, M.E.; Munné-Bosch, S. Salicylic Acid May Be Involved in the Regulation of Drought-Induced Leaf Senescence in Perennials: A Case Study in Field-Grown Salvia officinalis L. Plants. Environ. Exp. Bot. 2008, 64, 105–112. [Google Scholar] [CrossRef]
- Safari, M.; Mousavi-Fard, S.; Rezaei Nejad, A.; Sorkheh, K.; Sofo, A. Exogenous salicylic acid positively affects morpho-physiological and molecular responses of Impatiens walleriana plants grown under drought stress. Int. J. Environ. Sci. Technol. 2021, 19, 969–984. [Google Scholar] [CrossRef]
- Lee, B.-R.; Islam, M.T.; Park, S.-H.; Jung, H.-i.; Bae, D.-W.; Kim, T.-H. Characterization of salicylic acid-mediated modulation of the drought stress responses: Reactive oxygen species, proline, and redox state in Brassica napus. Environ. Exp. Bot. 2019, 157, 1–10. [Google Scholar] [CrossRef]
- Ansari, O.; Sharif-Zadeh, F. Does Gibberelic acid (GA), Salicylic acid (SA) and Ascorbic acid (ASc) improve Mountain Rye (Secale montanum) seeds germination and seedlings growth under cold stress. Int. Res. J. Appl. Basic Sci. 2012, 3, 1651–1657. [Google Scholar]
- Gharib, F.; Hegazi, A. Salicylic acid ameliorates germination, seedling growth, phytohormone and enzymes activity in bean (Phaseolus vulgaris L.) under cold stress. J. Am. Sci. 2010, 6, 675–683. [Google Scholar]
- Kaur, S.; Gupta, N. Effect of proline and salicylic acid on germination and antioxidant enzymes at different temperatures in Muskmelon (Cucumis melo L.) seeds. J. Appl. Nat. Sci. 2017, 9, 2165–2169. [Google Scholar] [CrossRef]
- Rajjou, L.; Belghazi, M.; Huguet, R.; Robin, C.; Moreau, A.; Job, C.; Job, D. Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 2006, 141, 910–923. [Google Scholar] [CrossRef]
- Vaño, M.S.; Nourimand, M.; MacLean, A.; Pérez-López, E. Getting to the root of a club—Understanding developmental manipulation by the clubroot pathogen. Semin. Cell Dev. Biol. 2023, 148–149, 22–32. [Google Scholar] [CrossRef]
- Wang, X.; Fu, M.; Qu, X.; Liu, J.; Bu, J.; Feng, S.; Zhao, H.; Jiao, W.; Sun, F. (E)-2-Hexenal-based coating induced acquired resistance in apple and its antifungal effects against Penicillium expansum. LWT 2022, 163, 113536. [Google Scholar] [CrossRef]
- Wang, D.; Dong, X. A Highway for War and Peace: The Secretory Pathway in Plant–Microbe Interactions. Mol. Plant 2011, 4, 581–587. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.D.C.; Fadiji, A.E.; Babalola, O.O.; Santoyo, G. Bacterial elicitors of the plant immune system: An overview and the way forward. Plant Stress 2023, 7, 100138. [Google Scholar] [CrossRef]
- Villadangos, S.; Munné-Bosch, S. Acclimation to a combination of water deficit and nutrient deprivation through simultaneous increases in abscisic acid and bioactive jasmonates in the succulent plant Sempervivum tectorum L. J. Plant Physiol. 2023, 287, 154040. [Google Scholar] [CrossRef]
- Kgang, I.E.; Klein, A.; Husselmann, L.; Nkomo, A.; Mathabe, P.M.K.; Belay, Z.A.; Caleb, O.J. Bioassays and proteomics as early detection tools in postharvest management of table grapes (Vitis vinifera L.) diseases—A Review. Food Biosci. 2023, 53, 102645. [Google Scholar] [CrossRef]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone Inhibition of Shoot Branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of Shoot Branching by New Terpenoid Plant Hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef]
- Kapulnik, Y.; Koltai, H. Strigolactone involvement in root development, response to abiotic stress, and interactions with the biotic soil environment. Plant Physiol. 2014, 166, 560–569. [Google Scholar] [CrossRef]
- Decker, E.L.; Alder, A.; Hunn, S.; Ferguson, J.; Lehtonen, M.T.; Scheler, B.; Kerres, K.L.; Wiedemann, G.; Safavi-Rizi, V.; Nordzieke, S.; et al. Strigolactone Biosynthesis Is Evolutionarily Conserved, Regulated by Phosphate Starvation, and Contributes to Resistance against Phytopathogenic Fungi in a Moss, Physcomitrella patens. New Phytol. 2017, 216, 455–468. [Google Scholar] [CrossRef]
- Djennane, S.; Oyant, L.H.; Kawamura, K.; Lalanne, D.; Laffaire, M.; Thouroude, T.; Chalain, S.; Sakr, S.; Boumaza, R.; Foucher, F.; et al. Impacts of light and temperature on shoot branching gradient and expression of strigolactone synthesis and signalling genes in rose. Plant Cell Environ. 2014, 37, 742–757. [Google Scholar] [CrossRef]
- Ma, N.; Hu, C.; Wan, L.; Hu, Q.; Xiong, J.L.; Zhang, C.L. Strigolactones improve plant growth, photosynthesis, and alleviate oxidative stress under salinity in rapeseed (Brassica napus L.) by regulating gene expression. Front. Plant Sci. 2017, 8, 1671. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef]
- Saeed, W.; Naseem, S.; Ali, Z. Strigolactones biosynthesis and their role in abiotic stress resilience in plants: A critical review. Front. Plant Sci. 2017, 8, 1487. [Google Scholar] [CrossRef] [PubMed]
- Ruyter-Spira, C.; Al-Babili, S.; van der Krol, S.; Bouwmeester, H. The biology of strigolactones. Trends Plant Sci. 2013, 18, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Stirnberg, P.; van De Sande, K.; Leyser, H.M. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.D.; de Saint Germain, A.; Pillot, J.P.; Pouvreau, J.B.; Chen, V.X.; Ramos, S.; Stévenin, A.; Simier, P.; Delavault, P.; Beau, J.M.; et al. Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: Molecule design for shoot branching. Plant Physiol. 2012, 159, 1524–1544. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Seto, Y.; Yamaguchi, S. Strigolactone biosynthesis, transport, and perception. Plant J. 2021, 105, 335–350. [Google Scholar] [CrossRef]
- Xie, X.; Mori, N.; Yoneyama, K.; Nomura, T.; Uchida, K.; Yoneyama, K.; Akiyama, K. Lotuslactone, a non-canonical strigolactone from Lotus japonicus. Phytochemistry 2019, 157, 200–205. [Google Scholar] [CrossRef]
- Soliman, S.; Wang, Y.; Han, Z.; Pervaiz, T.; El-Kereamy, A. Strigolactones in plants and their interaction with the ecological microbiome in response to abiotic stress. Plants 2022, 11, 3499. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Sasaki, E.; Shimada, Y.; Nagae, M.; Ueno, K.; Nakano, T.; Yoneyama, K.; Suzuki, Y.; Asami, T. Feedback-Regulation of Strigolactone Biosynthetic Genes and Strigolactone-Regulated Genes in Arabidopsis. Biosci. Biotechnol. Biochem. 2009, 73, 2460–2465. [Google Scholar] [CrossRef]
- Braun, N.; de Saint Germain, A.; Pillot, J.P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N.; et al. The Pea TCP Transcription Factor PsBRC1 Acts Downstream of Strigolactones to Control Shoot Branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 Acts as a Repressor of Strigolactone Signalling in Rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCFD3-Dependent Degradation of D53 Regulates Strigolactone Signalling. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef]
- Kong, C.C.; Ren, C.G.; Li, R.Z.; Xie, Z.H.; Wang, J.P. Hydrogen peroxide and strigolactones signaling are involved in alleviation of salt stress induced by arbuscular mycorrhizal fungus in Sesbania cannabina seedlings. J. Plant Growth Regul. 2017, 36, 734–742. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Yu, H.; Guo, H.; Lin, T.; Kou, L.; Wang, A.; Shao, N.; Ma, H.; Xiong, G.; et al. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 2020, 583, 277–281. [Google Scholar] [CrossRef]
- Lu, T.; Yu, H.; Li, Q.; Chai, L.; Jiang, W.J. Improving plant growth and alleviating photosynthetic inhibition and oxidative stress from low-light stress with exogenous GR24 in tomato (Solanum lycopersicum L.) seedlings. Front. Plant Sci. 2019, 10, 490. [Google Scholar] [CrossRef]
- Blake, S.N.; Barry, K.M.; Gill, W.M.; Reid, J.B.; Foo, E. The role of strigolactones and ethylene in disease caused by Pythium irregulare: Ethylene, but not strigolactones, affects pea disease. Mol. Plant Pathol. 2016, 17, 680–690. [Google Scholar] [CrossRef]
- Foo, E.; Ross, J.J.; Jones, W.T.; Reid, J.B. Plant hormones in arbuscular mycorrhizal symbioses: An emerging role for gibberellins. Ann. Bot. 2013, 111, 769–779. [Google Scholar] [CrossRef]
- Torres-Vera, R.; García, J.M.; Pozo, M.J.; López-Ráez, J.A. Do strigolactones contribute to plant defence?: Strigolactones in plant defence. Mol. Plant Pathol. 2014, 15, 211–216. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Huang, Q.; Li, Y.; Liu, Y.; Wang, Y. Understanding transcription factor regulation by integrating gene expression and DNase I hypersensitive sites. BioMed Res. Int. 2015, 2015, 757530. [Google Scholar] [CrossRef]
- Hong, J.C. General Aspects of Plant Transcription Factor Families. In Plant Transcription Factors: Evolutionary, Structural and Functional Aspects; Gonzalez, D., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 35–56. [Google Scholar] [CrossRef]
- Priya, P.; Jain, M. RiceSRTFDB: A database of rice transcription factors containing comprehensive expression, cis-regulatory element and mutant information to facilitate gene function analysis. Database J. Biol. Database Curation 2013, 2013, bat027. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiao, Y.; Jiao, H.; Zhao, H.; Zhu, Y.X. Two-Step Functional Innovation of the Stem-Cell Factors WUS/WOX5 during Plant Evolution. Mol. Biol. Evol. 2017, 34, 640–653. [Google Scholar] [CrossRef]
- Evans, C.E.B.; Arunkumar, R.; Borrill, P. Transcription factor retention through multiple polyploidization steps in wheat. G3 Genes|Genomes|Genet. 2022, 12, jkac147. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY Transcription Factors and Plant Defense Responses: Latest Discoveries and Future Prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Tompa, M.; Li, N.; Bailey, T.L.; Church, G.M.; De Moor, B.; Eskin, E.; Favorov, A.V.; Frith, M.C.; Fu, Y.; Kent, W.J. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005, 23, 137–144. [Google Scholar] [CrossRef]
- Li, S.; Gao, J.; Yao, L.; Ren, G.; Zhu, X.; Gao, S.; Qiu, K.; Zhou, X.; Kuai, B. The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 2016, 35, 1729–1741. [Google Scholar] [CrossRef]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Aida, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell Online 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Pengfei, D.; Chong, C.; Yuzhen, Z.; Qingwei, M.; Wei, L.; Nana, M. The Role of NAC Transcription Factor in Plant Cold Response. Front. Plant Sci. 2020, 15, 1785668. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC Transcription Factors: Structurally Distinct, Functionally Diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Ernst, H.A.; Olsen, A.N.; Larsen, S.; Lo Leggio, L. Structure of the Conserved Domain of ANAC, a Member of the NAC Family of Transcription Factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Shao, H.; Wang, H.; Tang, X. NAC Transcription Factors in Plant Multiple Abiotic Stress Responses: Progress and Prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- Sperotto, R.A.; Ricachenevsky, F.K.; Duarte, G.L.; Boff, T.; Lopes, K.L.; Sperb, E.R.; Grusak, M.A.; Fett, J.P. Identification of Up-Regulated Genes in Flag Leaves During Rice Grain Filling and Characterization of Os NAC5, a New ABA-Dependent Transcription Factor. Planta 2009, 230, 985–1002. [Google Scholar] [CrossRef]
- Duval, M.; Hsieh, T.F.; Kim, S.Y.; Thomas, L.T. Molecular Characterization of AtNAM: A Member of the Arabidopsis NAC Domain Super Family. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC Transcription Factors in Plant Immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) Transcription Factor Enhances Drought Resistance and Salt Tolerance in Rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Köhler, B.; Mueller-Roeber, B. A Gene Regulatory Network Controlled by the NAC Transcription Factor ANAC092/AtNAC2/ORE1 during Salt-Promoted Senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- Sindhu, A.; Chintamanani, S.; Brandt, A.S.; Zanis, M.; Scofield, S.R.; Johal, G.S. A Guardian of Grasses: Specific Origin and Conservation of a Unique Disease-Resistance Gene in the Grass Lineage. Proc. Natl. Acad. Sci. USA 2008, 105, 1762–1767. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.I.; Watanabe, Y.A.; Mochida, K.E.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Genome-Wide Survey and Expression Analysis of the Plant-Specific NAC Transcription Factor Family in Soybean during Development and Dehydration Stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Chuong, N.N.; Hoa, T.T.K.; Doan, H.; Van, P.H.P.; Trang, L.D.M.; Huyen, P.N.T.; Le, D.T.; Tran, L.S.P.; Thao, N.P. The Drought-Mediated Soybean GmNAC085 Functions as a Positive Regulator of Plant Response to Salinity. Int. J. Mol. Sci. 2021, 22, 8986. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Moumeni, A.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Attia, K.; Kikuchi, S. Comprehensive Gene Expression Analysis of the NAC Gene Family under Normal Growth Conditions, Hormone Treatment, and Drought Stress Conditions in Rice Using Near-Isogenic Lines (NILs) Generated from Crossing Aday Selection (Drought Tolerant) and IR64. Mol. Genet. Genom. 2012, 287, 389–410. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Tolerance to Various Environmental Stresses Conferred by the Salt-Responsive Rice Gene ONAC063 in Transgenic Arabidopsis. Planta 2009, 229, 1065–1075. [Google Scholar] [CrossRef]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC Transcription Factor JUNGBRUNNEN1 Affects Thermomemory-Associated Genes and Enhances Heat Stress Tolerance in Primed and Unprimed Conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef]
- Lu, M.; Ying, S.; Zhang, D.F.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. A Maize Stress-Responsive NAC Transcription Factor, ZmSNAC1, Confers Enhanced Tolerance to Dehydration in Transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-Type Transcription Factor OsNAC2 Regulates ABA-Dependent Genes and Abiotic Stress Tolerance in Rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Liu, X.Y.; Deng, L.; Cai, G.L.; Zhang, Y.; Wang, X.J.; Zhao, J.; Huang, L.L.; Kang, Z.S. Characterization of a Novel Wheat NAC Transcription Factor Gene Involved in Defense Response Against Stripe Rust Pathogen Infection and Abiotic Stresses. Mol. Biol. Rep. 2010, 37, 3703–3712. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, M.; Gao, S.; Zhang, Z.; Zhao, X.; Zhao, C.; Zhang, F.; Chen, X. Molecular Characterization of Novel TaNAC Genes in Wheat and Overexpression of TaNAC2a Confers Drought Tolerance in Tobacco. Physiol. Plant. 2012, 144, 210–224. [Google Scholar] [CrossRef]
- Paul, A.; Muoki, R.C.; Singh, K.; Kumar, S. CsNAM-like protein encodes a nuclear localized protein and responds to varied cues in tea (Camellia sinensis (L.) O. Kuntze. Gene. 2012, 502, 69–74. [Google Scholar] [CrossRef]
- Zhou, W.; Qian, C.; Li, R.; Zhou, S.; Zhang, R.; Xiao, J.; Wang, X.; Zhang, S.; Xing, L.; Cao, A. TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 2018, 277, 218–228. [Google Scholar] [CrossRef]
- Wang, B.; Wei, J.; Song, N.; Wang, N.; Zhao, J.; Kang, Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J. Integr. Plant Biol. 2018, 60, 432–443. [Google Scholar] [CrossRef]
- Feng, H.; Wang, X.; Sun, Y.; Wang, X.; Chen, X.; Guo, J.; Duan, Y.; Huang, L.; Kang, Z. Cloning and Characterization of a Calcium Binding EF-Hand Protein Gene TaCab1 from Wheat and Its Expression in Response to Puccinia striiformis f. sp. tritici and Abiotic Stresses. Mol. Biol. Rep. 2011, 38, 3857–3866. [Google Scholar] [CrossRef]
- Zhang, X.M.; Zhang, Q.; Pei, C.L.; Li, X.; Huang, X.L.; Chang, C.Y.; Wang, X.J.; Huang, L.L.; Kang, Z.S. TaNAC2 is a negative regulator in the wheat-stripe rust fungus interaction at the early stage. Physiol. Mol. Plant Pathol. 2018, 102, 144–153. [Google Scholar] [CrossRef]
- Hickman, R.; Hill, C.; Penfold, C.A.; Breeze, E.; Bowden, L.; Moore, J.D.; Zhang, P.; Jackson, A.; Cooke, E.; Bewicke-Copley, F.; et al. A Local Regulatory Network Around Three NAC Transcription Factors in Stress Responses and Senescence in Arabidopsis Leaves. Plant J. 2013, 75, 26–39. [Google Scholar] [CrossRef]
- Masri, R.; Kiss, E. The Role of NAC Genes in Response to Biotic Stresses in Plants. Physiol. Mol. Plant Pathol. 2023, 126, 102034. [Google Scholar] [CrossRef]
- Liu, Q.; Yan, S.; Huang, W.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; Zhu, X.; et al. NAC Transcription Factor ONAC066 Positively Regulates Disease Resistance by Suppressing the ABA Signaling Pathway in Rice. Plant Mol Biol. 2018, 98, 289–302. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, H.; Li, D.; Huang, L.; Hong, Y.; Ding, X.S.; Nelson, R.S.; Zhou, X.; Song, F. Functions of Rice NAC Transcriptional Factors, ONAC122 and ONAC131, in Defense Responses Against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef]
- Bian, Z.; Gao, H.; Wang, C. NAC Transcription Factors as Positive or Negative Regulators During Ongoing Battle Between Pathogens and Our Food Crops. Int. J. Mol. Sci. 2020, 22, 81. [Google Scholar] [CrossRef]
- Park, S.R.; Kim, H.S.; Lee, K.S.; Hwang, D.J.; Bae, S.C.; Ahn, I.P.; Lee, S.H.; Kim, S.T. Overexpression of Rice NAC Transcription Factor OsNAC58 on Increased Resistance to Bacterial Leaf Blight. J. Plant Biotechnol. 2017, 44, 149–155. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, C.; Shao, X.; Hu, Z.; Li, J.; Wang, P.; Wang, A.; Yu, J.; Shi, K. Transcriptomic and Genetic Approaches Reveal an Essential Role of the NAC Transcription Factor SlNAP1 in the Growth and Defense Response of Tomato. Hortic. Res. 2020, 7, 209. [Google Scholar] [CrossRef]
- Li, T.; Cheng, X.; Wang, X.; Li, G.; Wang, B.; Wang, W.; Zhang, N.; Han, Y.; Jiao, B.; Wang, Y.; et al. Glyoxalase I-4 Functions Downstream of NAC72 to Modulate Downy Mildew Resistance in Grapevine. Plant J. 2021, 108, 394–410. [Google Scholar] [CrossRef]
- Shan, W.E.I.; Chen, J.Y.; Kuang, J.F.; Lu, W.J. Banana Fruit NAC Transcription Factor MaNAC5 Cooperates with MaWRKYs to Enhance the Expression of Pathogenesis-Related Genes against Colletotrichum musae. Mol. Plant Pathol. 2016, 17, 330–338. [Google Scholar] [CrossRef]
- Bu, Q.; Jiang, H.; Li, C.B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC Transcription Factors ANAC019 and ANAC055 in Regulating Jasmonic Acid-Signaled Defense Responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef]
- Cai, W.; Yang, S.; Wu, R.; Cao, J.; Shen, L.; Guan, D.; Shuilin, H. Pepper NAC-Type Transcription Factor NAC2c Balances the Trade-Off between Growth and Defense Responses. Plant Physiol. 2021, 186, 2169–2189. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, T.; Lin, Z.; Gu, B.; Xing, C.; Zhao, L.; Dong, H.; Gao, J.; Xie, Z.; Zhang, S.; et al. A WRKY Transcription Factor PbrWRKY53 from Pyrus betulaefolia Is Involved in Drought Tolerance and AsA Accumulation. Plant Biotechnol. J. 2019, 17, 1770–1787. [Google Scholar] [CrossRef]
- Dong, B.; Sutton, R.T.; Shaffrey, L. Understanding the Rapid Summer Warming and Changes in Temperature Extremes Since the Mid-1990s over Western Europe. Clim. Dyn. 2017, 48, 1537–1554. [Google Scholar] [CrossRef]
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate Variation Explains a Third of Global Crop Yield Variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef]
- Wang, X.; Shi, X.; Chen, S.; Ma, C.; Xu, S. Evolutionary Origin, Gradual Accumulation and Functional Divergence of Heat Shock Factor Gene Family with Plant Evolution. Front. Plant Sci. 2018, 9, 71. [Google Scholar] [CrossRef]
- Shamovsky, I.; Nudler, E. New Insights into the Mechanism of Heat Shock Response Activation. Cell. Mol. Life Sci. 2008, 65, 855–861. [Google Scholar] [CrossRef]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat Shock Factors: Integrators of Cell Stress, Development, and Lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef]
- Pirkkala, L.; Nykänen, P.; Sistonen, L.E.A. Roles of the Heat Shock Transcription Factors in Regulation of the Heat Shock Response and Beyond. FASEB J. 2001, 15, 1118–1131. [Google Scholar] [CrossRef]
- Xin, C.; Wang, X.; Cai, J.; Zhou, Q.; Liu, F.; Dai, T.; Cao, W.; Jiang, D. Changes of Transcriptome and Proteome Are Associated with the Enhanced Post-Anthesis High Temperature Tolerance Induced by Pre-Anthesis Heat Priming in Wheat. Plant Growth Regul. 2016, 79, 135–145. [Google Scholar] [CrossRef]
- Guihur, A.; Fauvet, B.; Finka, A.; Quadroni, M.; Goloubinoff, P. Quantitative Proteomic Analysis to Capture the Role of Heat-Accumulated Proteins in Moss Plant Acquired Thermotolerance. Plant Cell Environ. 2020, 44, 2117–2133. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lu, Z.; Wang, L.; Jin, B. Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. Int. J. Mol. Sci. 2020, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Al-Whaibi, M.H. Plant Heat-Shock Proteins: A Mini Review. J. King Saud Univ.-Sci. 2011, 23, 139–150. [Google Scholar]
- Jee, H. Size Dependent Classification of Heat Shock Proteins: A Mini-Review. J. Exerc. Rehabil. 2016, 12, 255–259. [Google Scholar] [CrossRef]
- Finka, A.; Sharma, S.K.; Goloubinoff, P. Multi-Layered Molecular Mechanisms of Polypeptide Holding, Unfolding, and Disaggregation by HSP70/HSP110 Chaperones. Front. Mol. Biosci. 2015, 2, 29. [Google Scholar]
- Fragkostefanakis, S.; Röth, S.; Schleiff, E.; Scharf, K.D. Prospects of Engineering Thermotolerance in Crops through Modulation of Heat Stress Transcription Factor and Heat Shock Protein Networks. Plant Cell Environ. 2015, 38, 1881–1895. [Google Scholar] [CrossRef]
- Kijima, T.; Prince, T.L.; Tigue, M.L.; Yim, K.H.; Schwartz, H.; Beebe, K.; Lee, S.; Budzynski, M.A.; Williams, H.; Trepel, J.B.; et al. HSP90 Inhibitors Disrupt a Transient HSP90-HSF1 Interaction and Identify a Noncanonical Model of HSP90-Mediated HSF1 Regulation. Sci. Rep. 2018, 8, 6976. [Google Scholar] [CrossRef]
- Ali, A.; Bharadwaj, S.; O’Carroll, R.; Ovsenek, N. HSP90 Interacts with and Regulates the Activity of Heat Shock Factor 1 in Xenopus Oocytes. Mol. Cell. Biol. 1998, 18, 4949–4960. [Google Scholar] [CrossRef]
- Zou, J.; Guo, Y.; Guettouche, T.; Smith, D.F.; Voellmy, R. Repression of Heat Shock Transcription Factor HSF1 Activation by HSP90 (HSP90 Complex) that Forms a Stress-Sensitive Complex with HSF1. Cell 1998, 94, 471–480. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Xie, Y.; Wang, X.; Khaleque, M.A.; Chou, S.D.; Murshid, A.; Prince, T.; Zhang, Y. Signal Transduction Pathways Leading to Heat Shock Transcription. Signal Transduct. Insights 2010, 2, 13–24. [Google Scholar]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The Heat-Inducible Transcription Factor HsfA2 Enhances Anoxia Tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef]
- Pérez-Salamó, I.; Papdi, C.; Rigó, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horváth, B.; Domoki, M.; Darula, Z.; et al. The Heat Shock Factor A4A Confers Salt Tolerance and Is Regulated by Oxidative Stress and the Mitogen-Activated Protein Kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef]
- Huang, H.Y.; Chang, K.Y.; Wu, S.J. High Irradiance Sensitive Phenotype of Arabidopsis hit2/xpo1a Mutant Is Caused in Part by Nuclear Confinement of AtHsfA4a. Biol. Plant. 2018, 62, 69–79. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.D. In the Complex Family of Heat Stress Transcription Factors, HsfA1 Has a Unique Role as Master Regulator of Thermotolerance in Tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar]
- Zhu, B.; Ye, C.; Lü, H.; Chen, X.; Chai, G.; Chen, J.; Wang, C. Identification and Characterization of a Novel Heat Shock Transcription Factor Gene, GmHsfA1, in Soybeans (Glycine max). J. Plant Res. 2006, 119, 247–256. [Google Scholar] [CrossRef]
- Peng, S.; Zhu, Z.; Zhao, K.; Shi, J.; Yang, Y.; He, M.; Wang, Y. A Novel Heat Shock Transcription Factor, VpHsf1, from Chinese Wild Vitis pseudoreticulata Is Involved in Biotic and Abiotic Stresses. Plant Mol. Biol. Report. 2013, 31, 240–247. [Google Scholar] [CrossRef]
- Tian, X.; Wang, F.; Zhao, Y.; Lan, T.; Yu, K.; Zhang, L.; Qin, Z.; Hu, Z.; Yao, Y.; Ni, Z.; et al. Heat Shock Transcription Factor A1b Regulates Heat Tolerance in Wheat and Arabidopsis through OPR3 and Jasmonate Signaling Pathway. Plant Biotechnol. J. 2019, 18, 1109. [Google Scholar] [CrossRef]
- Shah, Z.; Shah, S.H.; Ali, G.S.; Munir, I.; Khan, R.S.; Iqbal, A.; Ahmed, N.; Jan, A. Introduction of Arabidopsis’s Heat Shock Factor HsfA1d Mitigates Adverse Effects of Heat Stress on Potato (Solanum tuberosum L.) Plant. Cell Stress Chaperones 2020, 25, 57–63. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The Role of Class A1 Heat Shock Factors (HSFA1s) in Response to Heat and Other Stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Ma, H.; Wang, C.; Yang, B.; Cheng, H.; Wang, Z.; Mijiti, A.; Ren, C.; Qu, G.; Zhang, H.; Ma, L. CarHSFB2, a Class B Heat Shock Transcription Factor, Is Involved in Different Developmental Processes and Various Stress Responses in Chickpea (Cicer arietinum L.). Plant Mol. Biol. Rep. 2016, 34, 1–14. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional Activation and Phosphorylation of OsCNGC9 Confer Enhanced Chilling Tolerance in Rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.L.; Liu, J.G.; Zhang, Z.; Peng, R.H.; Xiong, A.S.; Yao, Q.H.; Chen, J.M. Isolation, Optimization, and Functional Analysis of the cDNA Encoding Transcription Factor OsDREB1B in Oryza sativa L. Mol. Breed. 2007, 19, 329–340. [Google Scholar] [CrossRef]
- Li, S.; Khoso, M.A.; Xu, H.; Zhang, C.; Liu, Z.; Wagan, S.; Dinislam, K.; Liu, L. WRKY Transcription Factors (TFs) as Key Regulators of Plant Resilience to Environmental Stresses: Current Perspective. Agronomy 2024, 14, 2421. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA Encoding a Novel DNA-Binding Protein, SPF1, that Recognizes SP8 Sequences in the 5′ Upstream Regions of Genes Coding for Sporamin and β-Amylase from Sweet Potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Yuan, H.; Guo, W.; Zhao, L.; Yu, Y.; Chen, S.; Tao, L.; Cheng, L.; Kang, Q.; Song, X.; Wu, J.; et al. Genome-Wide Identification and Expression Analysis of the WRKY Transcription Factor Family in Flax (Linum usitatissimum L.). BMC Genom. 2021, 22, 375. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and Functional Analyses of the Rice WRKY Gene Superfamily Reveal Positive and Negative Regulators of Abscisic Acid Signaling in Aleurone Cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome Sequence of the Palaeopolyploid Soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY Transcription Factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Qin, Y.; Tian, Y.; Liu, X. A Wheat Salinity-Induced WRKY Transcription Factor TaWRKY93 Confers Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef]
- Rizhsky, L.; Davletova, S.; Liang, H.; Mittler, R. The Zinc Finger Protein Zat12 Is Required for Cytosolic Ascorbate Peroxidase 1 Expression During Oxidative Stress in Arabidopsis. J. Biol. Chem. 2004, 279, 11736–11743. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY Transcription Factors in Plant Responses to Stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The Evolution of WRKY Transcription Factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Y.; Liu, J.; Zhao, T.; Yang, C.; Ding, Q.; Zhang, Y.; Mu, J.; Wang, D. Identification of WRKY Transcription Factors Responding to Abiotic Stresses in Brassica napus L. Planta 2022, 255, 3. [Google Scholar] [CrossRef]
- Machens, F.; Becker, M.; Umrath, F.; Hehl, R. Identification of a Novel Type of WRKY Transcription Factor Binding Site in Elicitor-Responsive Cis-Sequences from Arabidopsis thaliana. Plant Mol. Biol. 2014, 84, 371–385. [Google Scholar] [CrossRef]
- Li, C.; Wu, J.; Hu, K.D.; Wei, S.W.; Sun, H.Y.; Hu, L.Y.; Han, Z.; Yao, G.F.; Zhang, H. PyWRKY26 and PybHLH3 Cotargeted the PyMYB114 Promoter to Regulate Anthocyanin Biosynthesis and Transport in Red-Skinned Pears. Hortic. Res. 2020, 7, 37. [Google Scholar] [CrossRef]
- Goyal, P.; Devi, R.; Verma, B.; Hussain, S.; Arora, P.; Tabassum, R.; Gupta, S. WRKY Transcription Factors: Evolution, Regulation, and Functional Diversity in Plants. Protoplasma 2023, 260, 331–348. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen Uptake, Assimilation, and Remobilization in Plants: Challenges for Sustainable and Productive Agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef]
- Kapoor, D.; Bhardwaj, S.; Landi, M.; Sharma, A.; Ramakrishnan, M.; Sharma, A. The Impact of Drought in Plant Metabolism: How to Exploit Tolerance Mechanisms to Increase Crop Production. Appl. Sci. 2020, 10, 5692. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY Superfamily of Plant Transcription Factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.L.; Guo, Z.J.; Wang, H.H.; Li, J. The WRKY Family of Transcription Factors in Rice and Arabidopsis and Their Origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, D.; Zhao, X.; Zhang, M.; Wang, Q.; Hou, X.; Di, D.; Su, B.; Wang, S.; Sun, P. Drought-Responsive WRKY Transcription Factor Genes IgWRKY50 and IgWRKY32 from Iris germanica Enhance Drought Resistance in Transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 983600. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.N.; Xu, S.W.; Sun, Y.L.; Liu, D.; Zhou, L.; Li, Y.; Li, X.B. The Cotton WRKY Transcription Factor (GhWRKY33) Reduces Transgenic Arabidopsis Resistance to Drought Stress. Sci. Rep. 2019, 9, 724. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Yan, Y.; Zhang, S.; Li, H.; Gao, Z.; Wang, C.; Guo, X. GhWRKY21 Regulates ABA-Mediated Drought Tolerance by Fine-Tuning the Expression of GhHAB in Cotton. Plant Cell Rep. 2021, 40, 2135–2150. [Google Scholar] [CrossRef]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating Transcription Factor OsWRKY5 Enhances Drought Tolerance through Abscisic Acid Signaling Pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef]
- Huang, K.; Wu, T.; Ma, Z.; Li, Z.; Chen, H.; Zhang, M.; Bian, M.; Bai, H.; Jiang, W.; Du, X. Rice Transcription Factor OsWRKY55 Is Involved in the Drought Response and Regulation of Plant Growth. Int. J. Mol. Sci. 2021, 22, 4337. [Google Scholar] [CrossRef]
- Fan, X.; Guo, Q.; Xu, P.; Gong, Y.; Shu, H.; Yang, Y.; Ni, W.; Zhang, X.; Shen, X. Transcriptome-Wide Identification of Salt-Responsive Members of the WRKY Gene Family in Gossypium aridum. PLoS ONE 2015, 10, e0126148. [Google Scholar] [CrossRef]
- Wu, M.; Liu, H.L.; Han, G.M.; Cai, R.H.; Pan, F.; Xiang, Y. A Moso Bamboo WRKY Gene PeWRKY83 Confers Salinity Tolerance in Transgenic Arabidopsis Plants. Sci. Rep. 2017, 7, 11721. [Google Scholar] [CrossRef]
- Jiang, Y.; Tong, S.; Chen, N.; Liu, B.; Bai, Q.; Chen, Y.; Bi, H.; Zhang, Z.; Lou, S.; Tang, H.; et al. The PalWRKY77 Transcription Factor Negatively Regulates Salt Tolerance and Abscisic Acid Signaling in Populus. Plant J. 2021, 105, 1258–1273. [Google Scholar] [CrossRef]
- Bo, C.; Cai, R.; Fang, X.; Wu, H.; Ma, Z.; Yuan, H.; Cheng, B.; Fan, J.; Ma, Q. Transcription Factor ZmWRKY20 Interacts with ZmWRKY115 to Repress Expression of ZmbZIP111 for Salt Tolerance in Maize. Plant J. 2022, 111, 1660–1675. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.B.; Cao, L.W.; Fan, J.B.; Ma, G.J.; Chen, L. CdWRKY2-Mediated Sucrose Biosynthesis and CBF-Signaling Pathways Coordinately Contribute to Cold Tolerance in Bermudagrass. Plant Biotechnol. J. 2022, 20, 660–675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T.; et al. The OsWRKY63–OsWRKY76–OsDREB1B Module Regulates Chilling Tolerance in Rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.T.; Xu, X.; Tang, Z.; Zhang, W.W.; Huang, X.Y.; Zhao, F.J. OsWRKY28 Regulates Phosphate and Arsenate Accumulation, Root System Architecture, and Fertility in Rice. Front. Plant Sci. 2018, 9, 1330. [Google Scholar] [CrossRef]
- Zhang, J.; Gu, M.; Liang, R.; Shi, X.; Chen, L.; Hu, X.; Wang, S.; Dai, X.; Qu, H.; Li, H.; et al. OsWRKY21 and OsWRKY108 Function Redundantly to Promote Phosphate Accumulation through Maintaining the Constitutive Expression of OsPHT1;1 under Phosphate-Replete Conditions. New Phytol. 2021, 229, 1598–1614. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhao, X.; Ji, G.; Zhang, X.; Qi, Q.; Xu, X.; Han, Z.; Qiu, C. MdWRKY39 Negatively Regulates Apple Phosphorus-Deficiency Tolerance by Inhibiting MdPHT1;7 Expression. Sci. Hortic. 2023, 310, 111715. [Google Scholar] [CrossRef]
- Chen, N.; Tong, S.; Yang, J.; Qin, J.; Wang, W.; Chen, K.; Shi, W.; Li, J.; Liu, J.; Jiang, Y. PtoWRKY40 Interacts with PtoPHR1-LIKE3 While Regulating the Phosphate Starvation Response in Poplar. Plant Physiol. 2022, 190, 2688–2705. [Google Scholar] [CrossRef]
- Shen, Q.H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ülker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear Activity of MLA Immune Receptors Links Isolate-Specific and Basal Disease-Resistance Responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.; van der Linden, C. The Role of Tomato WRKY Genes in Plant Responses to Combined Abiotic and Biotic Stresses. Front. Plant Sci. 2018, 9, 801. [Google Scholar] [CrossRef]
- Peng, Y.; Bartley, L.E.; Chen, X.; Dardick, C.; Chern, M.; Ruan, R.; Canlas, P.E.; Ronald, P.C. OsWRKY62 Is a Negative Regulator of Basal and Xa21-Mediated Defense against Xanthomonas oryzae pv. oryzae in Rice. Mol. Plant 2008, 1, 446–458. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Vinod, K.M.; Zheng, Z.; Fan, B.; Chen, Z. Roles of Arabidopsis WRKY3 and WRKY4 Transcription Factors in Plant Responses to Pathogens. BMC Plant Biol. 2008, 8, 68. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 Transcription Factor Is Required for Resistance to Necrotrophic Fungal Pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef]
- Bakshi, M.; Oelmuller, R. WRKY Transcription Factors: Jack of Many Trades in Plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef]
- Chen, X.; Liu, J.; Lin, G.; Wang, A.; Wang, Z.; Lu, G. Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis Enhances Resistance to Oxalic Acid and Sclerotinia sclerotiorum. Plant Cell Rep. 2013, 32, 1589–1599. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, D. The WRKY57 Transcription Factor Affects the Expression of Jasmonate ZIM-Domain Genes Transcriptionally to Compromise Botrytis cinerea Resistance. Plant Physiol. 2016, 171, 2771–2782. [Google Scholar] [CrossRef]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D.; et al. Overexpression of CaWRKY27, a Subgroup IIe WRKY Transcription Factor of Capsicum annuum, Positively Regulates Tobacco Resistance to Ralstonia solanacearum Infection. Physiol. Plant. 2014, 150, 397–411. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 Transcription Factor Functions in the TMV-Cg Defense Response by Mediating Both Abscisic Acid and Ethylene Signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Yu, D. Wounding-Induced WRKY8 Is Involved in Basal Defense in Arabidopsis. Mol. Plant-Microbe Interact. 2010, 23, 558–565. [Google Scholar] [CrossRef]
- Pandey, S.P.; Roccaro, M.; Schön, M.; Logemann, E.; Somssich, I.E. Transcriptional Reprogramming Regulated by WRKY18 and WRKY40 Facilitates Powdery Mildew Infection of Arabidopsis. Plant J. 2010, 64, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.C.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 Transcription Factors Interact with Histone Deacetylase 19 in Basal Defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef]
- Wang, J.; Tao, F.; An, F.; Zou, Y.; Tian, W.; Chen, X.; Xu, X.; Hu, X. Wheat Transcription Factor TaWRKY70 Is Positively Involved in High-Temperature Seedling Plant Resistance to Puccinia Striiformis F. Sp. Tritici. Mol. Plant Pathol. 2017, 18, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP Kinase Signalling Cascade in Arabidopsis Innate Immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Choi, H.W.; Hwang, I.S.; Choi, D.S.; Hwang, B.K. Functional Roles of the Pepper Pathogen-Induced bZIP Transcription Factor, CAbZIP1, in Enhanced Resistance to Pathogen Infection and Environmental Stresses. Planta 2006, 224, 1209–1225. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.Y.; Chung, E.; Joung, Y.H.; Pai, H.S.; Hur, C.G.; Choi, D. EST and Microarray Analyses of Pathogen-Responsive Genes in Hot Pepper (Capsicum annuum L.) Non-Host Resistance Against Soybean Pustule Pathogen (Xanthomonas axonopodis pv. glycines). Funct. Integr. Genomics 2004, 4, 196–205. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF Super-Family Transcription Factors in Plants. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Ma, L.; Shi, Q.; Ma, Q.; Wang, X.; Chen, X.; Han, P.; Luo, Y.; Hu, H.; Fei, X.; Wei, A. Genome-Wide Analysis of AP2/ERF Transcription Factors That Regulate Fruit Development of Chinese Prickly Ash. BMC Plant Biol. 2024, 24, 565. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 Domain of APETALA2 Defines a Large New Family of DNA Binding Proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP Family of Plant Transcription Factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar]
- Magnani, E.; Sjolander, K.; Hake, S. From Endonucleases to Transcription Factors: Evolution of the AP2 DNA Binding Domain in Plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A Novel Mode of DNA Recognition by a β-Sheet Revealed by the Solution Structure of the GCC-Box Binding Domain in Complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration- and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis Flower and Seed Development by the Homeotic Gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Zhang, C.H.; Shangguan, L.F.; Ma, R.J.; Sun, X.; Tao, R.; Guo, L.; Korir, N.K.; Yu, M.L. Genome-Wide Analysis of the AP2/ERF Superfamily in Peach (Prunus persica). Genet. Mol. Res. 2012, 11, 4789–4809. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S. Genome-Wide Identification and Phylogenetic Analysis of the ERF Gene Family in Cucumbers. Genet. Mol. Biol. 2011, 34, 624–633. [Google Scholar] [CrossRef]
- Cheng, F.; Liu, S.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Li, P.; Hua, W.; Wang, X. BRAD, the Genetics and Genomics Database for Brassica Plants. BMC Plant Biol. 2011, 11, 136. [Google Scholar] [CrossRef]
- Zhuang, J.; Xiong, A.S.; Peng, R.H.; Gao, F.; Zhu, B.; Zhang, J.; Fu, X.Y.; Jin, X.F.; Chen, J.M.; Zhang, Z.; et al. Analysis of Brassica rapa ESTs: Gene Discovery and Expression Patterns of AP2/ERF Family Genes. Mol. Biol. Rep. 2010, 37, 2485–2492. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.C.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, Gene Structures, and Expression Patterns of the ERF Gene Family in Soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef]
- Zhuang, J.; Anyia, A.; Vidmar, J.; Xiong, A.S.; Zhang, J. Discovery and Expression Assessment of the AP2-Like Genes in Hordeum vulgare. Acta Physiol. Plant. 2011, 33, 1639–1649. [Google Scholar] [CrossRef]
- Zhuang, J.; Chen, J.M.; Yao, Q.H.; Xiong, F.; Sun, C.C.; Zhou, X.R.; Zhang, J.; Xiong, A.S. Discovery and Expression Profile Analysis of AP2/ERF Family Genes from Triticum aestivum. Mol. Biol. Rep. 2011, 38, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic Expression of BABY BOOM Triggers a Conversion from Vegetative to Embryonic Growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Hirota, A.; Kato, T.; Fukaki, H.; Aida, M.; Tasaka, M. The Auxin-Regulated AP2/EREBP Gene PUCHI is Required for Morphogenesis in the Early Lateral Root Primordium of Arabidopsis. Plant Cell 2007, 19, 2156–2168. [Google Scholar] [CrossRef] [PubMed]
- Kitomi, Y.; Ito, H.; Hobo, T.; Aya, K.; Kitano, H.; Inukai, Y. The Auxin Responsive AP2/ERF Transcription Factor CROWN ROOTLESS5 is Involved in Crown Root Initiation in Rice Through the Induction of OsRR1, a Type-A Response Regulator of Cytokinin Signaling. Plant J. 2011, 67, 472–484. [Google Scholar] [CrossRef]
- Marsch-Martinez, N.; Greco, R.; Becker, J.D.; Dixit, S.; Bergervoet, J.H.; Karaba, A.; de Folter, S.; Pereira, A. BOLITA, an Arabidopsis AP2/ERF-Like Transcription Factor That Affects Cell Expansion and Proliferation/Differentiation Pathways. Plant Mol. Biol. 2006, 62, 825–843. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Wang, H.; Fu, Z.; Liu, J.; Yu, Y. Identification and Expression Analysis of ERF Transcription Factor Genes in Petunia During Flower Senescence and in Response to Hormone Treatments. J. Exp. Bot. 2011, 62, 825–840. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Omidyar, P.K.; Gee, Z.; Okamuro, J.K. Control of Seed Mass and Seed Yield by the Floral Homeotic Gene APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3117–3122. [Google Scholar] [CrossRef]
- Pietsch, C.; Sreenivasulu, N.; Wobus, U.; Röder, M.S. Linkage Mapping of Putative Regulator Genes of Barley Grain Development Characterized by Expression Profiling. BMC Plant Biol. 2009, 9, 4. [Google Scholar] [CrossRef]
- Chuck, G.; Meeley, R.B.; Hake, S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 1998, 12, 1145–1154. [Google Scholar] [CrossRef]
- Irish, V.F.; Sussex, I.M. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 1990, 2, 741–753. [Google Scholar]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal, and environmental signals in stress acclimation and retrograde signaling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.Q.; Shen, C.; Wu, L.H.; Tang, K.X.; Lin, J. CBF-Dependent Signaling Pathway: A Key Responder to Low Temperature Stress in Plants. Crit. Rev. Biotechnol. 2011, 31, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Li, J.; Long, S.; Wei, S. A DREB1 gene from zoysiagrass enhances Arabidopsis tolerance to temperature stresses without growth inhibition. Plant Sci. 2019, 278, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Hwang, J.E.; Lim, C.J.; Chen, H.; Je, J.; Song, C.; Lim, C.O. Overexpression of Arabidopsis dehydration-responsive element-binding protein 2C confers tolerance to oxidative stress. Mol. Cells 2012, 33, 135–140. [Google Scholar] [CrossRef]
- Mao, D.; Chen, C. Colinearity and Similar Expression Pattern of Rice DREB1s Reveal Their Functional Conservation in the Cold-Responsive Pathway. Plant Mol. Biol. 2012, 80, 243–257. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Z.L.; Park, J.; Tyler, L.; Yusuke, J.; Qiu, K.; Nam, E.A.; Lumba, S.; Desveaux, D.; McCourt, P. The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 2016, 171, 2760–2770. [Google Scholar] [CrossRef]
- Wang, K.; Guo, H.; Yin, Y. AP2/ERF Transcription Factors and Their Functions in Arabidopsis Responses to Abiotic Stresses. Environ. Exp. Bot. 2024, 222, 105763. [Google Scholar] [CrossRef]
- Ma, Z.; Jin, Y.M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 2022, 13, 1007811. [Google Scholar] [CrossRef]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. Int. J. Mol. Sci. 2022, 23, 12013. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.Y.; Zhang, J.F.; Zhang, H.W.; Zhang, Z.J.; Quan, R.D.; Zhou, S.R.; Huang, R.F. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 2011, 6, e25216. [Google Scholar] [CrossRef]
- Fu, J.; Zhu, C.; Wang, C.; Liu, L.; Shen, Q.; Xu, D.; Wang, Q. Maize transcription factor ZmEREB20 enhanced salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 159, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Li, X.; Zhang, Z.; Ai, S.; Liu, C.; Ma, F.; Li, C. MdERF114 Enhances the Resistance of Apple Roots to Fusarium solani by Regulating the Transcription of MdPRX63. Plant Physiol. 2023, 192, 2015–2029. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, D.; Kawamata, A.; Kato, H.; Saburi, W.; Mori, H.; Imai, R. The Rice Ethylene Response Factor OsERF83 Positively Regulates Disease Resistance to Magnaporthe oryzae. Plant Sci. 2019, 135, 263–271. [Google Scholar] [CrossRef]
- Zhao, Y.; Chang, X.; Qi, D.; Dong, L.; Wang, G.; Fan, S.; Jiang, L.; Cheng, Q.; Chen, X.; Han, D.; et al. A Novel Soybean ERF Transcription Factor, GmERF113, Increases Resistance to Phytophthora sojae Infection in Soybean. Front. Plant Sci. 2017, 8, 299. [Google Scholar] [CrossRef]
- Tian, Z.; He, Q.; Wang, H.; Liu, Y.; Zhang, Y.; Shao, F.; Xie, C. The Potato ERF Transcription Factor StERF3 Negatively Regulates Resistance to Phytophthora infestans and Salt Tolerance in Potato. Plant Cell Physiol. 2015, 56, 992–1005. [Google Scholar] [CrossRef]
- Maruyama, Y.; Yamoto, N.; Suzuki, Y.; Chiba, Y.; Yamazaki, K.I.; Sato, T.; Yamaguchi, J. The Arabidopsis Transcriptional Repressor ERF9 Participates in Resistance Against Necrotrophic Fungi. Plant Sci. 2013, 213, 79–87. [Google Scholar] [CrossRef]
- Wang, J.H.; Gu, K.D.; Han, P.L.; Yu, J.Q.; Wang, C.K.; Zhang, Q.Y.; You, C.X.; Hu, D.G.; Hao, Y.J. Apple Ethylene Response Factor MdERF11 Confers Resistance to Fungal Pathogen Botryosphaeria dothidea. Plant Sci. 2020, 291, 110351. [Google Scholar] [CrossRef]
- Zheng, X.; Xing, J.; Zhang, K.; Pang, X.; Zhao, Y.; Wang, G.; Zang, J.; Huang, R.; Dong, J. Ethylene Response Factor ERF11 Activates BT4 Transcription to Regulate Immunity to Pseudomonas syringae. Plant Physiol. 2019, 180, 1132–1151. [Google Scholar] [CrossRef]
- Moffat, C.S.; Ingle, R.A.; Wathugala, D.L.; Saunders, N.J.; Knight, H.; Knight, M.R. ERF5 and ERF6 Play Redundant Roles as Positive Regulators of JA/Et-Mediated Defense Against Botrytis cinerea in Arabidopsis. PLoS ONE 2012, 7, e35995. [Google Scholar] [CrossRef]
- Huang, L.J.; Zhang, J.; Lin, Z.; Yu, P.; Lu, M.; Li, N. The AP2/ERF Transcription Factor ORA59 Regulates Ethylene-Induced Phytoalexin Synthesis Through Modulation of an Acyltransferase Gene Expression. J. Cell Physiol. 2024, 239, e30935. [Google Scholar] [CrossRef] [PubMed]
- Zarei, A.; Körbes, A.P.; Younessi, P.; Montiel, G.; Champion, A.; Memelink, J. Two GCC Boxes and AP2/ERF-Domain Transcription Factor ORA59 in Jasmonate/Ethylene-Mediated Activation of the PDF1.2 Promoter in Arabidopsis. Plant Mol. Biol. 2011, 75, 321–331. [Google Scholar] [CrossRef]
- Lu, J.; Ju, H.; Zhou, G.; Zhu, C.; Erb, M.; Wang, X.; Wang, P.; Lou, Y. An EAR-Motif-Containing ERF Transcription Factor Affects Herbivore-Induced Signaling, Defense, and Resistance in Rice. Plant J. 2011, 68, 583–596. [Google Scholar] [CrossRef]
- Yang, C.; Liu, R.; Pang, J.; Ren, B.; Zhou, H.; Wang, G.; Wang, E.; Liu, J. Poaceae-Specific Cell Wall-Derived Oligosaccharides Activate Plant Immunity via OsCERK1 During Magnaporthe oryzae Infection in Rice. Nat. Commun. 2021, 12, 2178. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Wu, C.J.; Shan, W.; Liu, X.C.; Zhu, L.S.; Wei, W.; Yang, Y.Y.; Guo, Y.F.; Bouzayen, M.; Chen, J.Y.; Lu, W.J.; et al. Phosphorylation of Transcription Factor bZIP21 by MAP Kinase MPK6-3 Enhances Banana Fruit Ripening. Plant Physiol. 2022, 188, 1665–1685. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Wei, K.A.; Chen, J.U.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-Wide Analysis of bZIP-Encoding Genes in Maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef]
- Li, X.; Gao, S.; Tang, Y.; Li, L.; Zhang, F.; Feng, B.; Fang, Z.; Ma, L.; Zhao, C. Genome-Wide Identification and Evolutionary Analyses of bZIP Transcription Factors in Wheat and Its Relatives and Expression Profiles of Anther Development-Related TabZIP Genes. BMC Genom. 2015, 16, 976. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62, and GmbZIP78 Genes Function as Negative Regulators of ABA Signaling and Confer Salt and Freezing Tolerance in Transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wang, L.; Tie, W.; Yan, Y.; Ding, Z.; Liu, J.; Li, M.; Peng, M.; Xu, B.; Jin, Z. Genome-Wide Analyses of the bZIP Family Reveal Their Involvement in the Development, Ripening, and Abiotic Stress Response in Banana. Sci. Rep. 2016, 6, 30203. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Alber, T. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 1992, 2, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Pabo, C.O.; Sauer, R.T. Transcription factors: Structural families and principles of DNA recognition. Annu. Rev. Biochem. 1992, 61, 1053–1095. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP Transcription Factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Joo, H.; Baek, W.; Lim, C.W.; Lee, S.C. Post-translational modifications of bZIP transcription factors in abscisic acid signaling and drought responses. Curr. Genomics 2021, 22, 4–15. [Google Scholar] [CrossRef]
- Hwang, I.; Jung, H.J.; Park, J.I.; Yang, T.J.; Nou, I.S. Transcriptome analysis of newly classified bZIP transcription factors of Brassica rapa in cold stress response. Genomics 2014, 104, 194–202. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP transcription factors in plant salt stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Liu, J.; Jia, J. A Novel Wheat bZIP Transcription Factor, TabZIP60, Confers Multiple Abiotic Stress Tolerances in Transgenic Arabidopsis. Physiol. Plant. 2015, 153, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xia, C.; Gao, L.; Hao, C. A Novel Wheat C-bZIP Gene, TabZIP14-B, Participates in Salt and Freezing Tolerance in Transgenic Plants. Front. Plant Sci. 2017, 8, 710. [Google Scholar] [CrossRef]
- Chang, Y.; Nguyen, B.H.; Xie, Y.; Xiao, B.; Tang, N.; Zhu, W.; Mou, T.; Xiong, L. Co-Overexpression of the Constitutively Active Form of OsbZIP46 and ABA-Activated Protein Kinase SAPK6 Improves Drought and Temperature Stress Resistance in Rice. Front. Plant Sci. 2017, 8, 1102. [Google Scholar] [CrossRef]
- Alves, M.S.; Soares, Z.G.; Vidigal, P.M.; Barros, E.G.; Poddanosqui, A.M.; Aoyagi, L.N.; Abdelnoor, R.V.; Marcelino-Guimarães, F.C.; Fietto, L.G. Differential expression of four soybean bZIP genes during Phakopsora pachyrhizi infection. Funct. Integr. Genom. 2015, 15, 685–696. [Google Scholar] [CrossRef]
- Li, X.; Fan, S.; Hu, W.; Liu, G.; Wei, Y.; He, C.; Shi, H. Two Cassava Basic Leucine Zipper (bZIP) Transcription Factors (MebZIP3 and MebZIP5) Confer Disease Resistance Against Cassava Bacterial Blight. Front. Plant Sci. 2017, 8, 2110. [Google Scholar] [CrossRef]
- Chern, M.S.; Fitzgerald, H.A.; Yadav, R.C.; Canlas, P.E.; Dong, X.; Ronald, P.C. Evidence for a Disease-Resistance Pathway in Rice Similar to the NPR1-Mediated Signaling Pathway in Arabidopsis. Plant J. 2001, 27, 101–113. [Google Scholar] [CrossRef]
- Kuhlmann, M.; Horvay, K.; Strathmann, A.; Heinekamp, T.; Fischer, U.; Böttner, S.; Dröge-Laser, W. The α-Helical D1 Domain of the Tobacco bZIP Transcription Factor BZI-1 Interacts with the Ankyrin-Repeat Protein ANK1 and Is Important for BZI-1 Function, Both in Auxin Signaling and Pathogen Response. J. Biol. Chem. 2003, 278, 8786–8794. [Google Scholar] [CrossRef]
- Joo, J.; Hab, Y.; Sang, L.; Song, I. OsbZIP42 Is a Positive Regulator of ABA Signaling and Confers Drought Tolerance to Rice. Planta 2019, 249, 1521–1533. [Google Scholar] [CrossRef]
- Tak, H.; Mhatre, M. Cloning and Molecular Characterization of a Putative bZIP Transcription Factor VvbZIP23 from Vitis vinifera. Protoplasma 2012, 250, 333–345. [Google Scholar] [CrossRef]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef]
- Parenicova, L.; De Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and Phylogenetic Analyses of the Complete MADS-Box Transcription Factor Family in Arabidopsis: New Openings to the MADS World. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J.; Lee, S.; An, G.; Ma, H.; Nei, M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 2004, 101, 1910–1915. [Google Scholar] [CrossRef]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Agarwal, P.; Khurana, P. Overexpression of TaMADS from wheat promotes flowering by upregulating expression of floral promoters and provides protection against thermal stress. Plant Gene 2019, 17, 100168. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Callens, C.; Tucker, M.R.; Zhang, D.; Wilson, Z.A. Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 2018, 69, 2435–2459. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Yan, W.; Chen, D.; Kaufmann, K. Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 2016, 29, 154–162. [Google Scholar] [CrossRef]
- Bartlett, M.E. Changing MADS-box transcription factor protein–protein interactions as a mechanism for generating floral morphological diversity. Integr. Comp. Biol. 2017, 57, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, B.; Wu, Q.; Min, Q.; Zeng, R.; Xie, Z.; Huang, J. OsMADS23 Phosphorylated by SAPK9 Confers Drought and Salt Tolerance by Regulating ABA Biosynthesis in Rice. PLoS Genet. 2021, 17, e1009699. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Xu, Y.; Liu, H.; Mao, Z.; Zhang, C.; Ma, Y.; Zhang, Q.; Meng, Z.; Chong, K. The Interaction Between OsMADS57 and OsTB1 Modulates Rice Tillering via DWARF14. Nat. Commun. 2013, 4, 1566. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Liang, B.; Yang, L.; Hu, W.; Kuang, L.; Song, J.; Xie, J.; Huang, Y.; Liu, D.; Liu, Y. The MADS-Box Family Gene PtrANR1 Encodes a Transcription Activator Promoting Root Growth and Enhancing Plant Tolerance to Drought Stress. Plant Cell Rep. 2023, 43, 16. [Google Scholar] [CrossRef]
- Jia, J.; Zhao, P.; Cheng, L.; Yuan, G.; Yang, W.; Liu, S.; Chen, S.; Qi, D.; Liu, G.; Li, X. MADS-Box Family Genes in Sheepgrass and Their Involvement in Abiotic Stress Responses. BMC Plant Biol. 2018, 18, 42. [Google Scholar] [CrossRef]
- Zhang, H.; Teng, W.; Liang, J.; Liu, X.; Zhang, H.; Zhang, Z.; Zheng, X. MADS1, a Novel MADS-Box Protein, Is Involved in the Response of Nicotiana benthamiana to Bacterial Harpin (Xoo). J. Exp. Bot. 2016, 67, 131–141. [Google Scholar] [CrossRef]
- Khong, G.N.; Pati, P.K.; Richaud, F.; Parizot, B.; Bidzinski, P.; Mai, C.D.; Bès, M.; Bourrié, I.; Meynard, D.; Beeckman, T.; et al. OsMADS26 Negatively Regulates Resistance to Pathogens and Drought Tolerance in Rice. Plant Physiol. 2015, 169, 2935–2949. [Google Scholar] [CrossRef]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P.; et al. Genome-Wide Identification and Analysis of the MADS-Box Gene Family in Bread Wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef]
- Koops, P.; Pelser, S.; Ignatz, M.; Klose, C.; Marrocco-Selden, K.; Kretsch, T. EDL3 Is an F-Box Protein Involved in the Regulation of Abscisic Acid Signaling in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 5547–5560. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Paz-Ares, J. MYB Transcription Factors in Plants. Trends Genet. 1997, 13, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Lipsick, J.S. One Billion Years of Myb. Oncogene 1996, 13, 223–235. [Google Scholar] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Du, H.; Liang, Z.; Zhao, S.; Nan, M.G.; Tran, L.S.; Lu, K.; Huang, Y.B.; Li, J.N. The Evolutionary History of R2R3-MYB Proteins across 50 Eukaryotes: New Insights into Subfamily Classification and Expansion. Sci. Rep. 2015, 5, 11037. [Google Scholar] [CrossRef]
- Romero, I.; Fuertes, A.; Benito, M.J.; Malpical, J.M.; Leyva, A.; Paz-Ares, J. More than 80 R2R3-MYB Regulatory Genes in the Genome of Arabidopsis thaliana. Plant J. 1998, 14, 273–284. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research Advances of MYB Transcription Factors in Plant Stress Resistance and Breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Li, P.; Wen, J.; Chen, P.; Guo, P.; Ke, Y.; Wang, M.; Liu, M.; Tran, L.S.P.; Li, J.; Du, H. MYB Superfamily in Brassica napus: Evidence for Hormone-Mediated Expression Profiles, Large Expansion, and Functions in Root Hair Development. Biomolecules 2020, 10, 875. [Google Scholar] [CrossRef]
- Shi, M.Z.; Xie, D.Y. Biosynthesis and Metabolic Engineering of Anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar]
- Ogata, K.; Morikawa, S.; Nakamura, H.; Sekikawa, A.; Inoue, T.; Kanai, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution Structure of a Specific DNA Complex of the Myb DNA-Binding Domain with Cooperative Recognition Helices. Cell 1994, 79, 639–648. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yang, X.Y.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.X.; Qiu, X.; et al. The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef]
- Li, C.; Ng, C.K.Y.; Fan, L.M. MYB Transcription Factors, Active Players in Abiotic Stress Signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Wu, X.; Xia, M.; Su, P.; Zhang, Y.; Tu, L.; Zhao, H.; Gao, W.; Huang, L.; Hu, Y. MYB Transcription Factors in Plants: A Comprehensive Review of Their Discovery, Structure, Classification, Functional Diversity, and Regulatory Mechanism. Int. J. Biol. Macromol. 2024, 282, 136652. [Google Scholar] [CrossRef]
- Oshima, Y.; Shikata, M.; Koyama, T.; Ohtsubo, N.; Mitsuda, N.; Ohme-Takagi, M. MIXTA-like Transcription Factors and WAX INDUCER1/SHINE1 Coordinately Regulate Cuticle Development in Arabidopsis and Torenia fournieri. Plant Cell 2013, 25, 1609–1625. [Google Scholar] [CrossRef]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 Transcription Factor Regulates Cuticular Wax Biosynthesis under Drought Conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef]
- Liu, A.; Yu, Y.; Li, R.; Duan, X.; Zhu, D.; Sun, X.; Duanmu, H.; Zhu, Y. A Novel Hybrid Proline-Rich Type Gene GsEARLI17 from Glycine soja Participated in Leaf Cuticle Synthesis and Plant Tolerance to Salt and Alkali Stresses. Plant Cell Tissue Organ Cult. 2015, 121, 633–646. [Google Scholar] [CrossRef]
- Chen, D.; Chen, H.; Dai, G.; Zhang, H.; Liu, Y.; Shen, W.; Zhu, B.; Cui, C.; Tan, C. Genome-Wide Identification of R2R3-MYB Gene Family and Association with Anthocyanin Biosynthesis in Brassica Species. BMC Genom. 2022, 23, 441. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, J.; Hu, R.; Wu, P.; Hou, X.L.; Song, X.M.; Xiong, A.S. Genome-Wide Analysis of the R2R3-MYB Transcription Factor Genes in Chinese Cabbage (Brassica rapa ssp. pekinensis) Reveals Their Stress and Hormone Responsive Patterns. BMC Genom. 2015, 16, 17. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Chen, G.; Ye, X.; Zhang, L.; Zhu, S.; Yuan, L.; Hou, J.; Wang, C. Functional Analysis of a MYB Transcription Factor BrTDF1 in Tapetum Development of Wucai (Brassica rapa ssp.). Sci. Hortic. 2019, 257, 108684. [Google Scholar] [CrossRef]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome-Wide Analysis of the Apple MYB Transcription Factor Family Allows the Identification of MdoMYB121 Gene Conferring Abiotic Stress Tolerance in Plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef]
- Wang, R.K.; Cao, Z.H.; Hao, Y.J. Overexpression of an R2R3 MYB Gene MdSIMYB1 Increases Tolerance to Multiple Stresses in Transgenic Tobacco and Apples. Physiol. Plant. 2014, 150, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Mattana, M.; Biazzi, E.; Consonni, R.; Locatelli, F.; Vannini, C.; Provera, S.; Coraggio, I. Overexpression of OsMYB4 Enhances Compatible Solute Accumulation and Increases Stress Tolerance of Arabidopsis thaliana. Physiol. Plant. 2005, 125, 212–223. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB Gene, OsMYB3R-2, Increases Tolerance to Freezing, Drought, and Salt Stress in Transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Li, Z. Overexpression of OsMYB48-1, a Novel MYB-Related Transcription Factor, Enhances Drought and Salinity Tolerance in Rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef]
- Casaretto, J.A.; El-Kereamy, A.; Zeng, B.; Stiegelmeyer, S.M.; Chen, X.; Bi, Y.-M.; Rothstein, S.J. Expression of OsMYB55 in Maize Activates Stress-Responsive Genes and Enhances Heat and Drought Tolerance. BMC Genom. 2016, 17, 312. [Google Scholar] [CrossRef]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W.; et al. Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, Y. Genome-Wide Survey and Characterization of Greenbug-Induced NAC Transcription Factors in Sorghum (Sorghum bicolor (L.) Moench). In Proceedings of the XXI Annual International Plant & Animal Genome Conference, San Diego, CA, USA, 12–16 January 2013; p. 191. [Google Scholar]
- Ibraheem, F.; Gafoor, I.; Tan, Q.; Shyu, C.-R.; Chopra, S. A Sorghum MYB Transcription Factor Induces 3-Deoxyanthocyanidins and Enhances Resistance Against Leaf Blights in Maize. Molecules 2015, 20, 2388–2404. [Google Scholar] [CrossRef]
- Shan, T.; Rong, W.; Xu, H.; Du, L.; Liu, X.; Zhang, Z. The Wheat R2R3-MYB Transcription Factor TaRIM1 Participates in Resistance Response Against the Pathogen Rhizoctonia cerealis Infection Through Regulating Defense Genes. Sci. Rep. 2016, 6, 28777. [Google Scholar] [CrossRef]
- Segarra, G.; Van der Ent, S.; Trillas, I.; Pieterse, C.M.J. MYB72, a Node of Convergence in Induced Systemic Resistance Triggered by a Fungal and a Bacterial Beneficial Microbe. Plant Biol. 2009, 11, 90–96. [Google Scholar] [CrossRef]
- Li, C.; Chen, Q.; Gao, X.; Qi, B.; Chen, N.; Xu, S.; Chen, J.; Wang, X. AtHsfA2 Modulates Expression of Stress Responsive Genes and Enhances Tolerance to Heat and Oxidative Stress in Arabidopsis. Sci. China Life Sci. 2005, 48, 540–550. [Google Scholar] [CrossRef]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-Level Overexpression of the Arabidopsis HsfA2 Gene Confers Not Only Increased Thermotolerance but Also Salt/Osmotic Stress Tolerance and Enhanced Callus Growth. J. Exp. Bot. 2007, 58, 3373–3383. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, S.; Yu, Y.; Cui, N.; Yu, G.; Zhao, H.; Meng, X.; Fan, H. The Pivotal Role of MYB Transcription Factors in Plant Disease Resistance. Planta 2023, 258, 16. [Google Scholar] [CrossRef] [PubMed]
- Shi, H. Function Analysis of Transcription Factor AtMYB44 and AtMYB15 During Defense Response in Arabidopsis. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2010. [Google Scholar]
- Liu, R.; Chen, L.; Jia, Z.; Lü, B.; Shi, H.; Shao, W.; Dong, H. Transcription Factor AtMYB44 Regulates Induced Expression of the ETHYLENE INSENSITIVE2 Gene in Arabidopsis Responding to a Harpin Protein. Mol. Plant-Microbe Interact. 2011, 24, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, K.; Maki, H.; Itaya, T.; Suzuki, T.; Nomoto, M.; Sakaoka, S.; Morikami, A.; Higashiyama, T.; Tada, Y.; Busch, W.; et al. MYB30 Links ROS Signaling, Root Cell Elongation, and Plant Immune Responses. Proc. Natl. Acad. Sci. USA 2018, 115, E4710–E4719. [Google Scholar] [CrossRef]
- Shen, X.; Guo, X.; Guo, X.; Zhao, D.; Zhao, W.; Chen, J.; Li, T. PacMYBA, a Sweet Cherry R2R3-MYB Transcription Factor, Is a Positive Regulator of Salt Stress Tolerance and Pathogen Resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef]
- Kim, S.H.; Lam, P.Y.; Lee, M.H.; Jeon, H.S.; Tobimatsu, Y.; Park, O.K. The Arabidopsis R2R3 MYB Transcription Factor MYB15 Is a Key Regulator of Lignin Biosynthesis in Effector-Triggered Immunity. Front. Plant Sci. 2020, 11, 583153. [Google Scholar] [CrossRef]
- Xiao, S.; Hu, Q.; Shen, J.; Liu, S.; Yang, Z.; Chen, K.; Klosterman, S.J.; Javornik, B.; Zhang, X.; Zhu, L. GhMYB4 Downregulates Lignin Biosynthesis and Enhances Cotton Resistance to Verticillium dahliae. Plant Cell Rep. 2021, 40, 735–751. [Google Scholar] [CrossRef]
- Liu, T.; Chen, T.; Kan, J.; Yao, Y.; Guo, D.; Yang, Y.; Ling, X.; Wang, J.; Zhang, B. The GhMYB36 Transcription Factor Confers Resistance to Biotic and Abiotic Stress by Enhancing PR1 Gene Expression in Plants. Plant Biotechnol. J. 2022, 20, 722–735. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, X.; Wang, P.; Wang, H.; Ge, X.; Li, F.; Hou, Y. GhODO1, an R2R3-Type MYB Transcription Factor, Positively Regulates Cotton Resistance to Verticillium dahliae via the Lignin Biosynthesis and Jasmonic Acid Signaling Pathway. Int. J. Biol. Macromol. 2022, 201, 580–591. [Google Scholar] [CrossRef]
- Li, T.; Zhang, X.Y.; Huang, Y.; Xu, Z.S.; Wang, F.; Xiong, A.S. An R2R3-MYB Transcription Factor, SlMYB28, Involved in the Regulation of TYLCV Infection in Tomato. Sci. Hortic. 2018, 237, 192–200. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, Y.; Yang, C.; Geng, S.; Sun, Q.; Long, L.; Cai, C.; Chu, Z.; Liu, X.; Wang, G.; et al. Characterization, Expression, and Functional Analysis of a Novel NAC Gene Associated with Resistance to Verticillium Wilt and Abiotic Stress in Cotton. G3 Genes Genomes Genet. 2016, 6, 3951–3961. [Google Scholar] [CrossRef]
- Wang, X.; Goregaoker, S.P.; Culver, J.N. Interaction of the Tobacco mosaic virus Replicase Protein with a NAC Domain Transcription Factor Is Associated with the Suppression of Systemic Host Defenses. J. Virol. 2009, 83, 9720–9730. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Wu, J.G.; Lii, Y.F.; Li, Y.; Jin, H.L. Contribution of Small RNA Pathway Components in Plant Immunity. Mol. Plant-Microbe Interact. 2013, 26, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and Functional Analysis of Arabidopsis Stress-Inducible NAC Transcription Factors That Bind to a Drought-Responsive Cis-Element in the Early Responsive to Dehydration Stress 1 Promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Kim, H.S.; Park, H.C.; Kim, K.E.; Jung, M.S.; Han, H.J.; Kim, S.H.; Kwon, Y.S.; Bahk, S.; An, J.; Bae, D.W.; et al. A NAC Transcription Factor and SNI1 Cooperatively Suppress Basal Pathogen Resistance in Arabidopsis thaliana. Nucleic Acids Res. 2012, 40, 9182–9192. [Google Scholar] [CrossRef]
- Guo, T.; Mao, X.; Zhang, H.; Zhang, Y.; Fu, M.; Sun, Z.; Kuai, P.; Lou, Y.; Fang, Y. Lamin-Like Proteins Negatively Regulate Plant Immunity through NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Mol. Plant 2017, 10, 1334–1348. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC Transcription Factors Promote Abiotic Stress Tolerance and Lateral Root Formation in Transgenic Plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeon, H.S.; Kim, H.G.; Park, O.K. An Arabidopsis NAC Transcription Factor NAC4 Promotes Pathogen-Induced Cell Death Under Negative Regulation by microRNA164. New Phytol. 2017, 214, 343–360. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A Dehydration-Induced NAC Protein, RD26, Is Involved in a Novel ABA-Dependent Stress-Signaling Pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC Transcription Factor Enhances Rice Drought and Salt Tolerance. Biochem. Biophys. Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of a NAC-Type Transcription Factor OsNAC6 Involved in Abiotic- and Biotic-Stress-Responsive Gene Expression in Rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-Specific Expression of OsNAC10 Improves Drought Tolerance and Grain Yield in Rice under Field Drought Conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of Transcription Factor Gene SNAC2 Conferring Cold and Salt Tolerance in Rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Liu, B.; Ouyang, Z.; Zhang, Y.; Li, X.; Hong, Y.; Huang, L.; Liu, S.; Zhang, H.; Li, D.; Song, F. Tomato NAC Transcription Factor SlSRN1 Positively Regulates Defense Response against Biotic Stress but Negatively Regulates Abiotic Stress Response. PLoS ONE 2014, 9, e102067. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely Related NAC Transcription Factors of Tomato Differentially Regulate Stomatal Closure and Reopening during Pathogen Attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The Target Gene of tae-miR164, a Novel NAC Transcription Factor from the NAM Subfamily, Negatively Regulates Resistance of Wheat to Stripe Rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef]
- Wang, F.; Lin, R.; Feng, J.; Chen, W.; Qiu, D.; Xu, S. TaNAC1 Acts as a Negative Regulator of Stripe Rust Resistance in Wheat, Enhances Susceptibility to Pseudomonas syringae, and Promotes Lateral Root Development in Transgenic Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 108. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Su, P.H.; Li, H.M. Arabidopsis Stromal 70-kD Heat Shock Proteins Are Essential for Plant Development and Important for Thermotolerance of Germinating Seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Jia, H.; Li, Y.; Xu, X.; Wang, L.; Lu, M. The Populus trichocarpa PtHSP17.8 Involved in Heat and Salt Stress Tolerances. Plant Cell Rep. 2016, 35, 1587–1599. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Zhang, H.; Wang, H.; Wei, T.; Che, S.; Zhang, L.; Hu, B.; Long, H.; Song, W.; et al. Identification of MsHsp20 Gene Family in Malus sieversii and Functional Characterization of MsHsp16.9 in Heat Tolerance. Front. Plant Sci. 2017, 8, 1761. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Bi, Y.; Li, M.; Zhang, R.; Feng, S.; Ming, F. A Small Heat Shock Protein Gene (RcHSP17.8) from Chinese Rose Confers Resistance to Various Abiotic Stresses in Transgenic Tobacco. Plant Cell Tiss. Org. 2020, 141, 407–415. [Google Scholar] [CrossRef]
- Li, H.Y.; Chang, C.S.; Lu, L.S.; Liu, C.A.; Chan, M.T.; Charng, Y.Y. Over-Expression of Arabidopsis thaliana Heat Shock Factor Gene (AtHsfA1b) Enhances Chilling Tolerance in Transgenic Tomato. Bot. Bull. Acad. Sin. 2003, 44, 129–140. [Google Scholar]
- Jiang, Y.; Qiu, Y.; Hu, Y.; Yu, D. Heterologous Expression of AtWRKY57 Confers Drought Tolerance in Oryza sativa. Front. Plant Sci. 2016, 7, 145. [Google Scholar] [CrossRef]
- Babitha, K.C.; Ramu, S.V.; Pruthvi, V.; Mahesh, P.; Nataraja, K.N.; Udayakumar, M. Co-Expression of AtbHLH17 and AtWRKY28 Confers Resistance to Abiotic Stress in Arabidopsis. Transgenic Res. 2013, 22, 327–341. [Google Scholar] [CrossRef]
- Grunewald, W.; Karimi, M.; Wieczorek, K.; Van de Cappelle, E.; Wischnitzki, E.; Grundler, F.; Inzé, D.; Beeckman, T.; Gheysen, G. A Role for AtWRKY23 in Feeding Site Establishment of Plant-Parasitic Nematodes. Plant Physiol. 2008, 148, 358–368. [Google Scholar] [CrossRef]
- Liu, H.; Yang, W.; Liu, D.; Han, Y.; Zhang, A.; Li, S. Ectopic Expression of a Grapevine Transcription Factor VvWRKY11 Contributes to Osmotic Stress Tolerance in Arabidopsis. Mol. Biol. Rep. 2011, 38, 417–427. [Google Scholar] [CrossRef]
- Song, Y.; Ai, C.R.; Jing, S.J.; Yu, D.Q. Research Progress on Function Analysis of Rice WRKY Gene. Rice Sci. 2010, 17, 60–72. [Google Scholar] [CrossRef]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.S.; Park, S.R.; Lee, S.; Hwang, D.J. Rice WRKY11 Plays a Role in Pathogen Defense and Drought Tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef]
- Zhou, Q.-Y.; Tian, A.-G.; Zou, H.-F.; Xie, Z.-M.; Lei, G.; Huang, J.; Wang, C.-M.; Wang, H.-W.; Zhang, J.-S.; Chen, S.-Y. Soybean WRKY-Type Transcription Factor Genes, GmWRKY13, GmWRKY21, and GmWRKY54, Confer Differential Tolerance to Abiotic Stresses in Transgenic Arabidopsis Plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Wang, F.; Hou, X.; Tang, J.; Wang, Z.; Wang, S.; Jiang, F.; Li, Y. A Novel Cold-Inducible Gene from Pak-Choi (Brassica campestris ssp. chinensis), BcWRKY46, Enhances the Cold, Salt and Dehydration Stress Tolerance in Transgenic Tobacco. Mol. Biol. Rep. 2011, 39, 4553–4564. [Google Scholar] [CrossRef]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The Maize WRKY Transcription Factor ZmWRKY17 Negatively Regulates Salt Stress Tolerance in Transgenic Arabidopsis Plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef]
- Dong, Q.; Zheng, W.; Duan, D.; Huang, D.; Wang, Q.; Liu, C.; Li, C.; Gong, X.; Li, C.; Mao, K.; et al. MdWRKY30, a Group IIa WRKY Gene from Apple, Confers Tolerance to Salinity and Osmotic Stresses in Transgenic Apple Callus and Arabidopsis Seedlings. Plant Sci. 2020, 299, 110611. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The miR156/SPL Module Regulates Apple Salt Stress Tolerance by Activating MdWRKY100 Expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef]
- Song, C.P.; Galbraith, D.W. AtSAP18, an Orthologue of Human SAP18, Is Involved in the Regulation of Salt Stress and Mediates Transcriptional Repression in Arabidopsis. Plant Mol. Biol. 2006, 60, 241–257. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, L.; Hu, H.; Tang, N.; Shi, L.; Xu, F.; Wang, S. Arabidopsis ERF012 Is a Versatile Regulator of Plant Growth, Development and Abiotic Stress Responses. Int. J. Mol. Sci. 2022, 23, 6841. [Google Scholar] [CrossRef]
- Li, J.; Guo, X.; Zhang, M.; Wang, X.; Zhao, Y.; Yin, Z.; Zhang, Z.; Wang, Y.; Xiong, H.; Zhang, H.; et al. OsERF71 Confers Drought Tolerance via Modulating ABA Signaling and Proline Biosynthesis. Plant Sci. 2018, 270, 131–139. [Google Scholar] [CrossRef]
- Chen, J.Q.; Meng, X.P.; Zhang, Y.; Xia, M.; Wang, X.P. Over-Expression of OsDREB Genes Lead to Enhanced Drought Tolerance in Rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of Rice Transcription Factor OsERF19 in Response to ABA and Salt Stress Responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a Novel DNA-Binding Protein, Binds to Bipartite Recognition Sequence through Two Distinct DNA-Binding Domains Uniquely Found in Higher Plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a Family of ABA-Responsive Element Binding Factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozake, K. Arabidopsis Basic Leucine Zipper Transcription Factors Involved in an Abscisic Acid-Dependent Signal Transduction Pathway under Drought and High-Salinity Conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Iwata, Y.; Koizumi, N. An Arabidopsis Transcription Factor, AtbZIP60, Regulates the Endoplasmic Reticulum Stress Response in a Manner Unique to Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 5280–5285. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic Reticulum Stress Responses in Plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Kaiser, A.; Lindsay, W.P.; Halkier, B.A.; Loake, G.J.; Doerner, P.; Dixon, R.A.; Lamb, C. Rapid Stimulation of a Soybean Protein-Serine Kinase That Phosphorylates a Novel bZIP DNA-Binding Protein, G/HBF-1, during the Induction of Early Transcription-Dependent Defenses. EMBO J. 1997, 16, 726–738. [Google Scholar] [CrossRef]
- Nazri, A.Z.; Griffin, J.H.C.; Peaston, K.A.; Alexander-Webber, D.G.A.; Williams, L.E. F-Group bZIPs in Barley—A Role in Zn Deficiency. Plant Cell Environ. 2017, 40, 2754–2770. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a Key Player of the Basic Leucine Zipper Transcription Factor Family for Conferring Abscisic Acid Sensitivity and Salinity and Drought Tolerance in Rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a Positive Regulator of ABA Response and Drought Tolerance in Rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef]
- Gao, S.Q.; Chen, M.; Xu, Z.S.; Zhao, C.P.; Li, L.; Xu, H.J.; Tang, Y.M.; Zhao, X.; Ma, Y.Z. The Soybean GmbZIP1 Transcription Factor Enhances Multiple Abiotic Stress Tolerances in Transgenic Plants. Plant Mol. Biol. 2011, 75, 537–553. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 Enhances Tolerance to Drought and Salt Stresses in Soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef]
- Takeshima, R.; Nan, H.; Harigai, K.; Dong, L.; Zhu, J.; Lu, S.; Xu, M.; Yamagishi, N.; Yoshikawa, N.; Liu, B.; et al. Functional Divergence Between Soybean FLOWERING LOCUS T Orthologues FT2a and FT5a in Post-Flowering Stem Growth. J. Exp. Bot. 2019, 70, 3941–3953. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Cao, D.; Zhang, D.; Li, Y.; Lu, S.; Tang, L.; Yuan, X.; Liu, B.; Kong, F. GmFT2a and GmFT5a Redundantly and Differentially Regulate Flowering Through Interaction with and Upregulation of the bZIP Transcription Factor GmFDL19 in Soybean. PLoS ONE 2014, 9, e97669. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Pei, X.; Kong, F.; Zhao, L.; Lin, X. Divergence of Functions and Expression Patterns of Soybean bZIP Transcription Factors. Front. Plant Sci. 2023, 14, 1150363. [Google Scholar] [CrossRef]
- Gonçalves, A.B.; Fontes, P.P.; Dadalto, S.P.; de Souza, G.B.; Marcelino-Guimarães, F.C.; Alves, M.S.; Fietto, L.G. GmbZIP45 Binds to H-Box Cis-Element in Vitro and Overexpression in Soybean Protoplasts Induces the Expression of CHS8 Gene. Physiol. Mol. Plant Pathol. 2020, 112, 101556. [Google Scholar] [CrossRef]
- Sun, C.H.; Yu, J.-Q.; Duan, X.; Wang, J.-H.; Zhang, Q.-Y.; Gu, K.-D.; Hu, D.-G.; Zheng, C.-S. The MADS Transcription Factor CmANR1 Positively Modulates Root System Development by Directly Regulating CmPIN2 in Chrysanthemum. Hortic. Res. 2018, 5, e52. [Google Scholar] [CrossRef]
- Mizzotti, C.; Mendes, M.A.; Caporali, E.; Schnittger, A.; Kater, M.M.; Battaglia, R.; Colombo, L. The MADS Box Genes SEEDSTICK and Arabidopsis Bsister Play a Maternal Role in Fertilization and Seed Development. Plant J. 2012, 70, 409–420. [Google Scholar] [CrossRef]
- He, S.; Ma, R.; Liu, Z.; Zhang, D.; Wang, S.; Guo, Y.; Chen, M. Overexpression of BnaAGL11, a MADS-Box Transcription Factor, Regulates Leaf Morphogenesis and Senescence in Brassica Napus. J. Agric. Food Chem. 2022, 70, 3420–3434. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence Meristem Identity in Rice Is Specified by Overlapping Functions of Three AP1/FUL-like MADS Box Genes and PAP2, a SEP-ALLATA MADS Box Gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef]
- Yin, X.; Liu, X.; Xu, B.; Lu, P.; Dong, T.; Yang, D.; Ye, T.; Feng, Y.Q.; Wu, Y. OsMADS18, a Membrane-Bound MADS-Box Transcription Factor, Modulates Plant Architecture and the Abscisic Acid Response in Rice. J. Exp. Bot. 2019, 70, 3895–3909. [Google Scholar] [CrossRef]
- Ku, A.T.; Huang, Y.S.; Wang, Y.S.; Ma, D.; Yeh, K.W. IbMADS1 (Ipomoea batatas MADS-Box 1 Gene) Is Involved in Tuberous Root Initiation in Sweet Potato (Ipomoea batatas). Ann. Bot. 2008, 102, 57–67. [Google Scholar] [CrossRef]
- Noh, S.A.; Lee, H.S.; Huh, E.J.; Huh, G.H.; Paek, K.H.; Shin, J.S.; Bae, J.M. SRD1 Is Involved in the Auxin-Mediated Initial Thickening Growth of Storage Root by Enhancing Proliferation of Metaxylem and Cambium Cells in Sweetpotato (Ipomoea batatas). J. Exp. Bot. 2010, 61, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Vailleau, F.; Léger, A.; Joubès, J.; Miersch, O.; Huard, C.; Roby, D. A MYB Transcription Factor Regulates Very-Long-Chain Fatty Acid Biosynthesis for Activation of the Hypersensitive Cell Death Response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Cominelli, E.; Sala, T.; Calvi, D.; Gusmaroli, G.; Tonelli, C. Overexpression of the Arabidopsis AtMYB41 Gene Alters Cell Expansion and Leaf Surface Permeability. Plant J. 2008, 53, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Kosma, D.K.; Murmu, J.; Razeq, F.M.; Santos, P.; Bourgault, R.; Molina, I.; Rowland, O. AtMYB41 Activates Ectopic Suberin Synthesis and Assembly in Multiple Plant Species and Cell Types. Plant J. 2014, 80, 216–229. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Y.H.; Zheng, H.; Lu, W.; Wu, C.A.; Huang, J.G.; Zheng, C.C. Salt-Induced Transcription Factor MYB74 Is Regulated by the RNA-Directed DNA Methylation Pathway in Arabidopsis. J. Exp. Bot. 2015, 66, 5997–6008. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W. An R2R3-Type MYB Gene, OsMYB2, Is Involved in Salt, Cold, and Dehydration Tolerance in Rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, Z.; Jiang, H.; Zhang, Z.; Chen, X. A Feedback Loop Involving MdMYB108L and MdHY5 Controls Apple Cold Tolerance. Biochem. Biophys. Res. Commun. 2019, 512, 381–386. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Zhou, X.; Hou, X.; Yang, G.; Meng, J.; Luan, Y. Tomato MYB49 Enhances Resistance to Phytophthora infestans and Tolerance to Water Deficit and Salt Stress. Planta 2018, 248, 1487–1503. [Google Scholar] [CrossRef]
- Xue, C.; Yao, J.L.; Xue, Y.S.; Su, G.Q.; Wang, L.; Lin, L.K.; Allan, A.C.; Zhang, S.L.; Wu, J. PbrMYB169 Positively Regulates Lignification in Fruit Stone Cells of Pear (Pyrus bretschneideri). J. Exp. Bot. 2019, 70, 1801–1814. [Google Scholar] [CrossRef]
- Slesak, I.; Libik, M.; Karpinska, B.; Karpinski, S.; Miszalski, Z. The Role of Hydrogen Peroxide in Regulation of Plant Metabolism and Cellular Signalling in Response to Environmental Stresses. Acta Biochim. Pol. 2007, 54, 39–50. [Google Scholar] [CrossRef]
- Mateo, A.; Mühlenbock, P.; Restérucci, C.; Chi-Chang, C.; Miszalski, Z.; Karpinska, B.; Parker, J.E.; Mullineaux, P.M.; Karpinski, S. LESION SIMULATING DISEASE 1 Required for Acclimation to Conditions That Promote Excess Excitation Energy. Plant Physiol. 2004, 136, 2818–2830. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Schinkel, H.; Witzell, J.; Hertzberg, H.; Torp, M.; Srivastava, K.M.; Karpinska, B.; Melzer, M.; Wingsle, G. Down-Regulation of High Isoelectric Point Extracellular Superoxide Dismutase Mediates Alterations in Reactive Oxygen Species Metabolism and Developmental Disturbances in Hybrid Aspen. Plant J. 2006, 49, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Do, P.T.; Drechsel, O.; Heyer, A.G.; Hincha, D.K.; Zuther, E. Changes in Free Polyamine Levels, Expression of Polyamine Biosynthesis Genes, and Performance of Rice Cultivars under Salt Stress: A Comparison with Responses to Drought. Front. Plant Sci. 2014, 5, 87243. [Google Scholar] [CrossRef]
- Ebeed, H.T. Genome-Wide Analysis of Polyamine Biosynthesis Genes in Wheat Reveals Gene Expression Specificity and Involvement of STRE and MYB-Elements in Regulating Polyamines under Drought. BMC Genom. 2022, 23, 734. [Google Scholar] [CrossRef]
- Liu, M.; Chen, J.; Guo, Z.; Lu, S. Differential Responses of Polyamines and Antioxidants to Drought in a Centipedegrass Mutant in Comparison to Its Wild-Type Plants. Front. Plant Sci. 2017, 8, 792. [Google Scholar] [CrossRef]
- Wu, H.; Fu, B.; Sun, P.; Xiao, C.; Liu, J.-H. A NAC Transcription Factor Represses Putrescine Biosynthesis and Affects Drought Tolerance. Plant Physiol. 2016, 172, 1532–1547. [Google Scholar] [CrossRef]
- Zhu, M.D.; Zhang, M.; Gao, D.J.; Zhou, K.; Tang, S.J.; Zhou, B.; Lv, Y.M. Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels. Int. J. Mol. Sci. 2020, 21, 1857. [Google Scholar] [CrossRef]
- Chávez-Martínez, A.I.; Ortega-Amaro, M.A.; Torres, M.; Serrano, M.; Jiménez-Bremont, J.F. Arabidopsis adc-Silenced Line Exhibits Differential Defense Responses to Botrytis cinerea and Pseudomonas syringae Infection. Plant Physiol. Biochem. 2020, 156, 494–503. [Google Scholar] [CrossRef]
- Jia, Y.; Qin, D.; Zheng, Y.; Wang, Y. Finding Balance in Adversity: Nitrate Signaling as the Key to Plant Growth, Resilience, and Stress Response. Int. J. Mol. Sci. 2023, 24, 14406. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-Regulated Auxin Transport by NRT1.1 Defines a Mechanism for Nutrient Sensing in Plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [PubMed]
- Ye, J.Y.; Tian, W.H.; Zhou, M.; Zhu, Q.Y.; Du, W.X.; Zhu, Y.X.; Liu, X.X.; Lin, X.Y.; Zheng, S.J.; Jin, C.W. STOP1 Activates NRT1.1-Mediated Nitrate Uptake to Create a Favorable Rhizospheric pH for Plant Adaptation to Acidity. Plant Cell 2021, 33, 3658–3674. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhao, C.; Hu, S.; Zuo, K. Arabidopsis Calcium-Dependent Protein Kinase CPK6 Regulates Drought Tolerance under High Nitrogen by the Phosphorylation of NRT1.1. J. Exp. Bot. 2023, 74, 5682–5693. [Google Scholar] [CrossRef] [PubMed]
- Pande, A.; Mun, B.G.; Lee, D.S.; Khan, M.; Lee, G.M.; Hussain, A.; Yun, B.W. NO Network for Plant–Microbe Communication Underground: A Review. Front. Plant Sci. 2021, 12, 658679. [Google Scholar] [CrossRef]
- Pande, A.; Mun, B.G.; Khan, M.; Rahim, W.; Lee, D.S.; Lee, G.M.; Al Azawi, T.N.I.; Hussain, A.; Yun, B.W. Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants. Int. J. Mol. Sci. 2022, 23, 1657. [Google Scholar] [CrossRef]
- Khan, M.; Imran, Q.M.; Shahid, M.; Mun, B.G.; Lee, S.U.; Khan, M.A.; Hussain, A.; Lee, I.J.; Yun, B.W. Nitric Oxide-Induced AtAO3 Differentially Regulates Plant Defense and Drought Tolerance in Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 602. [Google Scholar] [CrossRef]
- Brouquisse, R. Multifaceted Roles of Nitric Oxide in Plants. J. Exp. Bot. 2019, 70, 4319–4322. [Google Scholar] [CrossRef]
- Mur, L.A.; Kumari, A.; Brotman, Y.; Zeier, J.; Mandon, J.; Cristescu, S.M.; Harren, F.; Kaiser, W.M.; Fernie, A.R.; Gupta, K.J. Nitrite and Nitric Oxide Are Important in the Adjustment of Primary Metabolism during the Hypersensitive Response in Tobacco. J. Exp. Bot. 2019, 70, 4571–4582. [Google Scholar] [CrossRef]
- Meister, A. Metabolism and Functions of Glutathione. Trends Biochem. Sci. 1981, 6, 231–234. [Google Scholar] [CrossRef]
- Sharma, K.G.; Sharma, V.; Bourbouloux, A.; Delrot, S.; Bachhawat, A.K. Glutathione Depletion Leads to Delayed Growth Stasis in Saccharomyces cerevisiae: Evidence of a Partially Overlapping Role for Thioredoxin. Curr. Genet. 2000, 38, 71–77. [Google Scholar] [CrossRef]
- Srivastava, M.K.; Dwivedi, U.N. Salicylic Acid Modulates Glutathione Metabolism in Pea Seedlings. J. Plant Physiol. 1998, 153, 409–414. [Google Scholar] [CrossRef]
- Mou, Z.; Fan, W.; Dong, X. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell 2003, 113, 935–944. [Google Scholar] [PubMed]
- Cao, S.; Chen, Z.; Liu, G.; Jiang, L.; Yuan, H.; Ren, G.; Bian, X.; Jian, H.; Ma, X. The Arabidopsis Ethylene-Insensitive 2 Gene Is Required for Lead Resistance. Plant Physiol. Biochem. 2009, 47, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Ghanta, S.; Bhattacharyya, D.; Sinha, R.; Banerjee, A.; Chattopadhyay, S. Nicotiana tabacum Overexpressing γ-ECS Exhibits Biotic Stress Tolerance Likely through NPR1-Dependent Salicylic Acid-Mediated Pathway. Planta 2011, 233, 895–910. [Google Scholar] [CrossRef]
- Deprost, D.; Yao, L.; Sormani, R.; Moreau, M.; Leterreux, G.; Nicolaï, M.; Bedu, M.; Robaglia, C.; Meyer, C. The Arabidopsis TOR Kinase Links Plant Growth, Yield, Stress Resistance, and mRNA Translation. EMBO Rep. 2007, 8, 864–870. [Google Scholar] [CrossRef]
- Baena-González, E.; Sheen, J. Convergent Energy and Stress Signaling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose Metabolism in Plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef]
- Eh, T.J.; Jiang, Y.; Jiang, M.; Li, J.; Lei, P.; Ji, X.; Kim, H.I.; Zhao, X.; Meng, F. The Role of Trehalose Metabolism in Plant Stress Tolerance. J. Adv. Res. 2024, in press. [Google Scholar] [CrossRef]
- Samtani, H.; Sharma, A.; Khurana, P. Overexpression of HVA1 Enhances Drought and Heat Stress Tolerance in Triticum aestivum Doubled Haploid Plants. Cells 2022, 11, 912. [Google Scholar] [CrossRef]
- Mbinda, W.; Dixelius, C.; Oduor, R. Induced Expression of Xerophyta viscosa XvSap1 Gene Enhances Drought Tolerance in Transgenic Sweet Potato. Front. Plant Sci. 2019, 10, 1119. [Google Scholar] [CrossRef]
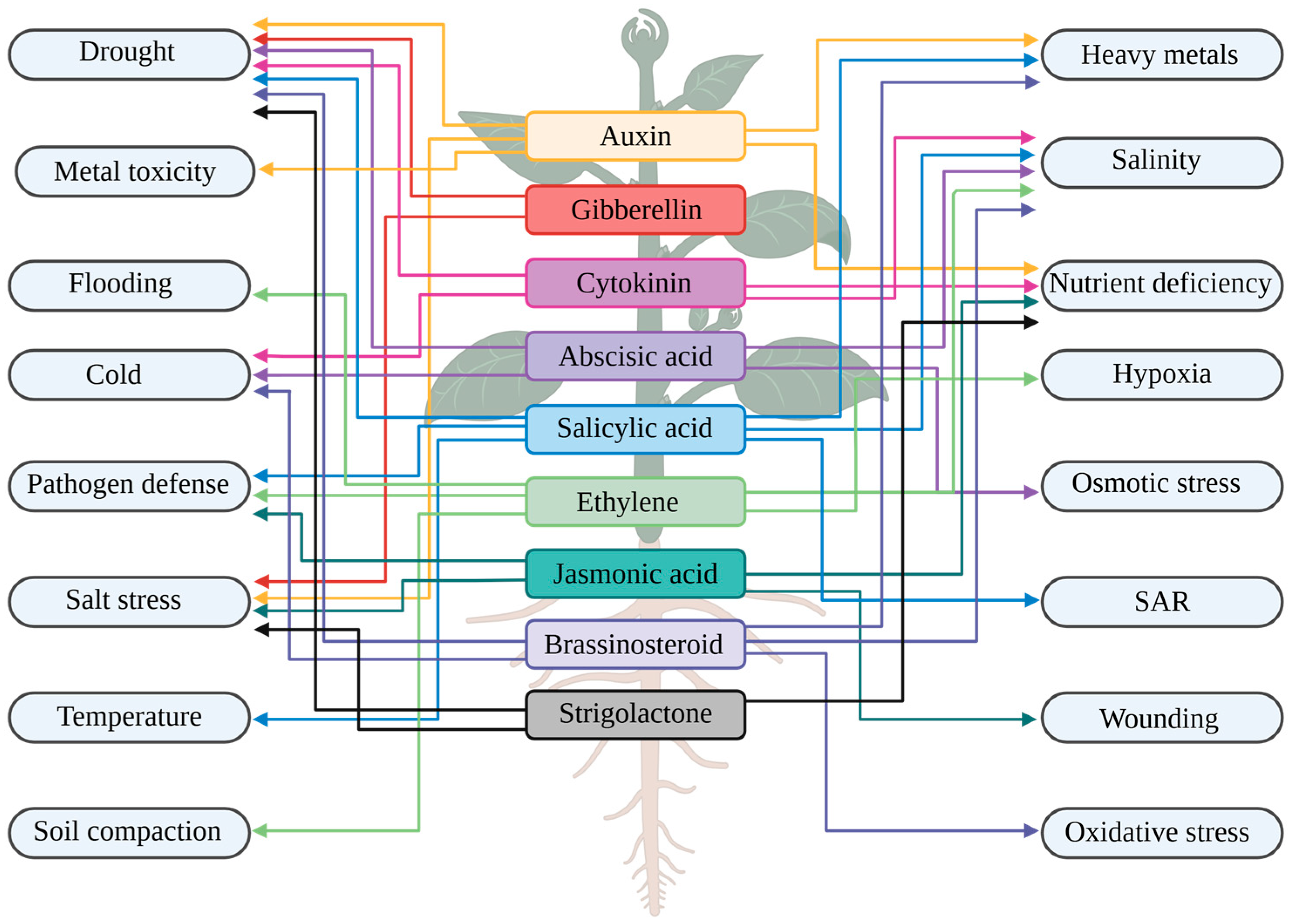
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thilakarathne, A.S.; Liu, F.; Zou, Z. Plant Signaling Hormones and Transcription Factors: Key Regulators of Plant Responses to Growth, Development, and Stress. Plants 2025, 14, 1070. https://doi.org/10.3390/plants14071070
Thilakarathne AS, Liu F, Zou Z. Plant Signaling Hormones and Transcription Factors: Key Regulators of Plant Responses to Growth, Development, and Stress. Plants. 2025; 14(7):1070. https://doi.org/10.3390/plants14071070
Chicago/Turabian StyleThilakarathne, Ayomi S., Fei Liu, and Zhongwei Zou. 2025. "Plant Signaling Hormones and Transcription Factors: Key Regulators of Plant Responses to Growth, Development, and Stress" Plants 14, no. 7: 1070. https://doi.org/10.3390/plants14071070
APA StyleThilakarathne, A. S., Liu, F., & Zou, Z. (2025). Plant Signaling Hormones and Transcription Factors: Key Regulators of Plant Responses to Growth, Development, and Stress. Plants, 14(7), 1070. https://doi.org/10.3390/plants14071070








