Mutation of ZmDIR5 Reduces Maize Tolerance to Waterlogging, Salinity, and Drought
Abstract
1. Introduction
2. Materials and Methods
2.1. Maize Materials and Stress Treatment Methods
2.2. Tobacco Materials and Subcellular Localization Analysis
2.3. Establishment of ZmDIR5-EMS Mutant Lines
2.4. Bioinformatics Analysis Methods
2.5. Measurement of Growth Indices
2.6. Measurement of Physiological Indices and Antioxidant Enzyme Activity
2.7. Measurement of Na+ and K+ Content
2.8. Measurement of Total Lignan Content
2.9. Measurement of ABA and Zeatin Content
2.10. Total RNA Extraction and Reverse Transcription
2.11. Quantitative Real-Time PCR
2.12. Statistical Analysis
3. Results
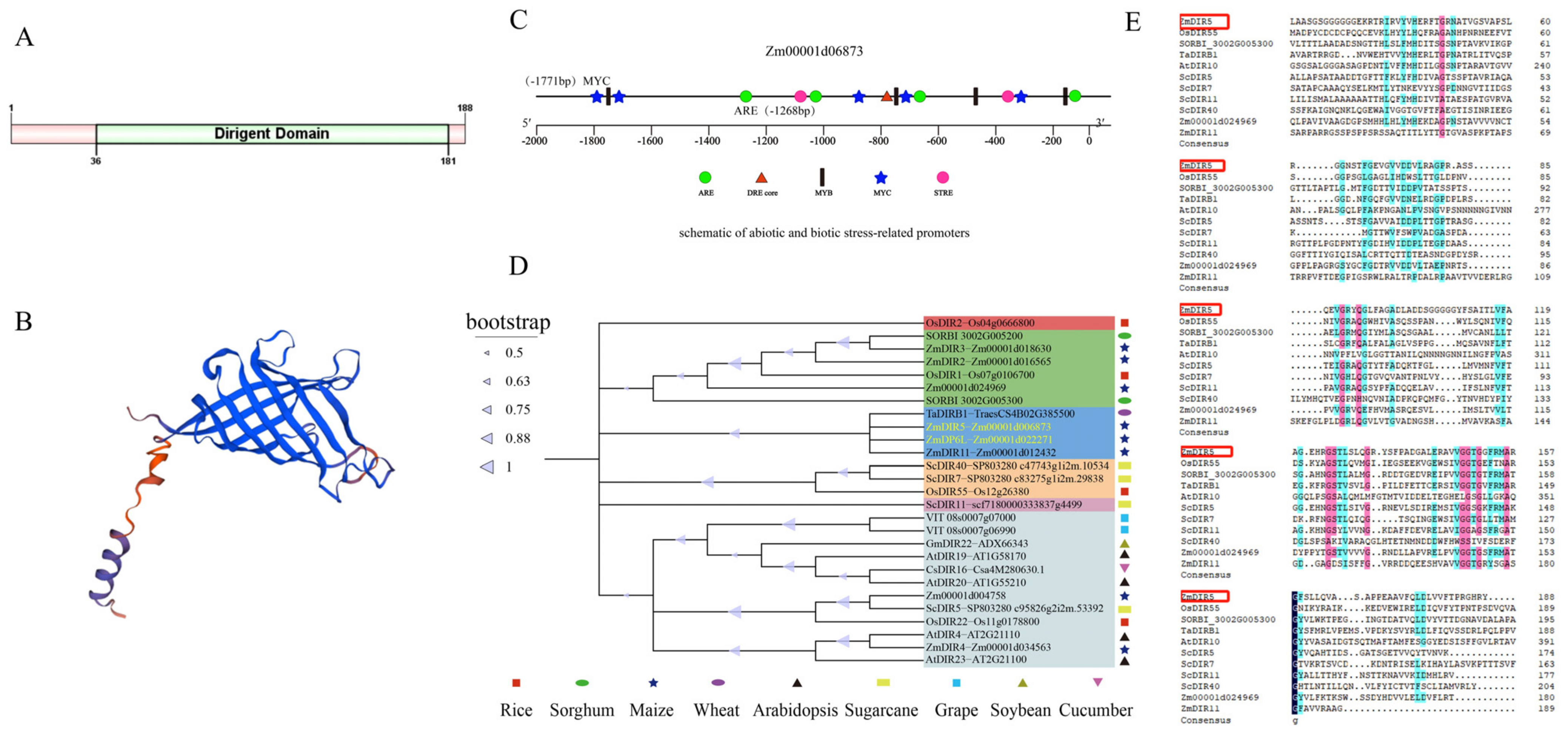
3.1. Identification and Bioinformatics Analysis of ZmDIR5
3.2. Expression Pattern Analysis and Subcellular Localization of ZmDIR5
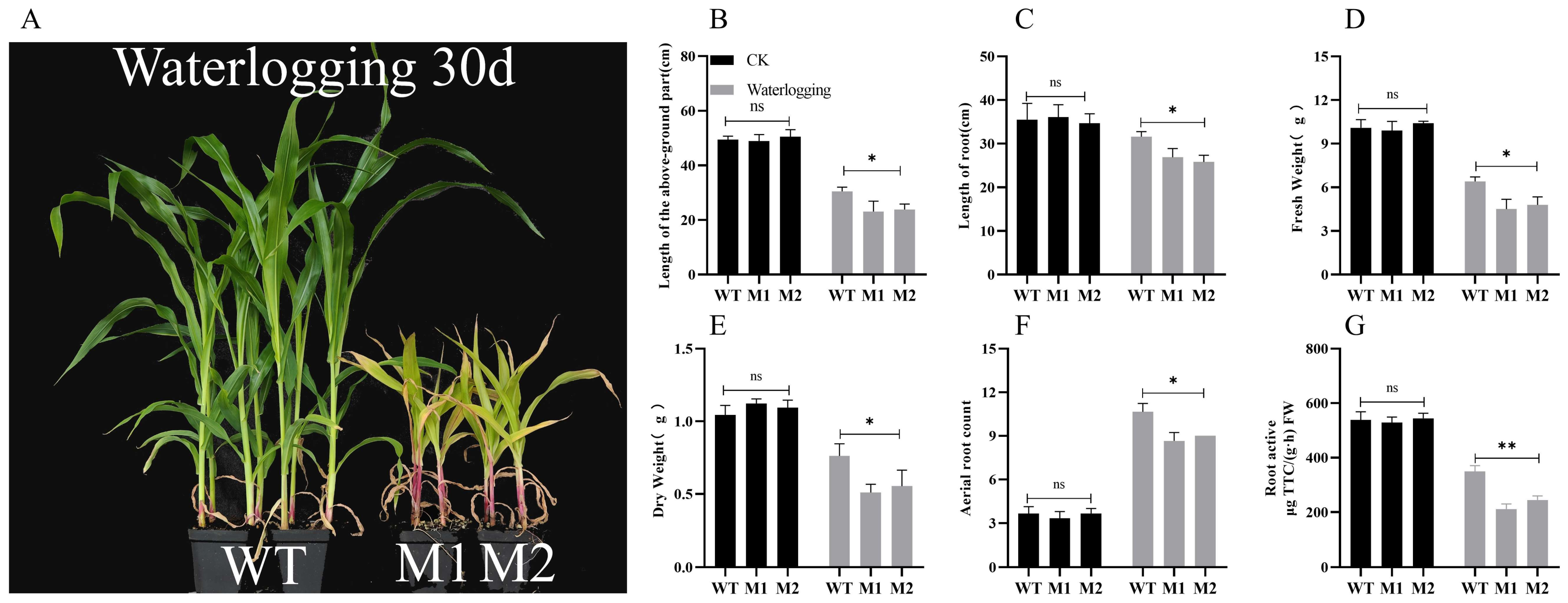
3.3. Mutation of ZmDIR5 Reduces the Ability of Maize Seedlings to Withstand Waterlogging Stress
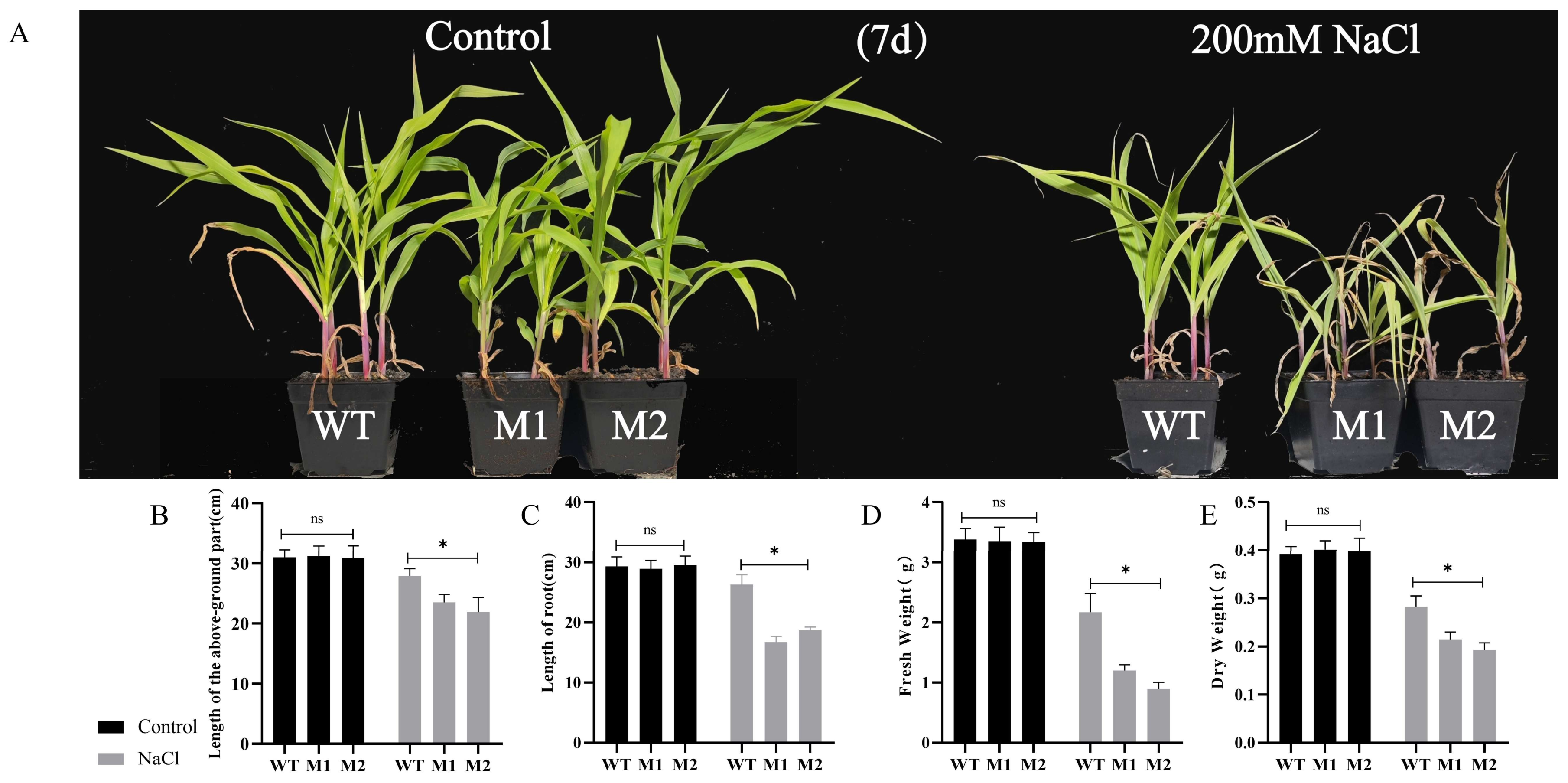
3.4. ZmDIR5 Positively Regulates Salt Tolerance in Maize Seedlings
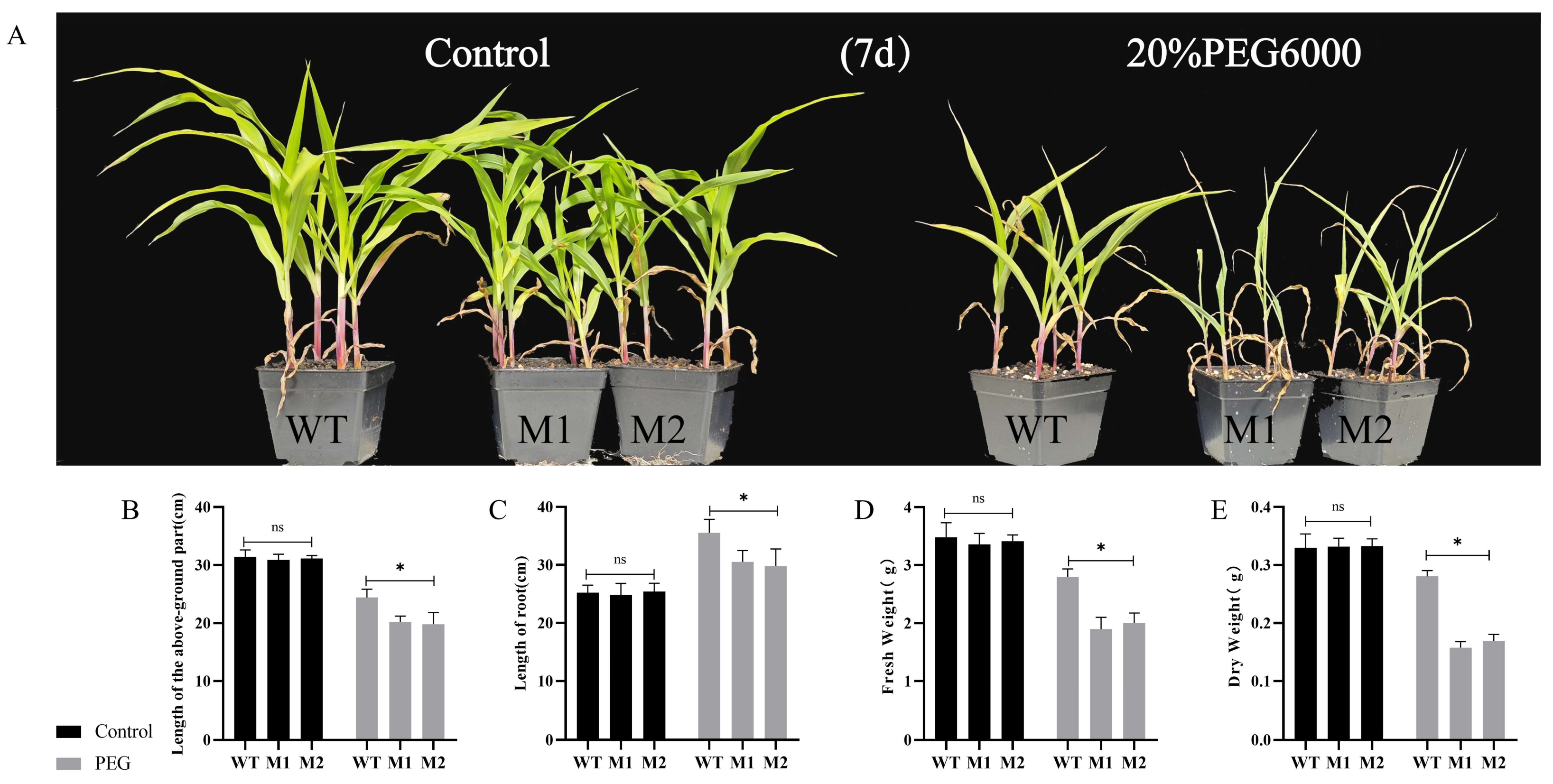
3.5. ZmDIR5 Enhances Drought Tolerance in Maize Seedlings
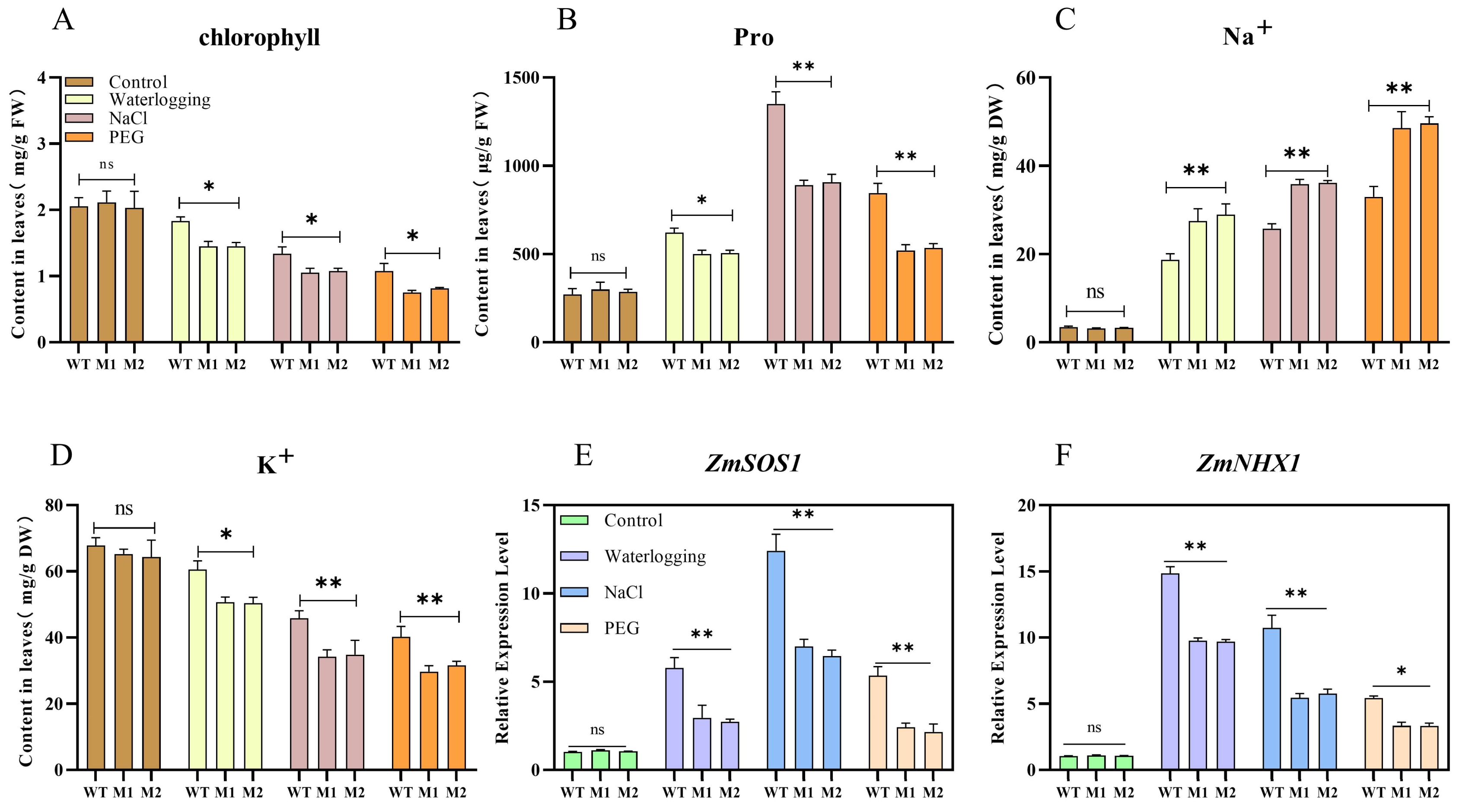
3.6. ZmDIR5 Enhances the Na+ and K+ Transport Capability of Plants Under Waterlogging, Salinity, and Drought Stress
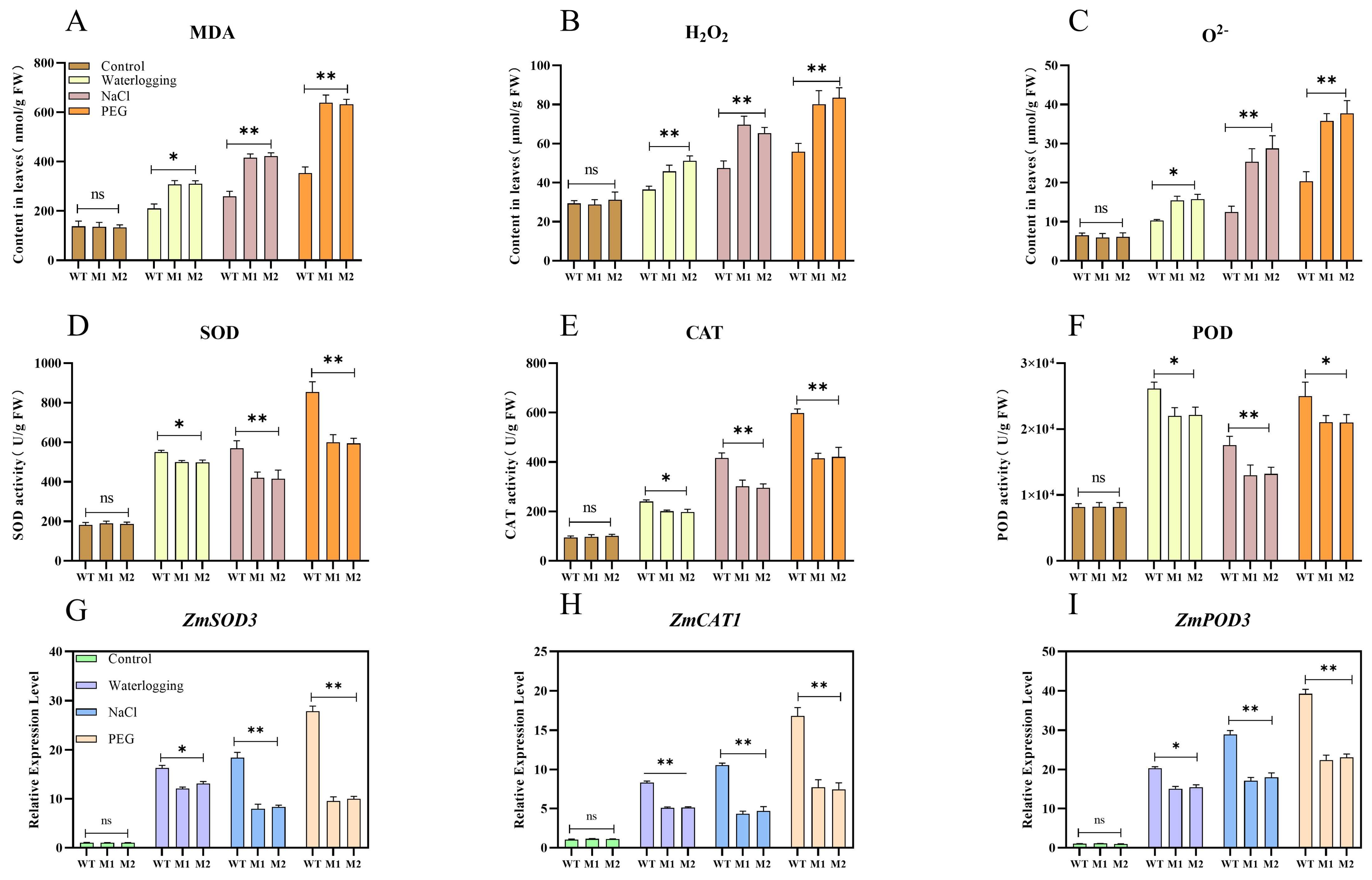
3.7. ZmDIR5 Enhances Antioxidant Capacity to Resist Three Types of Stress
3.8. Mutation of ZmDIR5 Reduces the Biosynthetic Capacity of ABA and Zeatin in Maize Seedlings
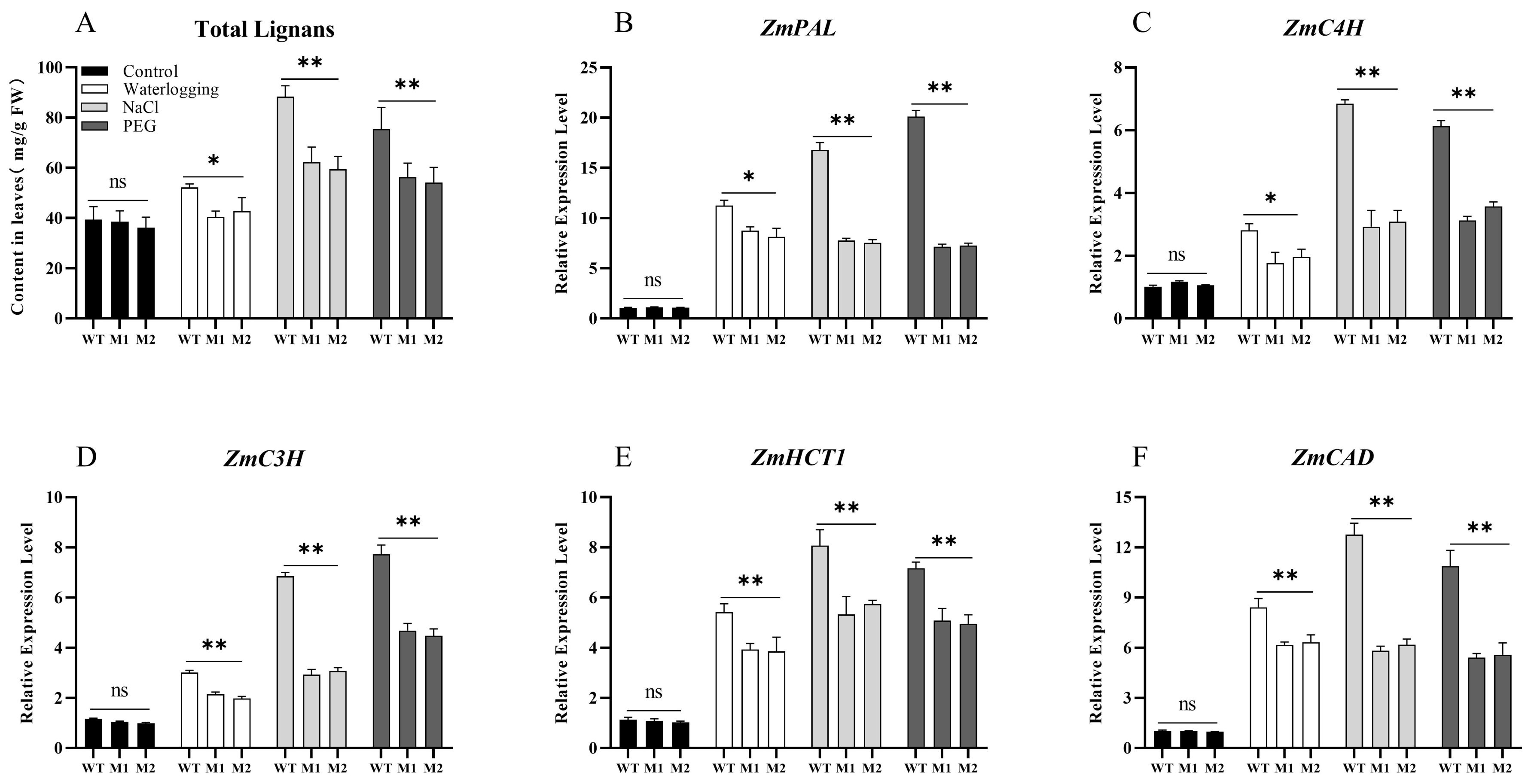
3.9. ZmDIR5 Mutation Affects Total Lignan Synthesis in Maize Seedlings Under Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abraham-Juárez, M.J.; Barnes, A.C.; Aragón-Raygoza, A.; Tyson, D.; Kur, A.; Strable, J.; Rellán-Álvarez, R. The arches and spandrels of maize domestication, adaptation, and improvement. Curr. Opin. Plant Biol. 2021, 64, 102124. [Google Scholar] [CrossRef] [PubMed]
- Abasi, F.; Raza, M.U.; Raja, N.I.; Mashwani, Z.-R.; Ehsan, M.; Ulfat, A.; Shahbaz, M.; Mehmood, A. Chapter 22—Physiological mechanism and adaptation of plants to abiotic stresses. In Improving Stress Resilience in Plants; Ahanger, M.A., Bhat, J.A., Ahmad, P., John, R., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 447–458. ISBN 978-0-443-18927-2. [Google Scholar]
- Muluneh, M.G. Impact of climate change on biodiversity and food security: A global perspective—A review article. Agric. Food Secur. 2021, 10, 36. [Google Scholar] [CrossRef]
- Ralph, S.G.; Jancsik, S.; Bohlmann, J. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 2007, 68, 1975–1991. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Li, R.-J.; Sun, J.-T.; Ma, F.; Zhang, H.-X.; Jin, J.-H.; Ali, M.; Haq, S.U.; Wang, J.-E.; Gong, Z.-H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhong, S.; Zhang, Q.; Ren, Y.; Sun, C.; Chen, F. A loss-of-function of the dirigent gene TaDIR-B1 improves resistance to Fusarium crown rot in wheat. Plant Biotechnol. J. 2021, 19, 866–868. [Google Scholar] [CrossRef]
- Li, X.; Liu, Z.; Zhao, H.; Deng, X.; Su, Y.; Li, R.; Chen, B. Overexpression of Sugarcane ScDIR Genes Enhances Drought Tolerance in Nicotiana benthamiana. Int. J. Mol. Sci. 2022, 23, 5340. [Google Scholar] [CrossRef]
- Li, L.; Sun, W.; Wang, P.; Li, H.; Rehman, S.; Li, D.; Zhuge, Q. Characterization, expression, and functional analysis of the pathogenesis-related gene PtDIR11 in transgenic poplar. Int. J. Biol. Macromol. 2022, 210, 182–195. [Google Scholar] [CrossRef]
- Zhang, Q.; Luo, N.; Dai, X.; Lin, J.; Ahmad, B.; Chen, Q.; Lei, Y.; Wen, Z. Ectopic and transient expression of VvDIR4 gene in Arabidopsis and grapes enhances resistance to anthracnose via affecting hormone signaling pathways and lignin production. BMC Genom. 2024, 25, 895. [Google Scholar] [CrossRef]
- Xue, B.; Duan, W.; Gong, L.; Zhu, D.; Li, X.; Li, X.; Liang, Y. The OsDIR55 gene increases salt tolerance by altering the root diffusion barrier. Plant J. 2024, 118, 1550–1568. [Google Scholar] [CrossRef]
- Song, M.; Peng, X. Genome-Wide Identification and Characterization of DIR Genes in Medicago truncatula. Biochem. Genet. 2019, 57, 487–506. [Google Scholar] [CrossRef]
- Xu, W.; Liu, T.; Zhang, H.; Zhu, H. Mungbean DIRIGENT Gene Subfamilies and Their Expression Profiles Under Salt and Drought Stresses. Front. Genet. 2021, 12, 658148. [Google Scholar] [CrossRef] [PubMed]
- Dokka, N.; Tyagi, S.; Ramkumar, M.K.; Rathinam, M.; Senthil, K.; Sreevathsa, R. Genome-wide identification and characterization of DIRIGENT gene family (CcDIR) in pigeonpea (Cajanus cajan L.) provide insights on their spatial expression pattern and relevance to stress response. Gene 2024, 914, 148417. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xiong, Y.; Li, M.; Zhang, S.; Han, Z.; Li, K. Genome-wide identification, characterization, evolution and expression analysis of the DIR gene family in potato (Solanum tuberosum). Front. Genet. 2023, 14, 1224015. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z. Functional analysis of a dirigent protein AtsDIR23 in Acorus tatarinowii. J. Plant Physiol. 2023, 290, 154098. [Google Scholar] [CrossRef]
- Osakabe, A. Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1. Nat. Commun. 2024, 15, 5187. [Google Scholar] [CrossRef]
- Verica, J.A.; He, Z.-H. The Cell Wall-Associated Kinase (WAK) and WAK -Like Kinase Gene Family. Plant Physiol. 2002, 129, 455–459. [Google Scholar] [CrossRef]
- Gomord, V.; Denmat, L.; Fitchette-Lainé, A.; Satiat-Jeunemaitre, B.; Hawes, C.; Faye, L. The C-terminal HDEL sequence is sufficient for retention of secretory proteins in the endoplasmic reticulum (ER) but promotes vacuolar targeting of proteins that escape the ER. Plant J. 1997, 11, 313–325. [Google Scholar] [CrossRef]
- Lu, X.; Liu, J.; Ren, W.; Yang, Q.; Chai, Z.; Chen, R.; Wang, L.; Zhao, J.; Lang, Z.; Wang, H.; et al. Gene-Indexed Mutations in Maize. Mol. Plant 2018, 11, 496–504. [Google Scholar] [CrossRef]
- Yao, Q. Crucial Waterlogging-Responsive Genes and Pathways Revealed by Comparative Physiology and Transcriptome in Tropical and Temperate Maize (Zea mays L.) inbred Lines. J. Plant Biol. 2021, 64, 313–325. [Google Scholar] [CrossRef]
- Asch, J.; Johnson, K.; Mondal, S.; Asch, F. Comprehensive assessment of extraction methods for plant tissue samples for determining sodium and potassium via flame photometer and chloride via automated flow analysis. J. Plant Nutr. Soil. Sci. 2022, 185, 308–316. [Google Scholar] [CrossRef]
- Verslues, P.E. Rapid Quantification of Abscisic Acid by GC-MS/MS for Studies of Abiotic Stress Response. In Plant Stress Tolerance: Methods and Protocols; Sunkar, R., Ed.; Springer: New York, NY, USA, 2017; Volume 1631, pp. 325–335. ISBN 978-1-4939-7136-7. [Google Scholar]
- Thippan, S.; Bunya-Atichart, K.; Ayuttaya, S.I.N.; Lerslerwong, L. Histological and biochemical aspects as potential markers for evaluation graft compatibility in durian at the early nursery stage. Sci. Hortic. 2024, 337, 113490. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Ma, X.; Xu, W.; Liu, T.; Chen, R.; Zhu, H.; Zhang, H.; Cai, C.; Li, S. Functional characterization of soybean (Glycine max) DIRIGENT genes reveals an important role of GmDIR27 in the regulation of pod dehiscence. Genomics 2021, 113, 979–990. [Google Scholar] [CrossRef]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Submergence and Waterlogging Stress in Plants: A Review Highlighting Research Opportunities and Understudied Aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef]
- Nawaz, M.; Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A review of plants strategies to resist biotic and abiotic environmental stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Irato, P.; Santovito, G. Enzymatic and Non-Enzymatic Molecules with Antioxidant Function. Antioxidants 2021, 10, 579. [Google Scholar] [CrossRef]
- Lim, C.; Baek, W.; Jung, J.; Kim, J.-H.; Lee, S. Function of ABA in Stomatal Defense against Biotic and Drought Stresses. Int. J. Mol. Sci. 2015, 16, 15251. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Wang, Y.-G.; Fu, F.-L.; Yu, H.-Q.; Hu, T.; Zhang, Y.-Y.; Tao, Y.; Zhu, J.-K.; Zhao, Y.; Li, W.-C. Interaction network of core ABA signaling components in maize. Plant Mol. Biol. 2018, 96, 245–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, L.; Chandrasekaran, U.; Luo, X.; Zheng, C.; Shu, K. ABA Biosynthesis and Signaling Cascades Under Hypoxia Stress. Front. Plant Sci. 2021, 12, 661228. [Google Scholar] [CrossRef] [PubMed]
- Gharib, A.; Marquez, C.; Meseguer-Beltran, M.; Sanchez-Sarasua, S.; Sanchez-Perez, A.M. Abscisic acid, an evolutionary conserved hormone: Biosynthesis, therapeutic and diagnostic applications in mammals. Biochem. Pharmacol. 2024, 229, 116521. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Antoniadi, I. IPT9, a cis-zeatin cytokinin biosynthesis gene, promotes root growth. Front. Plant Sci. 2022, 13, 932008. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Yun, L.; Zheng, Z.P.; Zhang, H.M.; Liu, X.H.; Cao, D.; Li, S.W. Cloning and Expression of Genes Related to Zeatin Synthesis Pathway. Seed 2022, 41, 55–60. [Google Scholar]
- Suzuki, S.; Umezawa, T. Biosynthesis of lignans and norlignans. J. Wood Sci. 2007, 53, 273–284. [Google Scholar] [CrossRef]
- Kolo, Z.; Majola, A.; Phillips, K.; Ali, A.E.E.; Sharp, R.E.; Ludidi, N. Water Deficit-Induced Changes in Phenolic Acid Content in Maize Leaves Is Associated with Altered Expression of Cinnamate 4-Hydroxylase and p-Coumaric Acid 3-Hydroxylase. Plants 2022, 12, 101. [Google Scholar] [CrossRef]
- Behr, M.; Sergeant, K.; Leclercq, C.C.; Planchon, S.; Guignard, C.; Lenouvel, A.; Renaut, J.; Hausman, J.-F.; Lutts, S.; Guerriero, G. Insights into the molecular regulation of monolignol-derived product biosynthesis in the growing hemp hypocotyl. BMC Plant Biol. 2018, 18, 1. [Google Scholar] [CrossRef]
- Davin, L.B.; Wang, H.-B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective Bimolecular Phenoxy Radical Coupling by an Auxiliary (Dirigent) Protein Without an Active Center. Science 1997, 275, 362–367. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, Y.; Liang, X.; Zhuang, J.; Wang, X.; Qin, F.; Jiang, C. A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 2022, 13, 2222. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.E.; Savouré, A.; Szabados, L. Proline metabolism as regulatory hub. Trends Plant Sci. 2022, 27, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Sensitivity and responses of chloroplasts to salt stress in plants. Front. Plant Sci. 2024, 15, 1374086. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, J.; Wang, Y.; Liang, X.; Zhang, M.; Lu, M.; Guo, Y.; Qin, F.; Jiang, C. The classical SOS pathway confers natural variation of salt tolerance in maize. New Phytol. 2022, 236, 479–494. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Noll, A.; Karl, S.; Leib, K.; Yan, F.; Schubert, S. Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress. J. Plant Physiol. 2005, 162, 55–66. [Google Scholar] [CrossRef]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Li, M.; Zhao, D.; Yang, J.-F.; Fu, J.-D.; Xu, Z.-S. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 Contributes to Stress Resistance in Maize by Regulating ABA Synthesis and Root Development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
- Wei, X.; Fan, X.; Zhang, H.; Jiao, P.; Jiang, Z.; Lu, X.; Liu, S.; Guan, S.; Ma, Y. Overexpression of ZmSRG7 Improves Drought and Salt Tolerance in Maize (Zea mays L.). Int. J. Mol. Sci. 2022, 23, 13349. [Google Scholar] [CrossRef]
- Meng, X.; Liu, S.; Zhang, C.; He, J.; Ma, D.; Wang, X.; Dong, T.; Guo, F.; Cai, J.; Long, T.; et al. The unique sweet potato NAC transcription factor IbNAC3 modulates combined salt and drought stresses. Plant Physiol. 2023, 191, 747–771. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Qin, T.; Zheng, H.; Guan, Y.; Gu, W.; Wang, H.; Yu, D.; Qu, J.; Wei, J.; Xu, W. Mutation of ZmDIR5 Reduces Maize Tolerance to Waterlogging, Salinity, and Drought. Plants 2025, 14, 785. https://doi.org/10.3390/plants14050785
Zhao Z, Qin T, Zheng H, Guan Y, Gu W, Wang H, Yu D, Qu J, Wei J, Xu W. Mutation of ZmDIR5 Reduces Maize Tolerance to Waterlogging, Salinity, and Drought. Plants. 2025; 14(5):785. https://doi.org/10.3390/plants14050785
Chicago/Turabian StyleZhao, Zhixiong, Tao Qin, Hongjian Zheng, Yuan Guan, Wei Gu, Hui Wang, Diansi Yu, Jingtao Qu, Jihui Wei, and Wen Xu. 2025. "Mutation of ZmDIR5 Reduces Maize Tolerance to Waterlogging, Salinity, and Drought" Plants 14, no. 5: 785. https://doi.org/10.3390/plants14050785
APA StyleZhao, Z., Qin, T., Zheng, H., Guan, Y., Gu, W., Wang, H., Yu, D., Qu, J., Wei, J., & Xu, W. (2025). Mutation of ZmDIR5 Reduces Maize Tolerance to Waterlogging, Salinity, and Drought. Plants, 14(5), 785. https://doi.org/10.3390/plants14050785





