DNA Damaging Agents Induce RNA Structural and Transcriptional Changes for Genes Associated with Redox Homeostasis in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
2.1. Library Quality Control
2.2. Transcriptional Changes After MMS Treatment
2.3. Plant NADPH Oxidases Play a Role in Plant Response to MMS
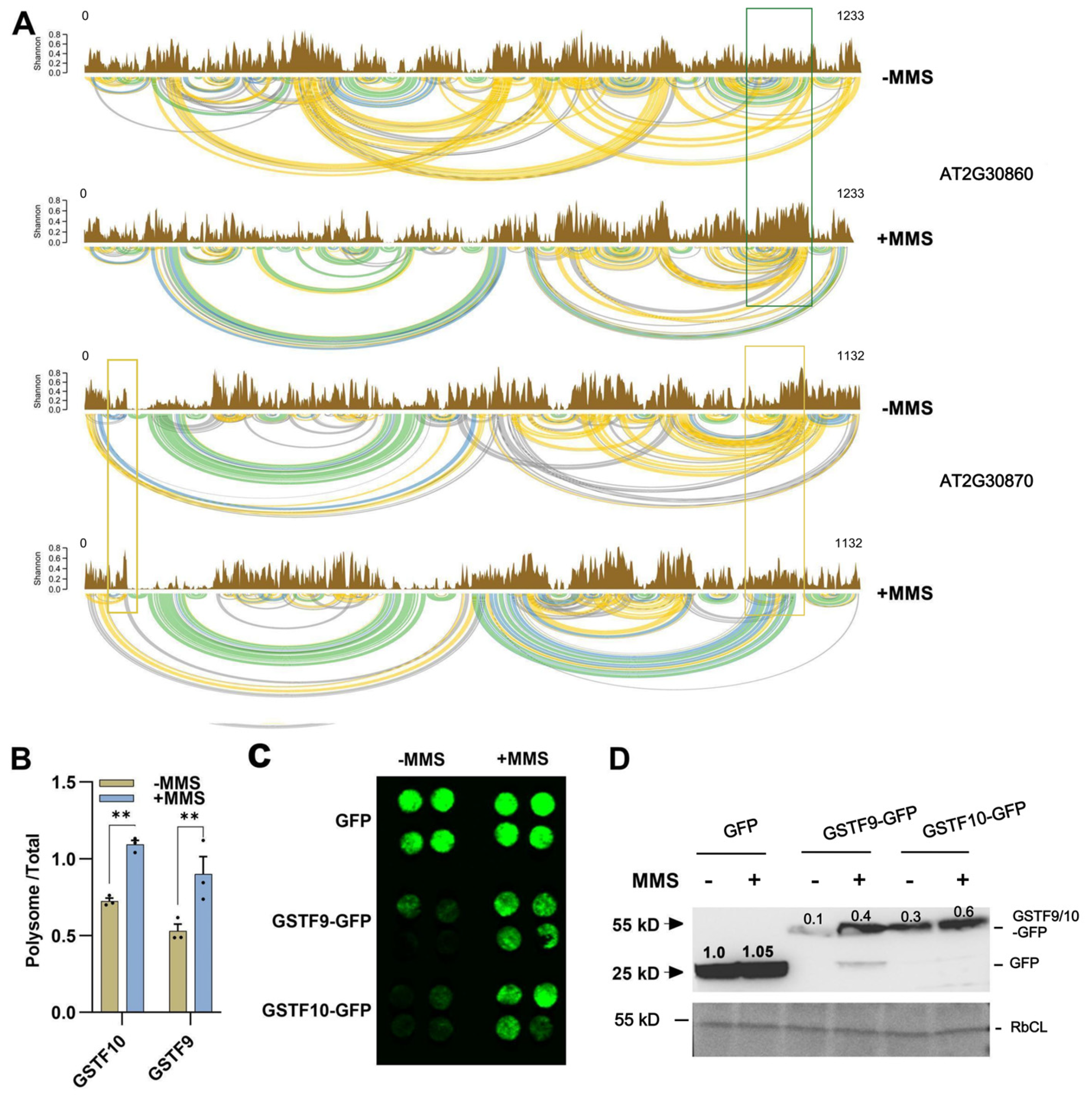
2.4. RNA Structure Analysis
2.5. RNA Structures for GSTF9 and GSTF10 Determine Their Translation
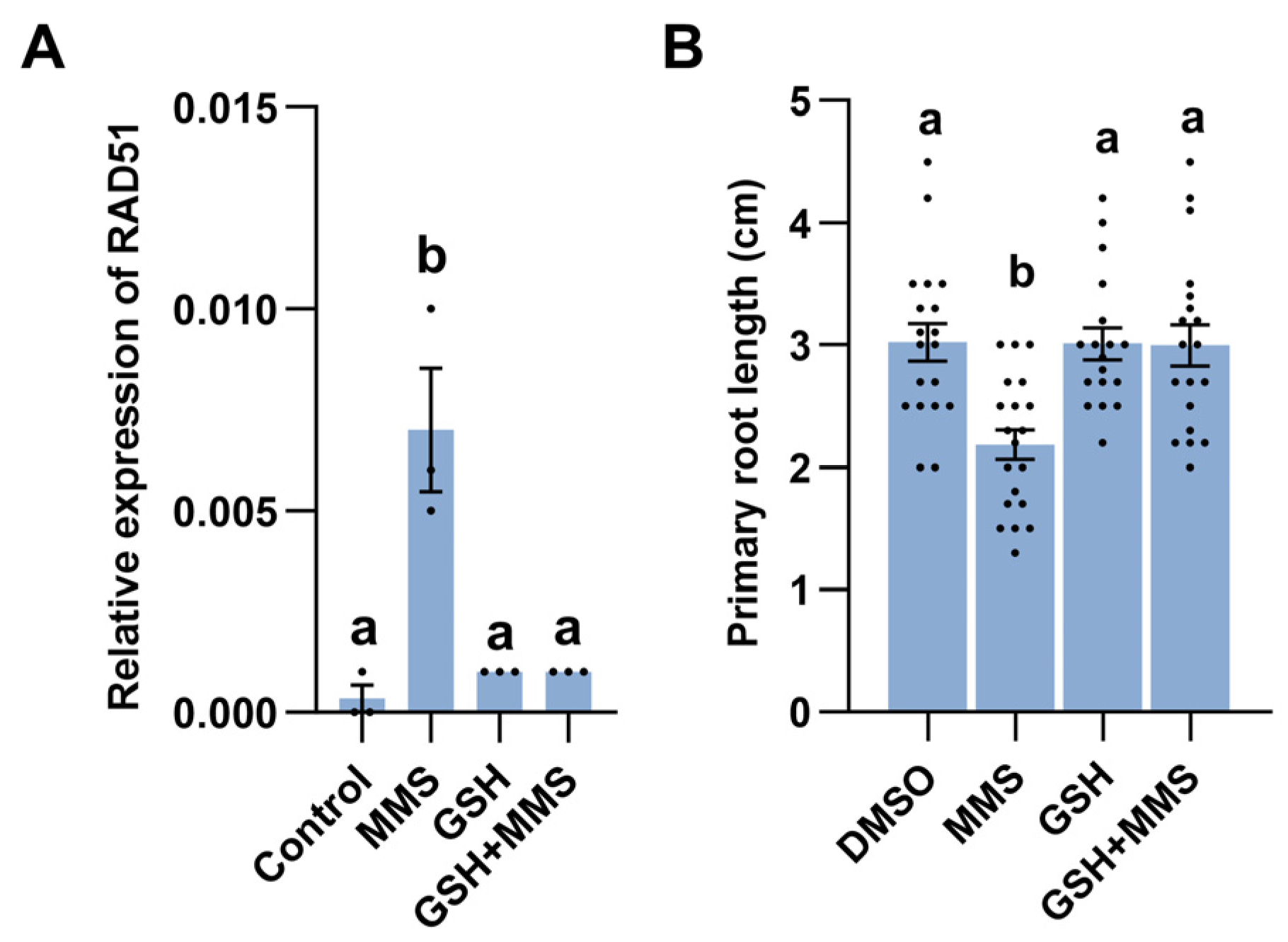
2.6. Redox Homeostasis Is Associated with MMS-Induced Response
3. Discussion
Limitation for the Study
4. Materials and Methods
4.1. The Plant Materials and Growth Conditions
4.2. Primary Root Growth Inhibition Assays
4.3. DMS-MaPseq Library Construction
4.4. DMS Data Analysis
4.5. RNA Isolation and qRT–PCR
4.6. RNA Translation Efficiency Detection
4.7. Protein Extraction and Immunoblot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahn, J.W.; Atwell, B.J.; Roberts, T.H. Serpin genes AtSRP2 and AtSRP3 are required for normal growth sensitivity to a DNA alkylating agent in Arabidopsis. BMC Plant Biol. 2009, 9, 52. [Google Scholar] [CrossRef]
- Lang, L.; Pettkó-Szandtner, A.; Tunçay Elbaşı, H.; Takatsuka, H.; Nomoto, Y.; Zaki, A.; Dorokhov, S.; De Jaeger, G.; Eeckhout, D.; Ito, M.; et al. The DREAM complex represses growth in response to DNA damage in Arabidopsis. Life Sci. Alliance 2021, 4, e202101141. [Google Scholar] [CrossRef]
- Dissmeyer, N.; Weimer, A.K.; Pusch, S.; De Schutter, K.; Alvim Kamei, C.L.; Nowack, M.K.; Novak, B.; Duan, G.L.; Zhu, Y.G.; De Veylder, L.; et al. Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 2009, 21, 3641–3654. [Google Scholar] [CrossRef]
- Hu, Y.; Li, J.; Yang, L.; Nan, W.; Cao, X.; Bi, Y. Inhibition of root growth by narciclasine is caused by DNA damage-induced cell cycle arrest in lettuce seedlings. Protoplasma 2014, 251, 1113–1124. [Google Scholar] [CrossRef]
- Cui, W.; Wang, H.; Song, J.; Cao, X.; Rogers, H.J.; Francis, D.; Jia, C.; Sun, L.; Hou, M.; Yang, Y.; et al. Cell cycle arrest mediated by Cd-induced DNA damage in Arabidopsis root tips. Ecotoxicol. Environ. Saf. 2017, 145, 569–574. [Google Scholar] [CrossRef]
- Garcia, V.; Bruchet, H.; Camescasse, D.; Granier, F.; Bouchez, D.; Tissier, A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 2003, 15, 119–132. [Google Scholar] [CrossRef]
- Bray, C.M.; West, C.E. DNA repair mechanisms in plants: Crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 2005, 168, 511–528. [Google Scholar] [CrossRef]
- Hu, Z.; Cools, T.; De Veylder, L. Mechanisms used by plants to cope with DNA damage. Annu. Rev. Plant Biol. 2016, 67, 439–462. [Google Scholar] [CrossRef]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef]
- Tamura, K.; Adachi, Y.; Chiba, K.; Oguchi, K.; Takahashi, H. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: Evidence for a role in the repair of DNA double-strand breaks. Plant J. 2002, 29, 771–781. [Google Scholar] [CrossRef]
- West, C.E.; Waterworth, W.M.; Story, G.W.; Sunderland, P.A.; Jiang, Q.; Bray, C.M. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 2002, 31, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Bundock, P.; van Attikum, H.; Hooykaas, P. Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 2002, 30, 3395–3400. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Kobayashi, J.; Ogita, N.; Ueda, M.; Kimura, S.; Maki, H.; Umeda, M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013, 14, 817–822. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Kimura, S. Ser-Gln sites of SOG1 are rapidly hyperphosphorylated in response to DNA double-strand breaks. Plant Signal. Behav. 2018, 13, e1477904. [Google Scholar] [CrossRef]
- Meschichi, A.; Zhao, L.; Reeck, S.; White, C.; Da Ines, O.; Sicard, A.; Pontvianne, F.; Rosa, S. The plant-specific DDR factor SOG1 increases chromatin mobility in response to DNA damage. EMBO Rep. 2022, 23, e54736. [Google Scholar] [CrossRef]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef]
- Barnes, D.E. Non-homologous end joining as a mechanism of DNA repair. Curr. Biol. 2001, 11, R455–R457. [Google Scholar] [CrossRef]
- Wyman, C.; Kanaar, R. DNA double-strand break repair: All’s well that ends well. Annu. Rev. Genet. 2006, 40, 363–383. [Google Scholar] [CrossRef]
- Dedon, P.C. Oxidation and Deamination of DNA by Endogenous Sources. In Chemical Carcinogenesis; Humana Press: Totowa, NJ, USA, 2010; pp. 209–225. [Google Scholar]
- Chakarov, S.; Petkova, R.; Russev, G.C.; Zhelev, N. DNA damage and mutation. Types of DNA damage. BioDiscovery 2014, 11, e8957. [Google Scholar] [CrossRef]
- Vandivier, L.E.; Anderson, S.J.; Foley, S.W.; Gregory, B.D. The conservation and function of RNA secondary structure in plants. Annu. Rev. Plant Biol. 2016, 67, 463–488. [Google Scholar] [CrossRef]
- Wang, X.W.; Liu, C.X.; Chen, L.L.; Zhang, Q.C. RNA structure probing uncovers RNA structure-dependent biological functions. Nat. Chem. Biol. 2021, 17, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Childs-Disney, J.L.; Yang, X.; Gibaut, Q.M.R.; Tong, Y.; Batey, R.T.; Disney, M.D. Targeting RNA structures with small molecules. Nat. Rev. Drug. Discov. 2022, 21, 736–762. [Google Scholar] [CrossRef]
- Bernetti, M.; Bussi, G. Integrating experimental data with molecular simulations to investigate RNA structural dynamics. Curr. Opin. Struct. Biol. 2023, 78, 102503. [Google Scholar] [CrossRef]
- Bevilacqua, P.C.; Ritchey, L.E.; Su, Z.; Assmann, S.M. Genome-wide analysis of RNA secondary structure. Annu. Rev. Genet. 2016, 50, 235–266. [Google Scholar] [CrossRef]
- Spitale, R.C.; Incarnato, D. Incarnato, Probing the dynamic RNA structurome and its functions. Nat. Rev. Genet. 2023, 24, 178–196. [Google Scholar] [CrossRef]
- Ding, Y.; Tang, Y.; Kwok, C.K.; Zhang, Y.; Bevilacqua, P.C.; Assmann, S.M. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 2014, 505, 696–700. [Google Scholar] [CrossRef]
- Mitchell, D.; Renda, A.J.; Douds, C.A.; Babitzke, P.; Assmann, S.M.; Bevilacqua, P.C. In vivo RNA structural probing of uracil and guanine base-pairing by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC). RNA 2019, 25, 147–157. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Q.; Yang, X.; Zhang, Y.; Norris, M.; Chen, X.; Cheema, J.; Zhang, H.; Ding, Y. In vivo nuclear RNA structurome reveals RNA-structure regulation of mRNA processing in plants. Genome Biol. 2021, 22, 11. [Google Scholar] [CrossRef]
- Wang, Q.; La, Y.; Xia, H.; Zhou, S.; Zhai, Z.; La, H. Roles of MEM1 in safeguarding Arabidopsis genome against DNA damage, inhibiting ATM/SOG1-mediated DNA damage response, and antagonizing global DNA hypermethylation. J. Integr. Plant Biol. 2022, 64, 87–104. [Google Scholar] [CrossRef]
- Kim, Y.S. Analysis of gene expression upon DNA damage in Arabidopsis. J. Plant Biol. 2006, 49, 5. [Google Scholar] [CrossRef]
- Zubradt, M.; Gupta, P.; Persad, S.; Lambowitz, A.M.; Weissman, J.S.; Rouskin, S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 2017, 14, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.M.; Muhlemann, J.K.; Gayomba, S.R.; Muday, G.K. RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem. Res. Toxicol. 2019, 32, 370–396. [Google Scholar] [CrossRef] [PubMed]
- Halim, V.A.; García-Santisteban, I.; Warmerdam, D.O.; van den Broek, B.; Heck, A.J.R.; Mohammed, S.; Medema, R.H. Doxorubicin-induced DNA damage causes extensive ubiquitination of ribosomal proteins associated with a decrease in protein translation. Mol. Cell Proteom. 2018, 17, 2297–2308. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Cabral, B.J.; Presnyak, V.; Su, S.V.; Reid, D.W.; Goodman, B.; Link, K.; Khatwani, N.; Reynders, J.; Moore, M.J.; et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA 2019, 116, 24075–24083. [Google Scholar] [CrossRef]
- Kawaguchi, D.; Shimizu, S.; Abe, N.; Hashiya, F.; Tomoike, F.; Kimura, Y.; Abe, H. Translational control by secondary-structure formation in mRNA in a eukaryotic system. Nucleosides Nucleotides Nucleic Acids 2020, 39, 195–203. [Google Scholar] [CrossRef]
- Wang, Z.; Schwacke, R.; Kunze, R. DNA damage-induced transcription of transposable elements and long non-coding RNAs in Arabidopsis is Rare and ATM-dependent. Mol. Plant 2016, 9, 1142–1155. [Google Scholar] [CrossRef]
- Bourbousse, C.; Vegesna, N.; Law, J.A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12453–E12462. [Google Scholar] [CrossRef]
- Nesnow, S.; Roop, B.C.; Lambert, G.; Kadiiska, M.; Mason, R.P.; Cullen, W.R.; Mass, M.J. DNA damage induced by methylated trivalent arsenicals is mediated by reactive oxygen species. Chem. Res. Toxicol. 2002, 15, 1627–1634. [Google Scholar] [CrossRef]
- Kang, M.A.; So, E.Y.; Simons, A.L.; Spitz, D.R.; Ouchi, T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012, 3, e249. [Google Scholar] [CrossRef]
- Weyemi, U.; Redon, C.E.; Aziz, T.; Choudhuri, R.; Maeda, D.; Parekh, P.R.; Bonner, M.Y.; Arbiser, J.L.; Bonner, W.M. Inactivation of NADPH oxidases NOX4 and NOX5 protects human primary fibroblasts from ionizing radiation-induced DNA damage. Radiat. Res. 2015, 183, 262–270. [Google Scholar] [CrossRef]
- Ren, G.; Luo, W.; Sun, W.; Niu, Y.; Ma, D.L.; Leung, C.H.; Wang, Y.; Lu, J.J.; Chen, X. Psoralidin induced reactive oxygen species (ROS)-dependent DNA damage and protective autophagy mediated by NOX4 in breast cancer cells. Phytomedicine 2016, 23, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Cooper, K.L.; Volk, L.B.; Dominguez, D.R.; Duran, A.D.; Ke Jian Liu, K.J.; Hudson, L.G. Contribution of NADPH oxidase to the retention of UVR-induced DNA damage by arsenic. Toxicol. Appl. Pharmacol. 2022, 434, 115799. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Floor, S.N. Dynamic regulation of messenger RNA structure controls translation. Nature 2023, 621, 259–260. [Google Scholar] [CrossRef]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef]
- Sieburth, L.E.; Vincent, J.N. Beyond transcription factors: Roles of mRNA decay in regulating gene expression in plants. F1000Research 2018, 7, 1940. [Google Scholar] [CrossRef]
- Hawkins, P.G.; Morris, K.V. RNA and transcriptional modulation of gene expression. Cell Cycle. 2008, 7, 602–607. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Hernández Estévez, I.; Rodríguez Hernández, M. Plant Glutathione S-transferases: An overview. Plant Gene 2020, 23, 100233. [Google Scholar] [CrossRef]
- Gao, J.; Chen, B.; Lin, H.; Liu, Y.; Wei, Y.; Chen, F.; Li, W. Identification and characterization of the glutathione -Transferase (GST) family in radish reveals a likely role in anthocyanin biosynthesis and heavy metal stress tolerance. Gene 2020, 743, 144484. [Google Scholar] [CrossRef]
- Li, X.; Pang, Y.; Zhong, Y.; Cai, Z.; Ma, Q.; Wen, K.; Nian, H. GmGSTU23 Encoding a Tau Class glutathione S-Transferase protein enhances the salt tolerance of Soybean (Glycine max L.). Int. J. Mol. Sci. 2023, 24, 5547. [Google Scholar] [CrossRef]
- Zhang, Y.; He, J.; Xiao, Y.; Zhang, Y.; Liu, Y.; Wan, S.; Liu, L.; Dong, Y.; Liu, H.; Yu, Y. CsGSTU8, a glutathione S-transferase from Camellia sinensis, is regulated by CsWRKY48 and plays a positive role in drought tolerance. Front. Plant Sci. 2021, 12, 795919. [Google Scholar] [CrossRef] [PubMed]
- Hahm, J.Y.; Park, J.; Jang, E.S.; Chi, S.W. 8-Oxoguanine: From oxidative damage to epigenetic and epitranscriptional modification. Exp. Mol. Med. 2022, 54, 1626–1642. [Google Scholar] [CrossRef] [PubMed]
- Volkova, N.V.; Meier, B.; González-Huici, V.; Bertolini, S.; Gonzalez, S.; Vöhringer, H.; Abascal, F.; Martincorena, I.; Campbell, P.J.; Gartner, A.; et al. Mutational signatures are jointly shaped by DNA damage and repair. Nat. Commun. 2020, 11, 2169. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.; Lee, H.; Lee, S.; Ahn, S.H.; Lee, H.S.; Kim, G.D.; Peak, J.; Park, J.; Cho, Y.K.; Jeong, Y.; et al. Position-specific oxidation of miR-1 encodes cardiac hypertrophy. Nature 2020, 584, 279–285. [Google Scholar] [CrossRef]
- Lee, W.L.; Sinha, A.; Lam, L.N.; Loo, H.L.; Liang, J.; Ho, P.; Cui, L.; Chan, C.S.C.; Begley, T.; Kline, K.A.; et al. An RNA modification enzyme directly senses reactive oxygen species for translational regulation in Enterococcus faecalis. Nat. Commun. 2023, 14, 4093. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, M.; Wang, T.; Zhang, Y.; Zhang, X. Genome-wide probing RNA structure with the modified DMS-MaPseq in Arabidopsis. Methods 2019, 155, 30–40. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Marinus, T.; Incarnato, D. RNA framework for assaying the structure of RNAs by high-throughput sequencing. Methods Mol. Biol. 2021, 2284, 63–76. [Google Scholar] [CrossRef]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization. Imeta 2024, 3, e228. [Google Scholar] [CrossRef]
- Incarnato, D.; Morandi, E.; Simon, L.M.; Oliviero, S. RNA Framework: An all-in-one toolkit for the analysis of RNA structures and post-transcriptional modifications. Nucleic Acids Res. 2018, 46, e97. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, N.A.; Busan, S.; Rice, G.M.; Nelson, J.A.; Weeks, K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 2014, 11, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Merchante, C.; Brumos, J.; Yun, J.; Hu, Q.; Spencer, K.R.; Enríquez, P.; Binder, B.M.; Heber, S.; Stepanova, A.N.; Alonso, J.M. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 2015, 163, 684–697. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Li, J.-Y.; Ma, Y.-J.; Wang, X.-W.; Chen, J.-P.; Li, Y.-Y. DNA Damaging Agents Induce RNA Structural and Transcriptional Changes for Genes Associated with Redox Homeostasis in Arabidopsis thaliana. Plants 2025, 14, 780. https://doi.org/10.3390/plants14050780
Li P, Li J-Y, Ma Y-J, Wang X-W, Chen J-P, Li Y-Y. DNA Damaging Agents Induce RNA Structural and Transcriptional Changes for Genes Associated with Redox Homeostasis in Arabidopsis thaliana. Plants. 2025; 14(5):780. https://doi.org/10.3390/plants14050780
Chicago/Turabian StyleLi, Ping, Jiong-Yi Li, Yu-Jiao Ma, Xiao-Wei Wang, Jian-Ping Chen, and Yi-Yuan Li. 2025. "DNA Damaging Agents Induce RNA Structural and Transcriptional Changes for Genes Associated with Redox Homeostasis in Arabidopsis thaliana" Plants 14, no. 5: 780. https://doi.org/10.3390/plants14050780
APA StyleLi, P., Li, J.-Y., Ma, Y.-J., Wang, X.-W., Chen, J.-P., & Li, Y.-Y. (2025). DNA Damaging Agents Induce RNA Structural and Transcriptional Changes for Genes Associated with Redox Homeostasis in Arabidopsis thaliana. Plants, 14(5), 780. https://doi.org/10.3390/plants14050780






