An Integrative Systematic Approach Reveals a New Species of Crocus Series Verni (Iridaceae) Endemic to Albania
Abstract
1. Introduction
2. Results
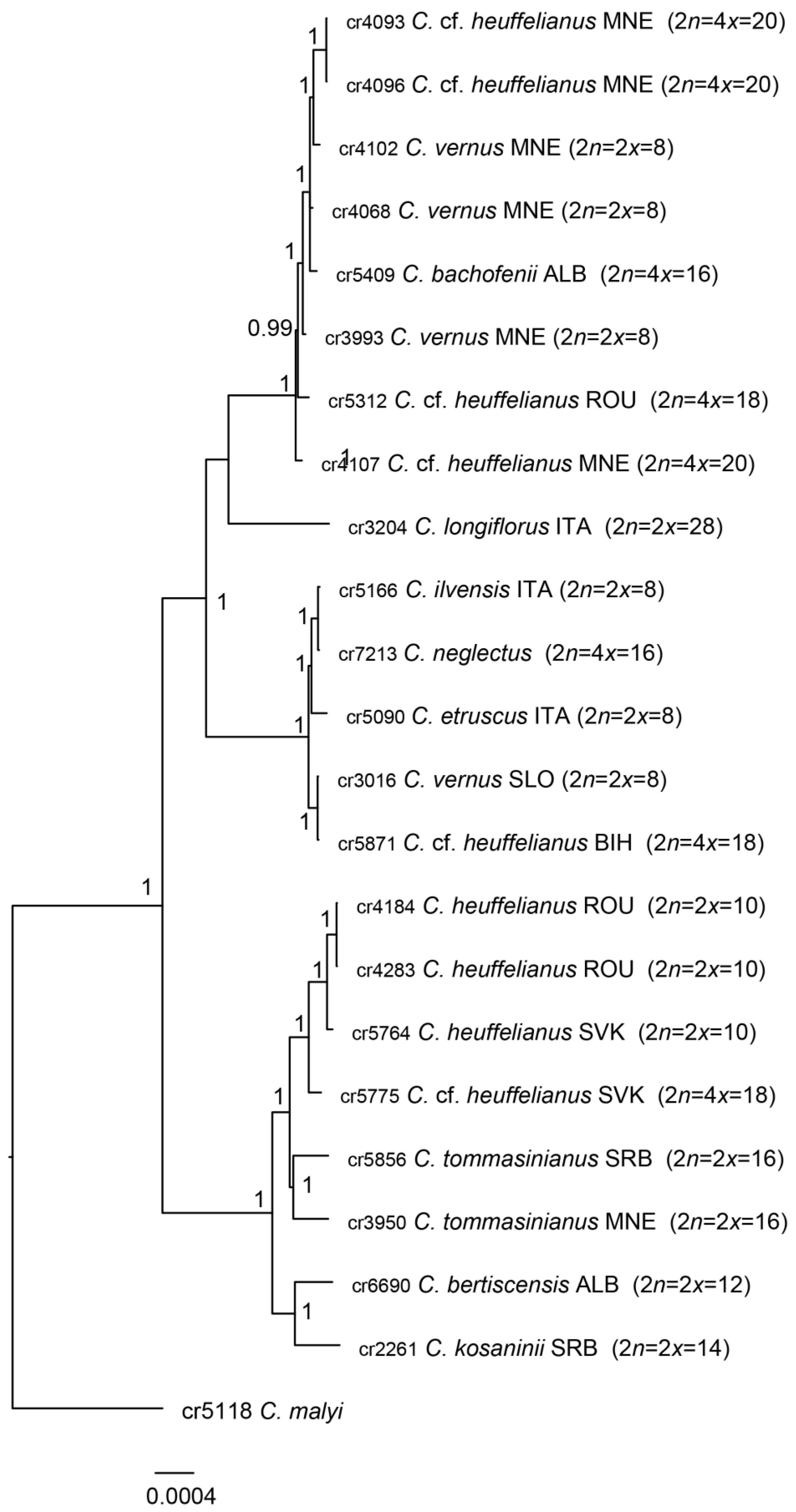
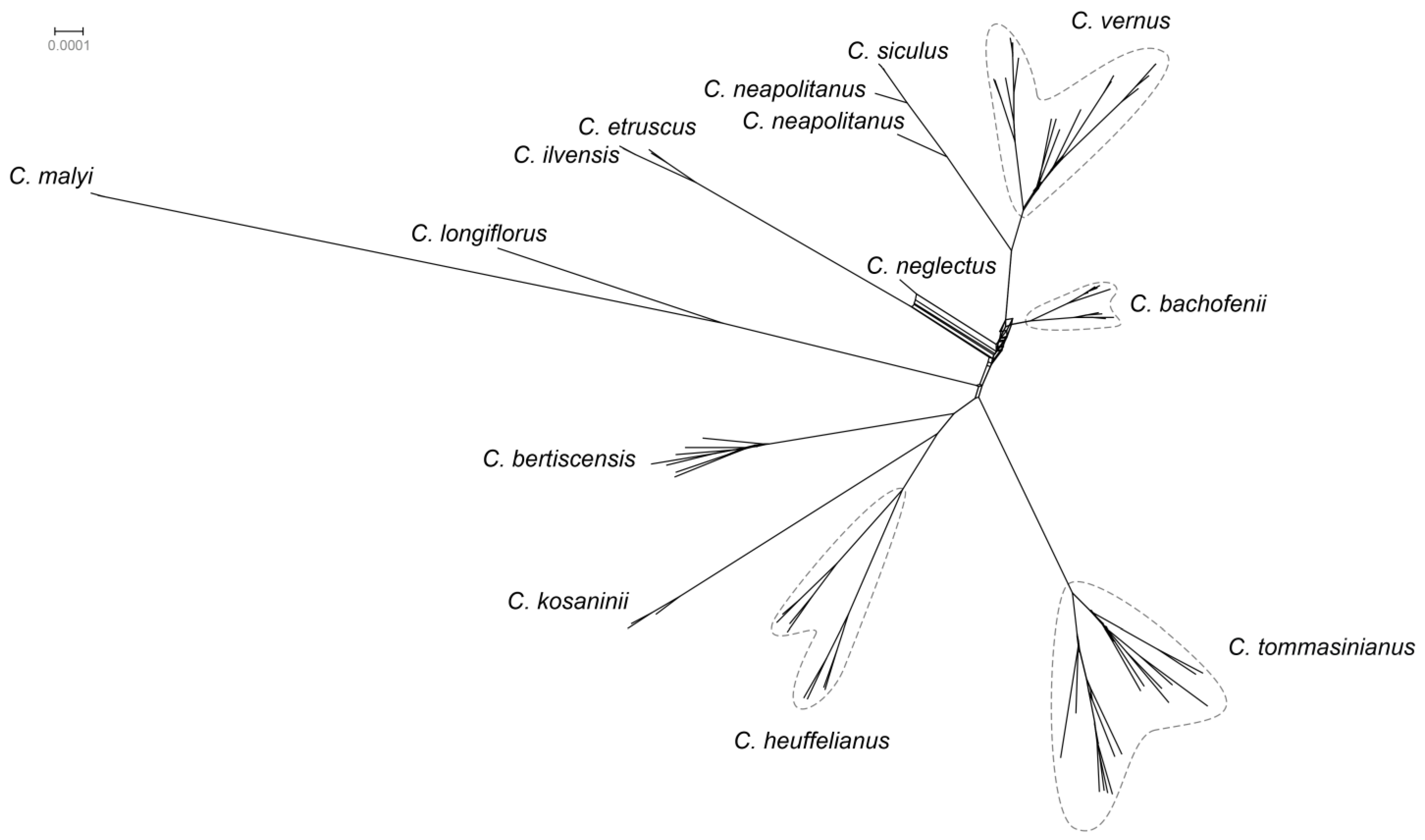
2.1. Phylogenetic Affiliation
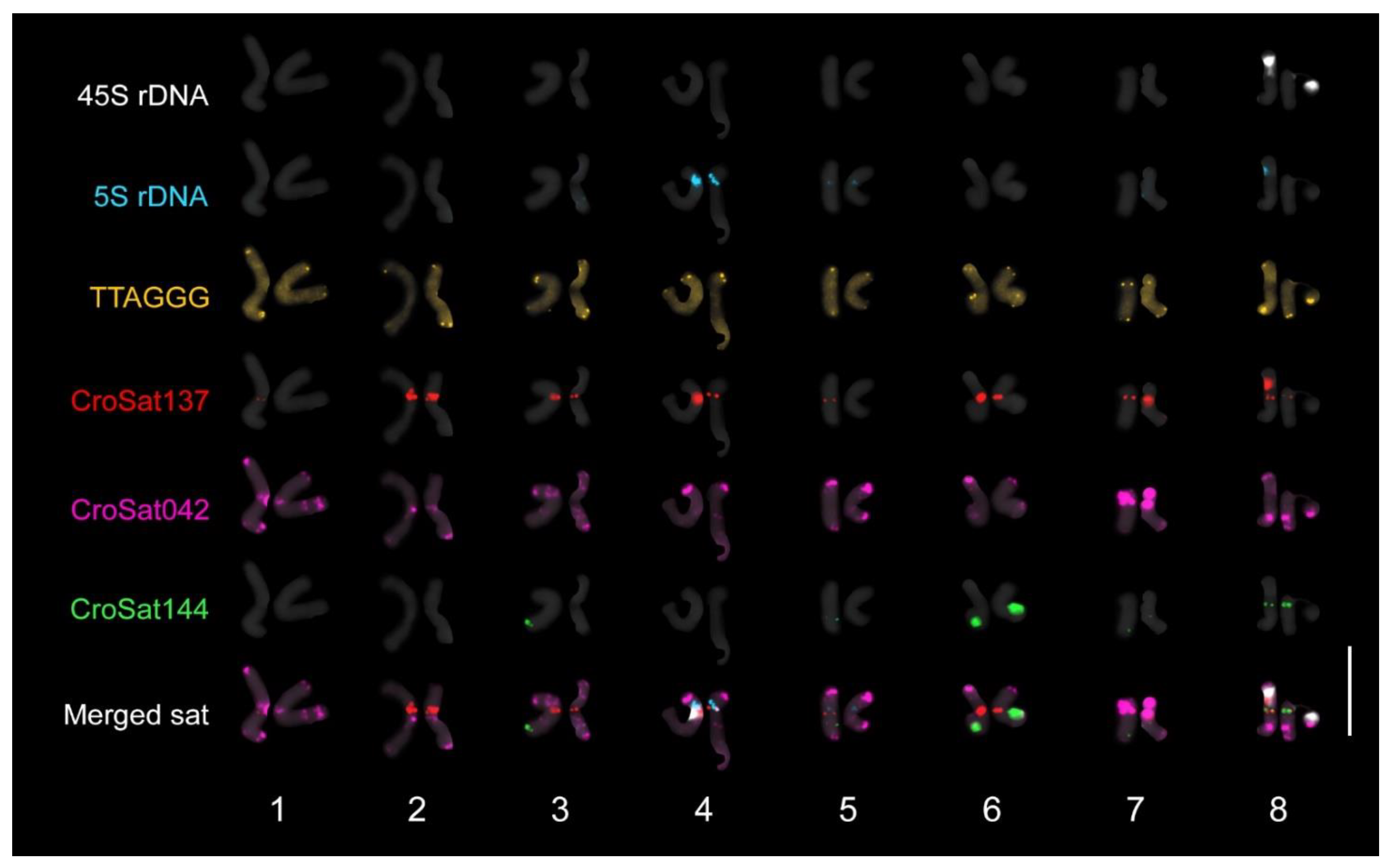
2.2. Karyological Analysis
2.3. Taxonomic Treatment—Crocus bachofenii D. Shuka, Raca, and Harpke sp. nov.
2.3.1. Type
2.3.2. Description
2.3.3. Etymology
2.3.4. Phenology
2.3.5. Ecology and Distribution
3. Materials and Methods
3.1. Plant Material
3.2. DNA Extraction
3.3. Library Preparation and Sequencing
3.4. Assembly of Chloroplast Genomes
3.5. Assembly of GBS Data
3.6. Phylogenetic Inference
3.7. Origin of Crocus bachofenii
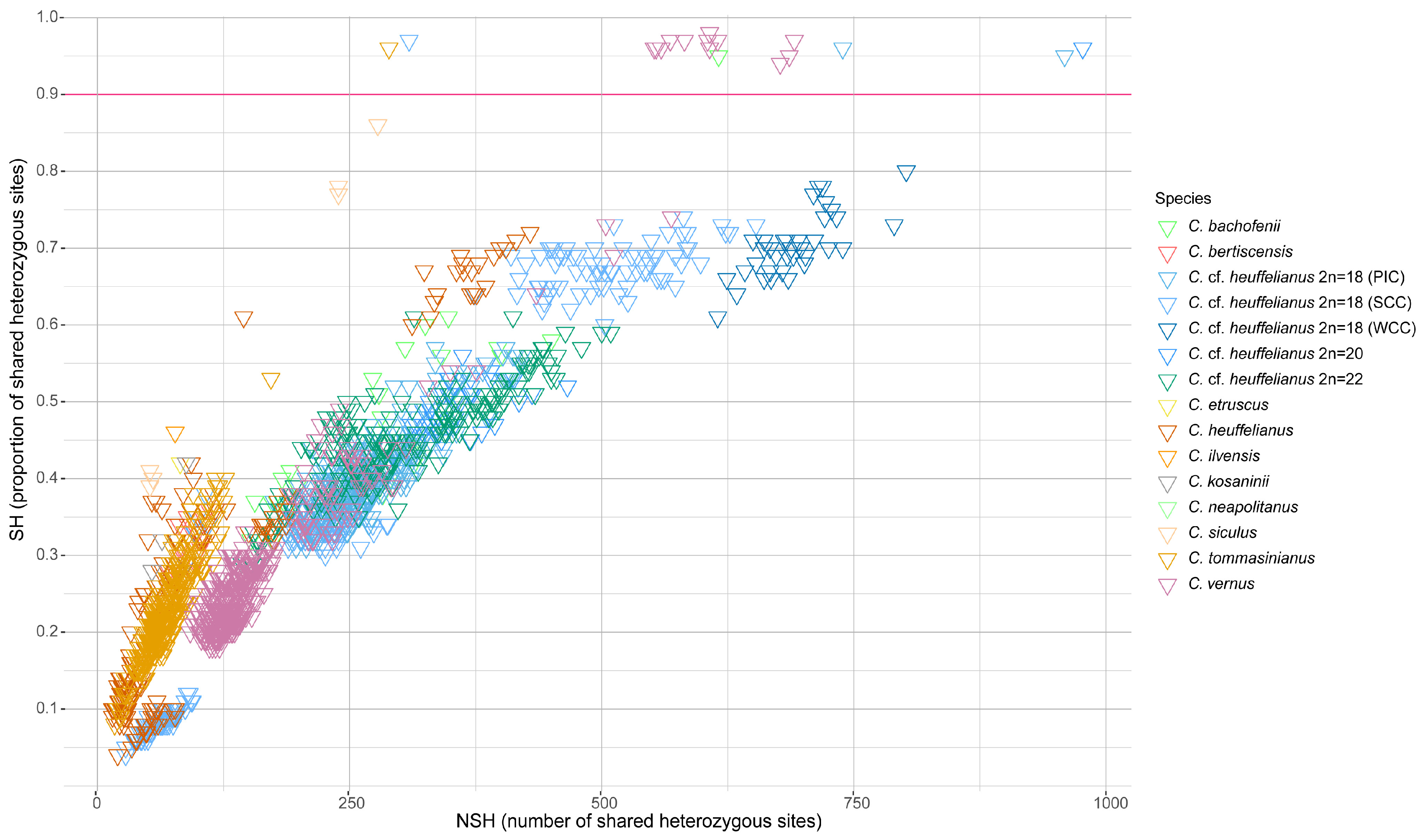
3.8. Inference of Clonal Reproduction
3.9. Cytological Analysis
3.10. Morphological Analysis
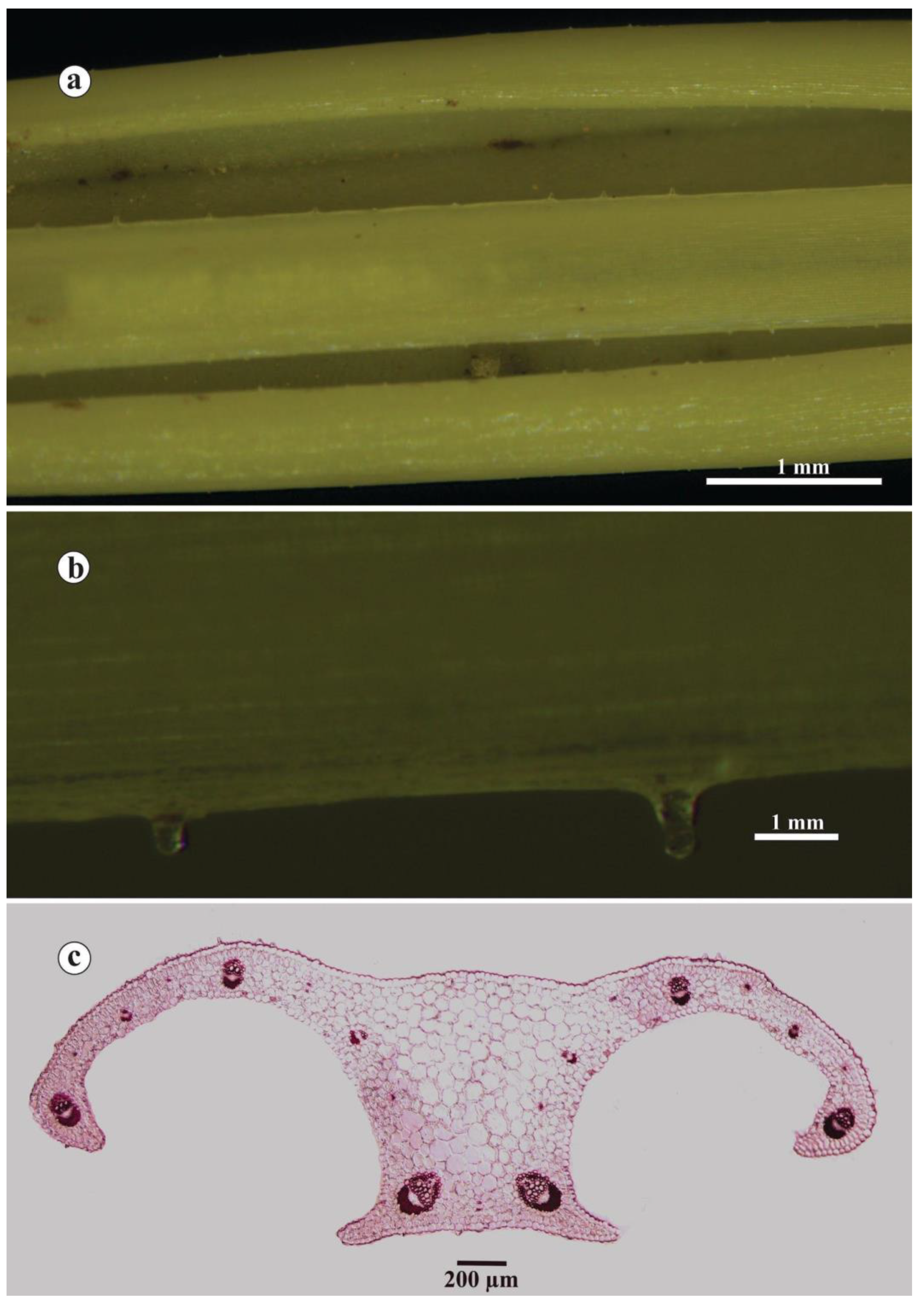
3.11. Anatomical Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyle, J.J.; Sherman-Broyles, S. Double trouble: Taxonomy and definitions of polyploidy. New Phytol. 2017, 213, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Żabicka, J.; Kirschey, T.; Migdałek, G.; Słomka, A.; Kuta, E. Genetic variation versus morphological variability in European peatland violets (Viola epipsila—V. palustris group). Biology 2023, 12, 362. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; McKain, M.R.; Bird, K.A.; Vanburen, R. Subgenome assignment in allopolyploids: Challenges and future directions. Curr. Opin. Plant Biol. 2018, 42, 76–80. [Google Scholar] [CrossRef]
- Raca, I.; Blattner, F.R.; Waminal, N.E.; Kerndorff, H.; Ranđelović, V.; Harpke, D. Disentangling Crocus series Verni and its polyploids. Biology 2023, 12, 303. [Google Scholar] [CrossRef]
- Harpke, D.; Carta, A.; Tomović, G.; Ranđelović, V.; Ranđelović, N.; Blattner, F.R.; Peruzzi, L. Phylogeny, karyotype evolution and taxonomy of Crocus series Verni (Iridaceae). Plant Syst. Evol. 2015, 301, 309–325. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Le Comber, S.C.; Ainouche, M.L.; Kovarik, A.; Leitch, A.R. Making a functional diploid: From polysomic to disomic inheritance. New Phytol. 2010, 186, 113–122. [Google Scholar] [CrossRef]
- Lv, Z.; Addo Nyarko, C.; Ramtekey, V.; Behn, H.; Mason, A.S. Defining autopolyploidy: Cytology, genetics, and taxonomy. Am. J. Bot. 2024, 111, e16292. [Google Scholar] [CrossRef]
- Shuka, L.; Zekaj, Z.; Mullaj, A. Biogeographical records of species of the genus Crocus L. in Albania. In Proceedings of the 5th Balkan Botanical Congress, Belgrade, Serbia, 7–11 September 2009; pp. 52–53. [Google Scholar]
- Raca, I.; Harpke, D.; Shuka, L.; Ranđelović, V. A new species of Crocus ser. Verni (Iridaceae) with 2n = 12 chromosomes from the Balkans. Plant Biosyst.-Int. J. Deal. All Asp. Plant Biol. 2022, 156, 36–42. [Google Scholar] [CrossRef]
- Meyer, F.K. Beitrage zur Flora von Albanien; The Thuringian Botanical: Jena, Germany, 2011. [Google Scholar]
- Waminal, N.E.; Blattner, F.; Harpke, D. The Crocus panrepeatome reveals the links between whole-genome duplications, repeat bursts, and descending dysploidy. Res. Sq. 2024. [Google Scholar] [CrossRef]
- Buzo, K. Bimësia e Kullotave dhe e Livadheve Natyrore të Shqiperisë; Publishing House of the Universities Books: Tirana, Albania, 1991. [Google Scholar]
- Shuka, L.; Çullaj, A.; Shumka, S.; Miho, A.; Duka, S.; Bachofen, R. The spatial and temporal variability of limnological properties of Bovilla Reservoir (Albania). Water Resour. Manag. 2011, 25, 3027–3039. [Google Scholar] [CrossRef]
- Meyer, M.; Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5448. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 February 2025).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Eaton, D.A.R.; Overcast, I. ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 2020, 36, 2592–2594. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; Depamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; Depristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Knaus, B.J.; Grünwald, N.J. VCFR: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Thia, J.A. genomalicious: Serving up a smorgasbord of R functions for performing and teaching population genomic analyses. BioRxiv 2019. [Google Scholar] [CrossRef]
- Ortiz, E.M. vcf2phylip v2.0: Convert a VCF Matrix into Several Matrix Formats for Phylogenetic Analysis. 2019. Available online: https://zenodo.org/records/2540861 (accessed on 24 February 2025).
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Barrett, T.; Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Hocking, T.; Schwendinger, B.; Krylov, I. data.table: Extension of ‘data.frame’, R Package Version 1.15.99. 2024. Available online: https://Rdatatable.gitlab.io/data.table (accessed on 24 February 2025).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation, R Package Version 1.1.4. 2023. Available online: https://github.com/tidyverse/dplyr (accessed on 24 February 2025).
- Wickham, H.; Vaughan, D.; Girlich, M. tidyr: Tidy Messy Data, R Package Version 1.3.1. 2024. Available online: https://tidyr.tidyverse.org (accessed on 24 February 2025).
- Müller, K.; Wickham, H. tibble: Simple Data Frames. 2023. Available online: https://tibble.tidyverse.org/ (accessed on 24 February 2025).
- Yu, L.; Stachowicz, J.J.; Dubois, K.; Reusch, T.B.H. Detecting clonemate pairs in multicellular diploid clonal species based on a shared heterozygosity index. Mol. Ecol. Resour. 2023, 23, 592–600. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; pp. 1–189. [Google Scholar]
- Waminal, N.E.; Pellerin, R.J.; Kang, S.-H.; Kim, H.H. Chromosomal mapping of tandem repeats revealed massive chromosomal rearrangements and insights into Senna tora dysploidy. Front. Plant Sci. 2021, 12, 629898. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef] [PubMed]
- Waminal, N.E.; Pellerin, R.J.; Kim, N.-S.; Jayakodi, M.; Park, J.Y.; Yang, T.-J.; Kim, H.H. Rapid and efficient FISH using pre-labeled oligomer probes. Sci. Rep. 2018, 8, 8224. [Google Scholar] [CrossRef] [PubMed]
- Milović, M. Rod Crocus L. (Iridaceae) u flori Hrvatske. J. Croat. Bot. Soc. 2016, 4, 4–20. [Google Scholar]
- Gligorijević, S.; Pejčinović, D. Contribution to the methodology of anatomical sections preparation. Acta Biol. Et Med. Exp. 1983, 8, 43–45. [Google Scholar]
- Raca, I.; Ljubisavljević, I.; Jušković, M.; Ranđelović, N.; Ranđelović, V. Comparative anatomical study of the taxa from series Verni Mathew (Crocus L.) in Serbia. Biol. Nyssana 2017, 8, 15–22. [Google Scholar]
- Raca, I.; Jovanovic, M.; Ljubisavljevic, I.; Juskovic, M.; Randelovic, V. Morphological and leaf anatomical variability of Crocus cf. heuffelianus Herb.(Iridaceae) populations from the different habitats of the Balkan Peninsula. Turk. J. Bot. 2019, 43, 645–658. [Google Scholar] [CrossRef]
- Raca, I. Taxonomy and Phylogeny of Series Verni Mathew (Crocus L.) in Southeastern Europe—Morpho-Anatomical, Cytological and Molecular Approach. Ph.D. Thesis, University of Nis, Niš, Serbia, 2021. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Keller, E.R.J.; Schubert, I.; Fuchs, J.; Meister, A.J.T.; Genetics, A. Interspecific crosses of onion with distant Allium species and characterization of the presumed hybrids by means of flow cytometry, karyotype analysis and genomic in situ hybridization. Theor. Appl. Genet. 1996, 92, 417–424. [Google Scholar] [CrossRef]
- Session, A.M.; Rokhsar, D.S. Transposon signatures of allopolyploid genome evolution. Nat. Commun. 2023, 14, 3180. [Google Scholar] [CrossRef] [PubMed]
- Brighton, C.A. Cytological problems in the genus Crocus (Iridaceae): I. Crocus vernus aggregate. Kew Bull. 1976, 31, 33. [Google Scholar] [CrossRef]
- Shuka, D.; Tan, K.; Hallaçi, B.; Shuka, L. Additions to the flora of North Albania. Phytol. Balc. 2020, 26, 517–522. [Google Scholar]
- Shumka, S.; Shumka, L.; Špoljar, M.; Shuka, L. Evidence of climate change and the conservation needed to halt the further deterioration of small glacial lakes. Climate 2024, 12, 124. [Google Scholar] [CrossRef]

| Species | No. of Shared Alleles | No. of Exclusively Shared Alleles | ||
|---|---|---|---|---|
| C. vernus (32) | 25,851 | (84.77%) | 1777 | (5.83%) |
| C. heuffelianus (18) | 21,187 | (69.47%) | 550 | (1.80%) |
| C. tommasinianus (29) | 18,412 | (60.38%) | 289 | (0.95%) |
| C. bertiscensis (7) | 15,078 | (49.44%) | 155 | (0.51%) |
| C. neapolitanus (2) | 13,391 | (43.91%) | 66 | (0.22%) |
| C. siculus (4) | 11,193 | (36.70%) | 39 | (0.13%) |
| C. kosaninii (3) | 10,328 | (33.87%) | 85 | (0.28%) |
| C. etruscus (2) | 9247 | (30.32%) | 18 | (0.06%) |
| C. ilvensis (2) | 8695 | (28.51%) | 4 | (0.01%) |
| C. longiflorus (1) | 3829 | (12.56%) | 47 | (0.15%) |
| Clades | No. of shared alleles | No. of exclusively shared alleles | ||
| CladeI: C. vernus—C. neapolitanus—C. siculus (38) | 26,483 | (86.84%) | 3276 | (10.74%) |
| CladeII: C. heuffelianus—C. bertiscensis—C. tommasinianus—C. kosaninii (57) | 26,671 | (87.46%) | 2690 | (8.82%) |
| CladeIII: C. etruscus—C. ilvensis (4) | 10,909 | (35.77%) | 70 | (0.23%) |
| CladeI + CladeII | 30,378 | (99.61%) | 17,482 | (57.33%) |
| C. bachofenii | C. vernus | C. bertiscensis | C. tommasinianus | C. neapolitanus | C. neglectus | |
| Corm width (mm) | 10.60 ± 2.77 | 12.80 ± 2.70 | 6.80 ± 0.80 | 10.40 ± 1.70 | 11–13 | 13.9 ± 1.9 |
| Fibre width (mm) | 0.10 ± 0.04 | 0.08 ± 0.03 | 0.08 ± 0.03 | 0.11 ± 0.02 | 0.06–0.02 | 0.10 ± 0.02 |
| Number of leaves | 2–5, usually 3 | 2–4, usually 3 | 1–3, usually 2 | 2–5, usually 3 | 2–4, usually 3 | – |
| Leaf width (mm) | 2.93 ± 0.30 | 1.83 ± 0.50 | 3.20 ± 0.50 | 1.76 ± 0.90 | (2–) 4–8 | – |
| Number of cataphylls | 3–5, usually 4 | 3–4, usually 3 | 2–4, usually 3 | 3–4, usually 3 | 2–3 | – |
| Perigone tube length (mm) | 44.30 ± 7.50 | 43.60 ± 10.60 | 33.10 ± 5.90 | 62.30 ± 10.60) | 40–100 | 60.7 ± 21.9 |
| Perigone tube color | Deep violet-to-violet | Lilac or white | Lilac or deep purple | White or whitish | Lilac in upper part | Concolor |
| Number of flowers | 1, rarely 2 | 1, rarely 2 | 1 | 1, rarely 2 | 1 (–2) | 1 (–2) |
| Perigone segment length (mm) | 38.50 ± 1.00 | 21.90 ± 0.50 | 27.22 ± 3.14 | 26.7 ± 2.00 | 25–55 | 34.8 ± 4.7 |
| Perigone segment width (mm) | 14.50 ± 1.50 | 6.40 ± 1.00 | 8.88 ± 1.25 | 11.27 ± 1.50 | 9–20 | – |
| Outer perigone segment color | Lilac-to-deep lilac at the base and lilac-to-violet above | White, white with lilac stripes, or completely lilac | Lilac at the base, with a dark violet heart- or V-shaped patch at the apex of perigone segments | Pale lilac-to-lilac | Purple-to-deep purple | Lilac, rarely white |
| Position of the stigma | Longer than anthers (rarely equal to) | Shorter than anthers | Equal to or longer than anthers (rarely shorter) | Equal to or longer than anthers (rarely shorter) | Equal to or longer than anthers | Longer than anthers (rarely equal to) |
| Stigma/anther ratio (mm) | 1.47 ± 2.50 | −8.40 ± 3.90 | 1.11 ± 3.80 | 1.20 ± 2.30 | 1.10 ± 3.90 | 3.7 ± 2.1 |
| Anther length (mm) | 15.67 ± 3.00 | 9.30 ± 1.60 | 8.13 ± 1.11 | 10.40 ± 1.50 | 11.80 ± 0.29 | 12.6 ± 1.7 |
| Anther color (mm) | Yellow-to-orange | Yellow | Yellow | Yellow | Yellow | Yellow |
| Stigma lobes length (mm) | 2.34 ± 0.53 | 1.38 ± 0.75 | 1.62 ± 0.55 | 1.00 ± 0.09 | – | – |
| Stigma sublobes length (mm) | Incised for 0.24 ± 0.01 | Incised for 0.23 ± 0.22 | Incised for 0.31 ± 0.30 | Incised for 0.90 ± 0.32 | Incised for 0.06 ± 0.02 | Incised for 0.13 ± 0.08 |
| Stigma color | Orange (rarely yellow) | Orange | Orange | Orange | Orange | Orange |
| Name | Length (bp) | Sequence | 5′ Modification | 3′ Modification | Reference |
|---|---|---|---|---|---|
| CroSat137a | 30 | RAGCTGCTTAGAGTGCTATAGRAATGCAAC | Cy3 | Cy3 | This study |
| Crosat137b | 30 | GCTTSGATCAAACTGGGCYAAGATGCTCCC | Cy3 | Cy3 | This study |
| CroSat042a | 30 | CATGACTTGTAAAAACRGAGTYTTGCAAGC | Cy5 | Cy5 | This study |
| CroSat042b | 30 | GAAGATATCTACTCAAATCACACTAAAATA | Cy5 | Cy5 | This study |
| CroSat144a | 30 | TCTCCCGGAAACTCGTTTCGAGGCCTACCT | FAM | FITC | This study |
| CroSat144b | 30 | CATCGCGGACGGTTTCGGTCGTTACGTTGA | FAM | FITC | This study |
| TTAGGG | 29 | TTAGGGTTAGGGTTAGGGTTAGGGTTAGG | Cy3 | Cy3 | This study |
| 18SrDNA_UniOP_1 | 28 | CCGGAGAGGGAGCCTGAGAAACGGCTAC | Cy5 | Cy5 | Waminal et al. (2018) [45] |
| 18SrDNA_UniOP_2 | 26 | ATCCAAGGAAGGCAGCAGGCGCGCAA | Cy5 | Cy5 | Waminal et al. (2018) [45] |
| 18SrDNA_UniOP_3 | 28 | GGGCAAGTCTGGTGCCAGCAGCCGCGGT | Cy5 | Cy5 | Waminal et al. (2018) [45] |
| 18SrDNA_UniOP_4 | 27 | TCGAAGACGATYAGATACCGTCSTAGT | Cy5 | Cy5 | Waminal et al. (2018) [45] |
| 5SrDNA_UniOP_1 | 30 | GGGTGCGATCATACCAGCACTAGAGCACCG | FAM | FITC | Waminal et al. (2018) [45] |
| 5SrDNA_UniOP_2 | 29 | CCCATCAGAACTCCGAAGTTAAGCGTGCT | FAM | FITC | Waminal et al. (2018) [45] |
| 5SrDNA_UniOP_3 | 24 | GCGAGAGTAGTACTAGGATGGGTG | FAM | FITC | Waminal et al. (2018) [45] |
| 5SrDNA_UniOP_4 | 26 | CCTGGGAAGTMCTCGTGTTGCAYYCC | FAM | FITC | Waminal et al. (2018) [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raca, I.; Shuka, D.; Shuka, L.; Waminal, N.E.; Harpke, D. An Integrative Systematic Approach Reveals a New Species of Crocus Series Verni (Iridaceae) Endemic to Albania. Plants 2025, 14, 741. https://doi.org/10.3390/plants14050741
Raca I, Shuka D, Shuka L, Waminal NE, Harpke D. An Integrative Systematic Approach Reveals a New Species of Crocus Series Verni (Iridaceae) Endemic to Albania. Plants. 2025; 14(5):741. https://doi.org/10.3390/plants14050741
Chicago/Turabian StyleRaca, Irena, Donald Shuka, Lulëzim Shuka, Nomar Espinosa Waminal, and Dörte Harpke. 2025. "An Integrative Systematic Approach Reveals a New Species of Crocus Series Verni (Iridaceae) Endemic to Albania" Plants 14, no. 5: 741. https://doi.org/10.3390/plants14050741
APA StyleRaca, I., Shuka, D., Shuka, L., Waminal, N. E., & Harpke, D. (2025). An Integrative Systematic Approach Reveals a New Species of Crocus Series Verni (Iridaceae) Endemic to Albania. Plants, 14(5), 741. https://doi.org/10.3390/plants14050741






