RNA-Seq Analysis Reveals Potential Genes Involved in Plant Growth Regulator-Induced Ovary Development in Male Kiwifruit (Actinidia eriantha)
Abstract
1. Introduction
2. Results
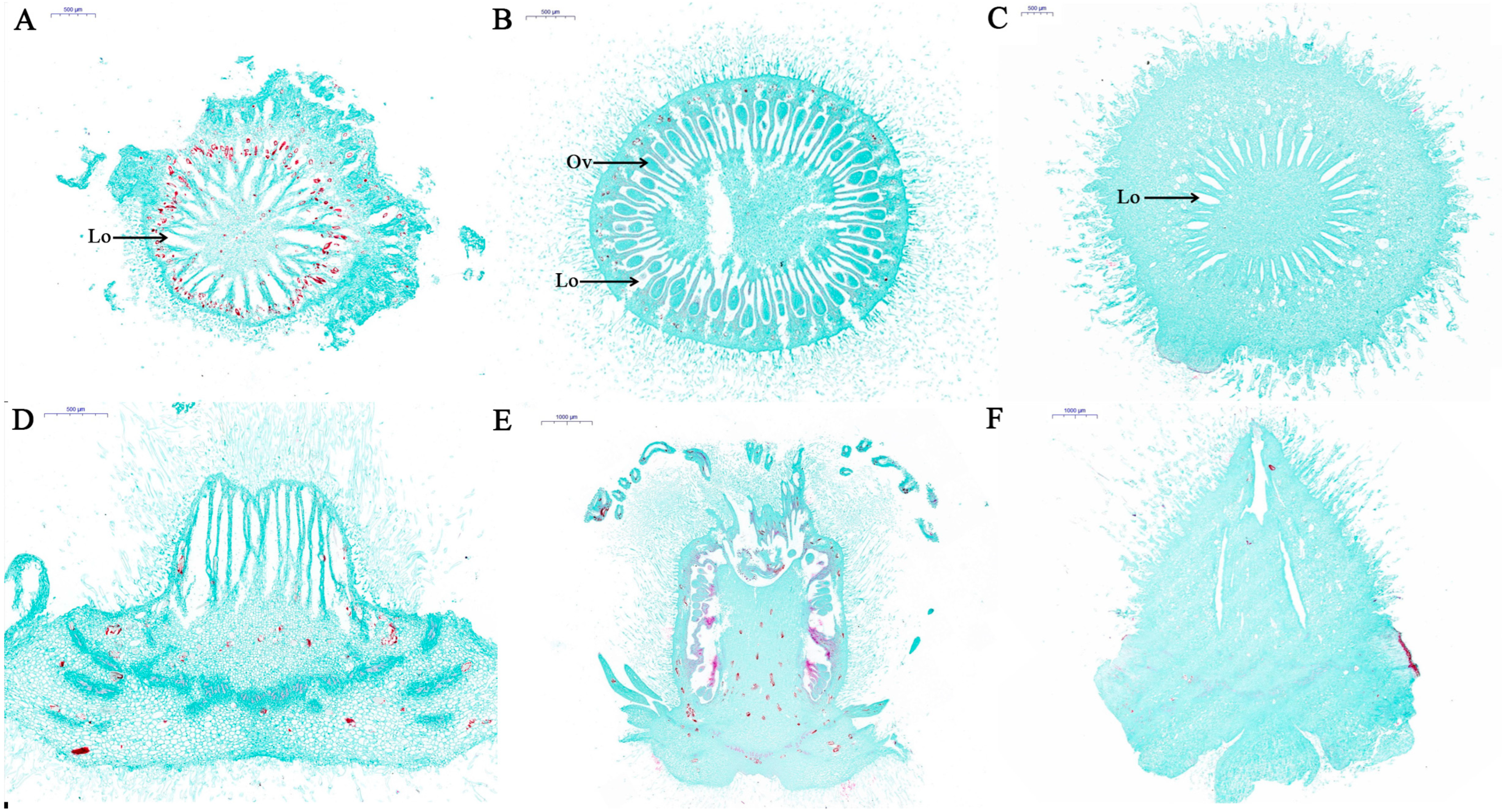
2.1. Histological Characterization of Con, Fem and PT
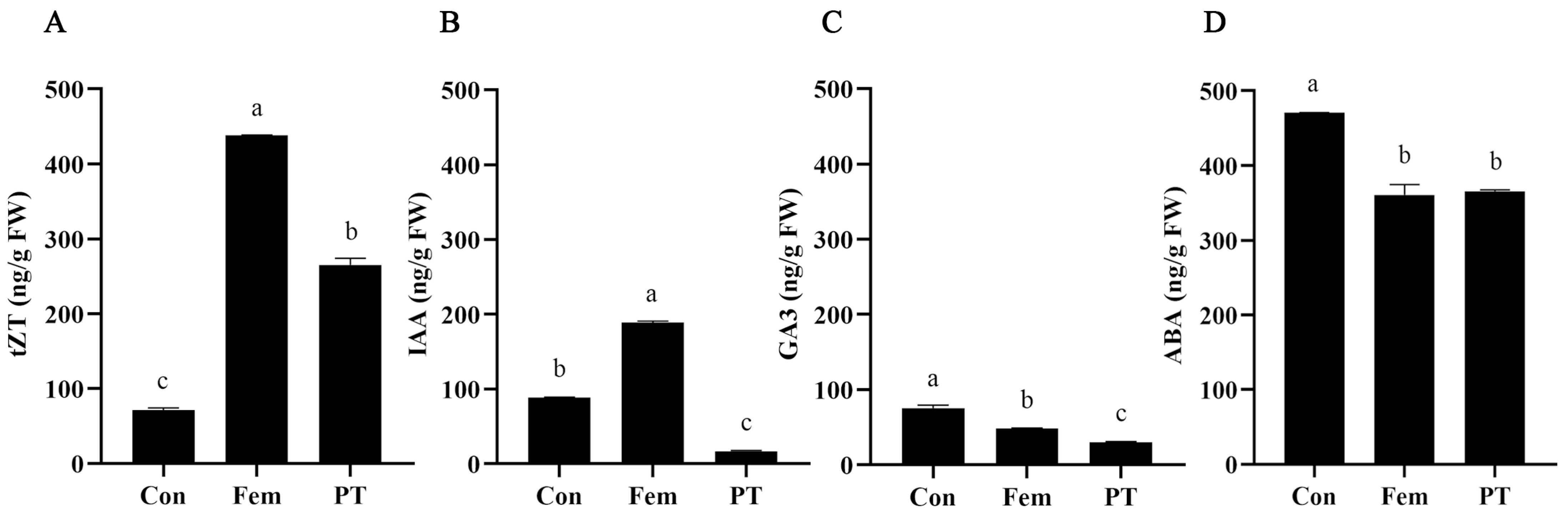
2.2. Phytohormone Level Detection
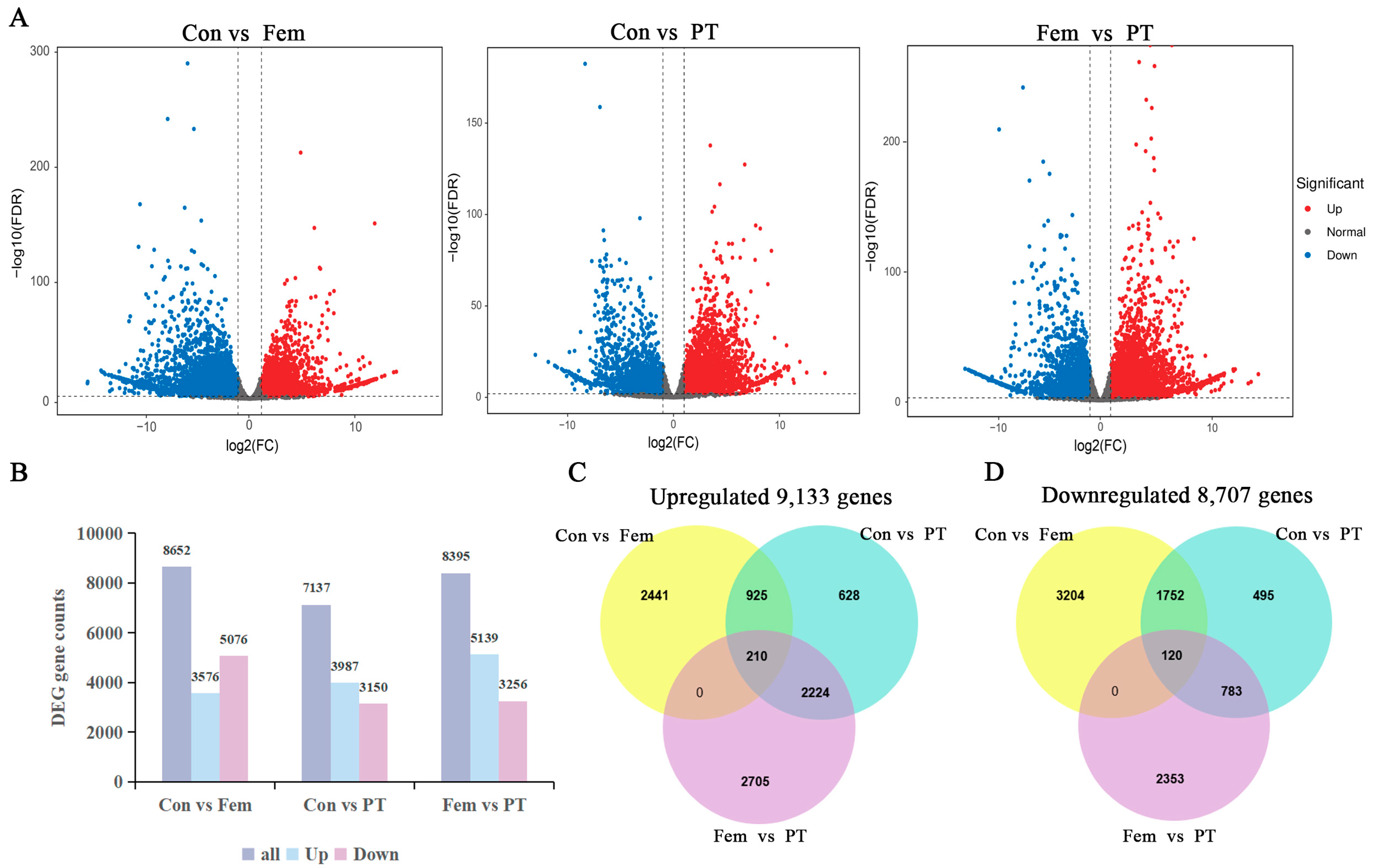
2.3. RNA-Seq and Differential Gene Expression Analyses
2.4. GO and KEGG Enrichment Analysis of DEGs
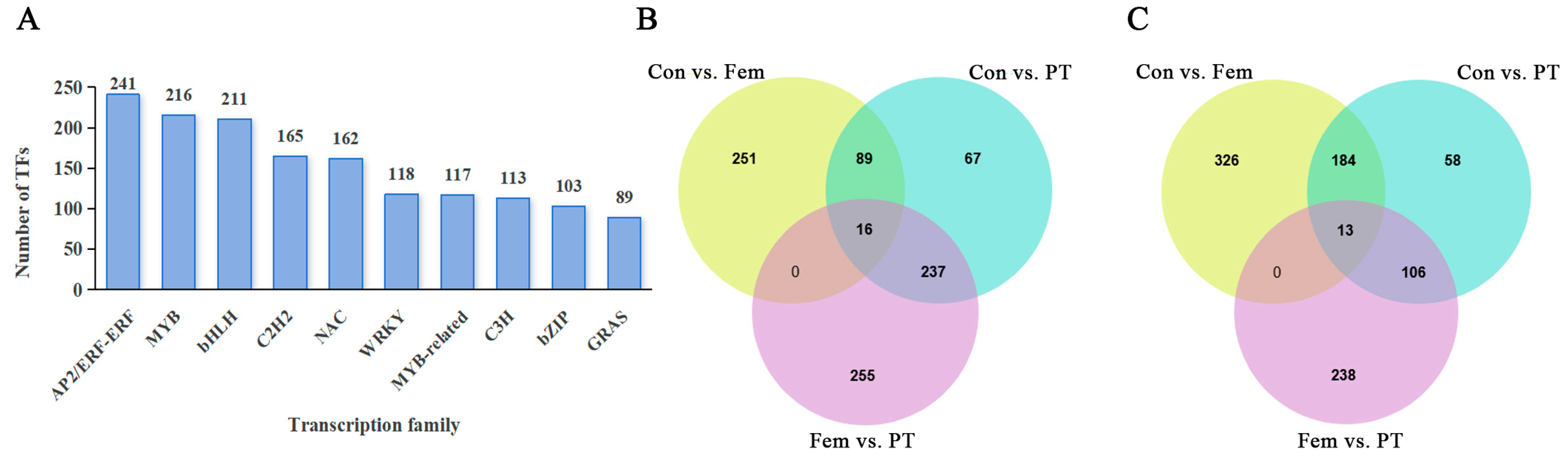
2.5. Identification of Unigenes or DEGs Encoding TFs
2.6. Key DEGs Related to Phytohormone Biosynthesis and Signaling Pathways
2.7. Analysis of DEGs Participating in Ovary Development
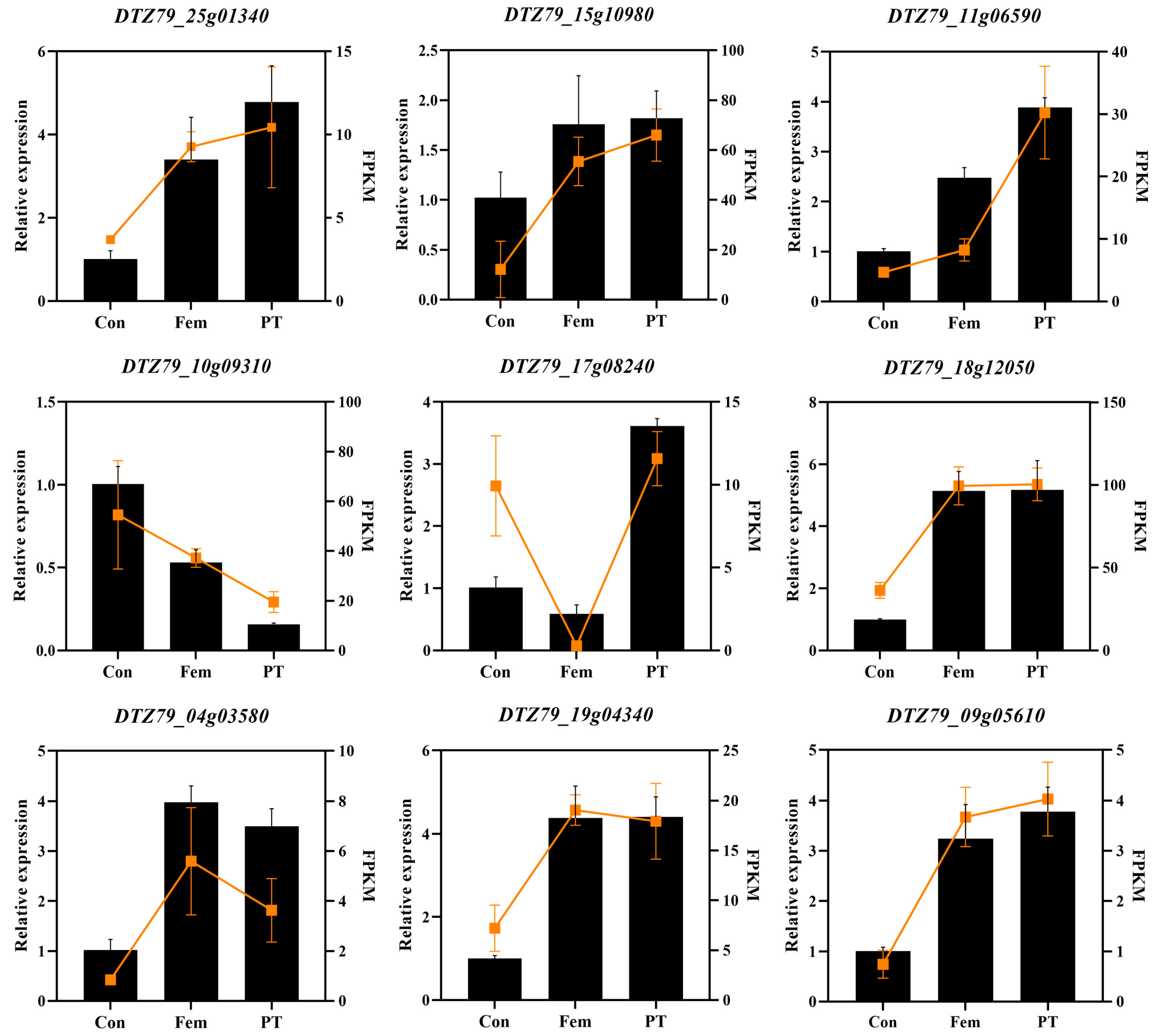
2.8. RNA-Seq Data Validation of Differentially Expressed Transcripts by Real-Time Quantitative PCR
3. Discussion
4. Materials and Methods
4.1. Plant Material and PGR Treatment
4.2. Histological Observation
4.3. Phytohormone Analysis
4.4. RNA Extraction and Illumina Sequencing
4.5. Analysis of the RNA-Seq Data
4.6. Functional Annotation of Transcripts and Prediction TFs
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, S.; Qiu, Y.; Zhu, F. Kiwifruit (Actinidia spp.): A review of chemical diversity and biological activities. Food Chem. 2021, 350, 128469. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gao, X.; Sun, Q.; Zhu, Y.; Qin, L.; Zhu, B. The phytochemical properties, pharmacological effects and traditional uses of Actinidia eriantha Benth: A review. Front. Pharmacol. 2022, 13, 959900. [Google Scholar] [CrossRef] [PubMed]
- Caporali, E.; Testolin, R.; Pierce, S.; Spada, A. Sex change in kiwifruit (Actinidia chinensis Planch): A developmental framework for the bisexual to unisexual floral transition. Plant Reprod. 2019, 32, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.R. Kiwifruit: The wild and the cultivated plants. Adv. Food Nutr. Res. 2013, 68, 15–32. [Google Scholar] [PubMed]
- Akagi, T.; Henry, I.M. A Y-Encoded Suppressor of Feminization Arose via Lineage-Specific Duplication of a Cytokinin Response Regulator in Kiwifruit. Plant Cell 2018, 30, 780–795. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Pilkington, S.M.; Varkonyi-Gasic, E.; Henry, I.M. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plants 2019, 5, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D. Plant contributions to our understanding of sex chromosome evolution. New Phytol. 2015, 208, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wang, T.; Cooney, J. Shy Girl, a kiwifruit suppressor of feminization, restricts gynoecium development via regulation of cytokinin metabolism and signalling. New Phytol. 2021, 230, 1461–1475. [Google Scholar] [CrossRef]
- Vanstraelen, M.; Benková, E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487. [Google Scholar] [CrossRef] [PubMed]
- El-Showk, S.; Ruonala, R.; Helariutta, Y. Crossing paths: Cytokinin signalling and crosstalk. Development 2013, 140, 1373–1383. [Google Scholar] [CrossRef]
- Pernisová, M.; Kuderová, A.; Hejátko, J. Cytokinin and auxin interactions in plant development: Metabolism, signalling, transport and gene expression. Curr. Protein Pept. Sci. 2011, 12, 137–147. [Google Scholar] [CrossRef]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; Zuñiga-Mayo, V.M.; Chávez Montes, R.A.; Marsch-Martínez, N.; de Folter, S. Inside the gynoecium: At the carpel margin. Trends Plant Sci. 2013, 18, 644–655. [Google Scholar] [CrossRef]
- Marsch-Martínez, N.; Ramos-Cruz, D.; Irepan Reyes-Olalde, J.; Lozano-Sotomayor, P.; Zúñiga-Mayo, V.M.; de Folter, S. The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. Cell Mol. Biol. 2012, 72, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Pischke, M.S.; Jones, L.G.; Otsuga, D.; Fernandez, D.E.; Drews, G.N.; Sussman, M.R. An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 15800–15805. [Google Scholar] [CrossRef]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.C.; Liang, Y.; Zuo, J. Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 2010, 22, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, C.; Liu, Z. A model for an early role of auxin in Arabidopsis gynoecium morphogenesis. Front. Plant Sci. 2014, 5, 327. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Feldman, L.J.; Zambryski, P.C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000, 127, 3877–3888. [Google Scholar] [CrossRef]
- Zúñiga-Mayo, V.M.; Reyes-Olalde, J.I.; Marsch-Martinez, N.; de Folter, S. Cytokinin treatments affect the apical-basal patterning of the Arabidopsis gynoecium and resemble the effects of polar auxin transport inhibition. Front. Plant Sci. 2014, 5, 191. [Google Scholar]
- Marsch-Martínez, N.; de Folter, S. Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 2016, 29, 104–114. [Google Scholar] [CrossRef]
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. Cell Mol. Biol. 2021, 105, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Schubert, R.; Dobritzsch, S. Tomato MYB21 Acts in Ovules to Mediate Jasmonate-Regulated Fertility. Plant Cell 2019, 31, 1043–1062. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Paponov, M.; Teale, W.; Menges, M.; Chakrabortee, S.; Murray, J.A.; Palme, K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 2008, 1, 321–337. [Google Scholar] [CrossRef]
- Wu, L.Y.; Shang, G.D.; Wang, F.X.; Gao, J.; Wan, M.C.; Xu, Z.G.; Wang, J.W. Dynamic chromatin state profiling reveals regulatory roles of auxin and cytokinin in shoot regeneration. Dev. Cell 2022, 57, 526–542.e7. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef]
- Higuchi, M.; Pischke, M.S.; Mähönen, A.P.; Miyawaki, K.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Shinozaki, K.; Kato, T.; Tabata, S.; et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 2004, 101, 8821–8826. [Google Scholar] [CrossRef]
- Inoue, T.; Higuchi, M.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Kato, T.; Tabata, S.; Shinozaki, K.; Kakimoto, T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 2001, 409, 1060–1063. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, C.; Ohashi, Y.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 2004, 16, 1365–1377. [Google Scholar] [CrossRef] [PubMed]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef]
- Kim, H.J.; Ryu, H.; Hong, S.H.; Woo, H.R.; Lim, P.O.; Lee, I.C.; Sheen, J.; Nam, H.G.; Hwang, I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Yamashino, T.; Yokoyama, A.; Mizuno, T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 47–57. [Google Scholar] [CrossRef]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.H.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schaller, G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 2008, 20, 2102–2116. [Google Scholar] [CrossRef]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. Int. J. Exp. Plant Biol. 2012, 190, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Wang, X.J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef] [PubMed]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 2015, 27, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Sessions, R.A.; Zambryski, P.C. Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 1995, 121, 1519–1532. [Google Scholar] [CrossRef]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Xin, R.; Kim, D.H.; Sung, S.; Lange, T.; Huq, E. NO FLOWERING IN SHORT DAY (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 2016, 143, 682–690. [Google Scholar] [PubMed]
- Li, K.; Yu, R.; Fan, L.M.; Wei, N.; Chen, H. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 2016, 7, 11868. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Olalde, J.I.; Zúñiga-Mayo, V.M.; Serwatowska, J. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 2017, 13, e1006726. [Google Scholar] [CrossRef]
- Bianchi, M.W.; Guivarc’h, D.; Thomas, M.; Woodgett, J.R.; Kreis, M. Arabidopsis homologs of the shaggy and GSK-3 protein kinases: Molecular cloning and functional expression in Escherichia coli. Mol. Gen. Genet. MGG 1994, 242, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Dornelas, M.C.; Lejeune, B.; Dron, M.; Kreis, M. The Arabidopsis SHAGGY-related protein kinase (ASK) gene family: Structure, organization and evolution. Gene 1998, 212, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Guo, H.; Lv, B.; Feng, J.; Wang, H.; Zhang, Z.; Chai, S. Gibberellin biosynthesis is required for CPPU-induced parthenocarpy in melon. Hortic. Res. 2023, 10, uhad084. [Google Scholar] [CrossRef]
- Yue, J.; Liu, J.; Tang, W.; Wu, Y.Q.; Tang, X.; Li, W.; Yang, Y.; Wang, L.; Huang, S.; Fang, C.; et al. Kiwifruit Genome Database (KGD): A comprehensive resource for kiwifruit genomics. Hortic. Res. 2020, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]

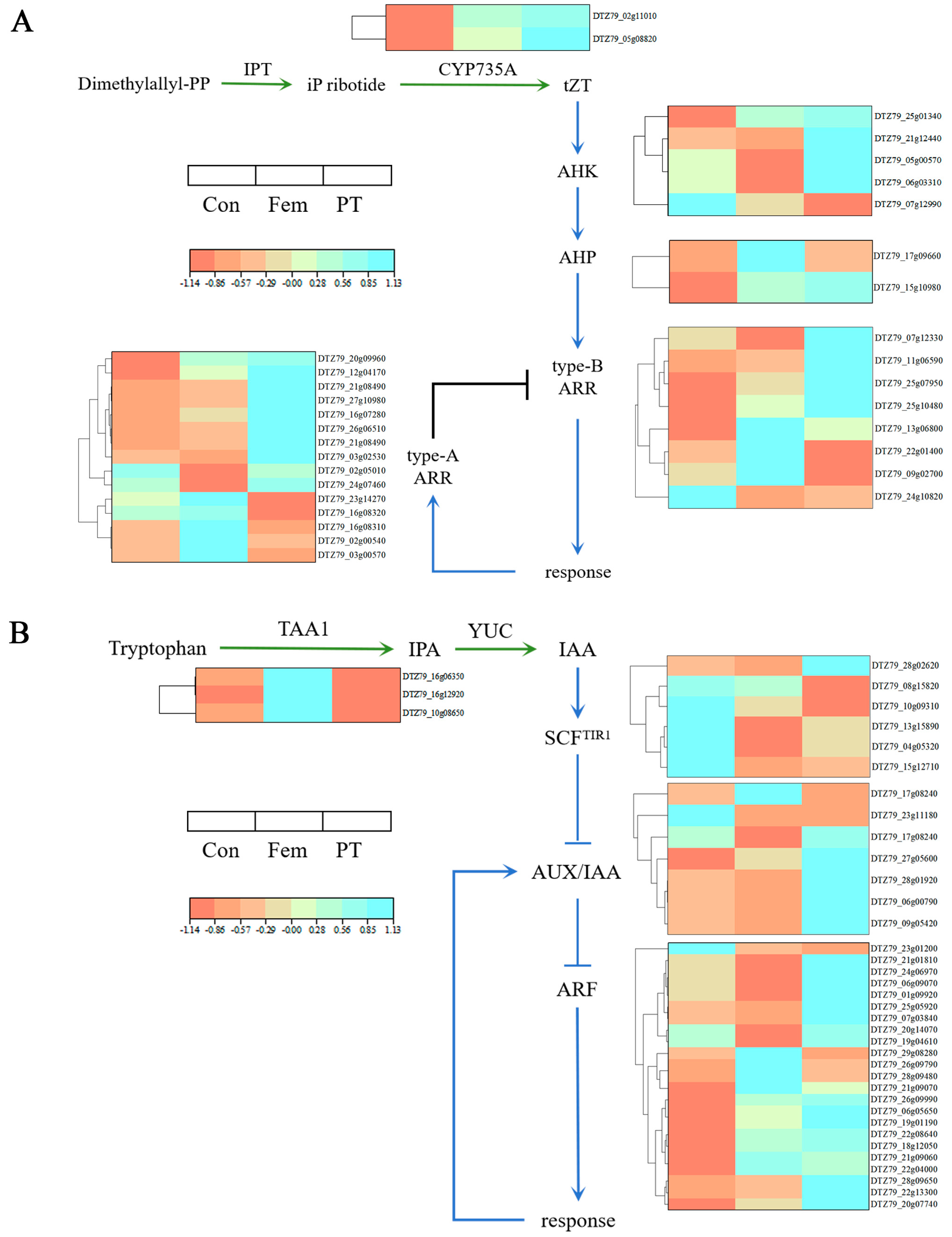
| Group | Gene ID | log2FC | Description |
|---|---|---|---|
| Con vs. Fem | DTZ79_04g03580 | 2.49 | Transcription factor SPATULA |
| DTZ79_19g04340 | 1.68 | Shaggy-related protein kinase | |
| Con vs. PT | DTZ79_04g03580 | 1.93 | Transcription factor SPATULA |
| DTZ79_19g04340 | 1.18 | Shaggy-related protein kinase | |
| Fem vs. PT | DTZ79_04g03580 | −0.58 | Transcription factor SPATULA |
| DTZ79_19g04340 | −0.02 | Shaggy-related protein kinase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, R.; Zhang, M.; Wei, F.; Lin, M.; Fang, J.; Wang, R.; Li, Y.; Chen, J.; Sun, L.; Qi, X. RNA-Seq Analysis Reveals Potential Genes Involved in Plant Growth Regulator-Induced Ovary Development in Male Kiwifruit (Actinidia eriantha). Plants 2025, 14, 703. https://doi.org/10.3390/plants14050703
Fu R, Zhang M, Wei F, Lin M, Fang J, Wang R, Li Y, Chen J, Sun L, Qi X. RNA-Seq Analysis Reveals Potential Genes Involved in Plant Growth Regulator-Induced Ovary Development in Male Kiwifruit (Actinidia eriantha). Plants. 2025; 14(5):703. https://doi.org/10.3390/plants14050703
Chicago/Turabian StyleFu, Rong, Min Zhang, Feng Wei, Miaomiao Lin, Jinbao Fang, Ran Wang, Yukuo Li, Jinyong Chen, Leiming Sun, and Xiujuan Qi. 2025. "RNA-Seq Analysis Reveals Potential Genes Involved in Plant Growth Regulator-Induced Ovary Development in Male Kiwifruit (Actinidia eriantha)" Plants 14, no. 5: 703. https://doi.org/10.3390/plants14050703
APA StyleFu, R., Zhang, M., Wei, F., Lin, M., Fang, J., Wang, R., Li, Y., Chen, J., Sun, L., & Qi, X. (2025). RNA-Seq Analysis Reveals Potential Genes Involved in Plant Growth Regulator-Induced Ovary Development in Male Kiwifruit (Actinidia eriantha). Plants, 14(5), 703. https://doi.org/10.3390/plants14050703






