Resistance Spectrum Analysis and Breeding Utilization of Rice Blast Resistance Gene Pigm-1
Abstract
1. Introduction
2. Results
2.1. Creation of Single Gene Lines Containing Pigm-1 and Pigm
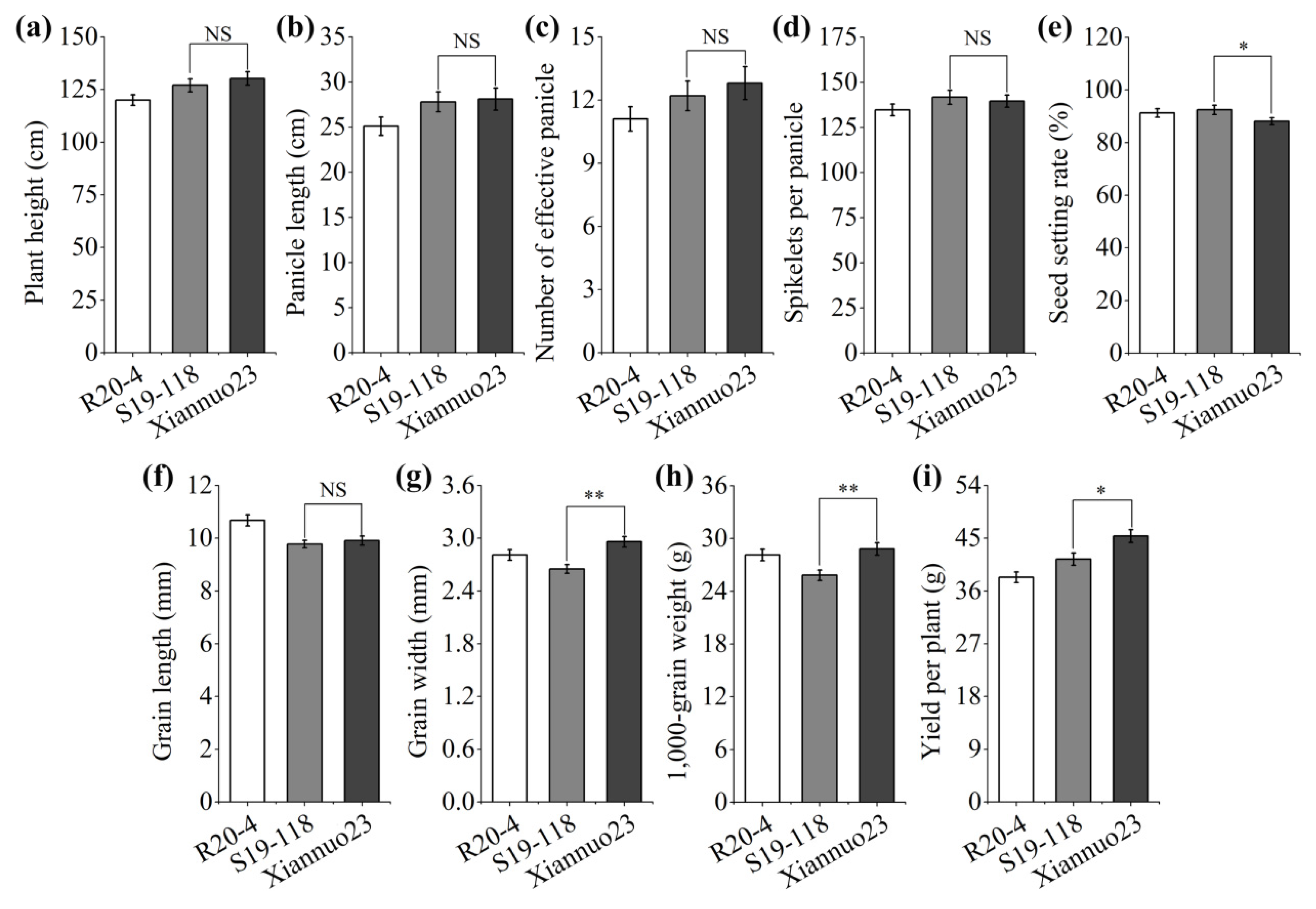
2.2. Analysis of the Main Agronomic Traits of Pigm-1 and Pigm Single Gene Lines
2.3. Analysis of Single Gene Line Pigm-1 and Pigm Blast Resistance Spectrum
2.4. An Improved Breeding Line Containing Pigm-1 Gene Was Obtained
2.5. Analysis of Rice Blast Resistance of Xiannuo 23 Improved Line
2.6. Analysis of Main Agronomic Characters of Xiannuo 23
3. Discussion
3.1. Pigm-1 Antispectral Analysis
3.2. Analysis of Pigm-1 Breeding and Utilization Prospects
3.3. Further Study on the Relationship Between Pigm-1 and Waxy Genes
4. Materials and Methods
4.1. Isolation of Rice Blast Fungus
4.2. Rice Blast Fungus Incubation
4.3. Detection and Analysis of Pigm-1 and Pigm Functional Markers
4.4. Investigation of Agronomic Traits
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elert, E. Rice by the numbers: A good grain. Nature 2014, 514, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Kamarudin, S.A.A.; Ahmad, F.; Hasan, N.; Hisham, S.N.; Yusof, S.N.; Abu, H.A.; Hussein, S.; Harun, A.R.; Wan, C.W.D.; Md, S.M.; et al. Whole genome resequencing data and grain quality traits of the rice cultivar Mahsuri and its blast disease resistant mutant line, Mahsuri Mutant. Data Brief 2023, 52, 109974. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.P.; Zhang, F.; Chen, K.; Shen, C.C.; Zhu, S.B.; Qiu, X.J.; Xu, J.L. Identification of rice blast resistance in xian and geng germplasms by genomewide association study. Acta Agron. Sin. 2023, 49, 1170–1183. [Google Scholar]
- He, F.; Zhang, H.; Liu, J.L.; Wang, Z.L.; Wang, G.L. Recent advances in understanding the innate immune mechanisms and developing new disease resistance breeding strategies against the rice blast fungus Magnaporthe oryzae in rice. Yi Chuan 2014, 36, 756–765. [Google Scholar] [PubMed]
- Kou, Y.J.; Shi, H.B.; Qiu, J.H.; Tao, Z.; Wang, W.M. Effectors and environment modulating rice blast disease: From understanding to effective control. Trends Microbiol. 2024, 32, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Annegowda, D.C.; Prasannakumar, M.K.; Mahesh, H.B.; Siddabasappa, C.B.; Devanna, P.; Banakar, S.N.; Manojkumar, H.B.; Prasad, S.R. Rice blast disease in India: Present status and future challenges. Integr. Adv. Rice Res. 2021, 98847, 157–197. [Google Scholar]
- An, Z.S.; Liu, G.L.; Mei, H.W.; Li, L.T.; Wang, J.H.; Luo, L.J. Improving blast resistance of parental lines of drought resistant hybrid rice by marker-assisted selection. Mol. Plant Breed. 2010, 8, 1172–1176. [Google Scholar]
- Greenwood, J.R.; Lacorte-Apostol, V.; Kroj, T.; Padilla, J.; Telebanco-Yanoria, M.J.; Glaus, A.N.; Roulin, A.; Padilla, A.; Zhou, B.; Keller, B.; et al. Genome-wide association analysis uncovers rice blast resistance alleles of Ptr and Pia. Commun. Biol. 2024, 7, 607. [Google Scholar] [CrossRef] [PubMed]
- Mutiga, S.K.; Orwa, P.; Nganga, E.M.; Kyallo, M.M.; Rotich, F.; Gichuhi, E.; Kimani, J.M.; Mwongera, D.T.; Were, V.M.; Yanoria, M.J.; et al. Characterization of blast resistance in a diverse rice panel from sub-saharan africa. Phytopathology 2023, 113, 1278–1288. [Google Scholar] [CrossRef]
- Xiao, N.; Wu, Y.Y.; Li, A.H. Strategy for use of rice blast resistance genes in rice molecular breeding. Rice Sci. 2020, 4, 263–277. [Google Scholar]
- Wu, Y.Y.; Xiao, N.; Yu, L.; Pan, C.H.; Li, Y.H.; Zhang, X.X.; Liu, G.Q.; Dai, Z.Y.; Pan, X.B.; Li, A.H. Combination patterns of major R genes determine the level of resistance to the M. oryzae in rice (Oryza sativa L.). PLoS ONE 2015, 10, e126130. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.W.; Zhai, K.R.; Xie, Z.; Yang, D.Y.; Zhu, X.D.; Liu, J.Z.; Wang, X.; Qin, P.; Yang, Y.Z.; Zhang, G.M.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Zhou, Y.; Wan, T.; Yuan, B.; Lei, F.; Chen, M.J.; Wang, Q.; Huang, P.; Kou, S.Y.; Qiu, W.X.; Liu, L. Improving rice blast resistance by mining broad-spectrum resistance genes at the Pik locus. Rice Sci. 2022, 29, 133–142. [Google Scholar]
- Yoshida, K.; Saitoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Yoshida, K.; Saitoh, H.; Fujisaki, K.; Hirabuchi, A.; Alaux, L.; Fournier, E.E.; Tharreau, D.; Terauchi, R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 2012, 72, 894–907. [Google Scholar] [CrossRef]
- Yang, D.W.; Li, S.P.; Lu, L.; Fang, J.B.; Wang, W.; Cui, H.T.; Tang, D.Z. Identification and application of the Pigm-1 gene in rice disease-resistance breeding. Plant Biol. 2020, 22, 1022–1029. [Google Scholar] [CrossRef]
- Feng, Z.M.; Li, M.Y.; Xu, Z.W.; Gao, P.; Wu, Y.Y.; Wu, K.T.; Zhao, J.H.; Wang, X.Q.; Wang, J.N.; Li, M.C.; et al. Development of rice variety with durable and broad-spectrum resistance to blast disease through marker-assisted introduction of Pigm gene. Front. Plant Sci. 2022, 13, 937767. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xiao, N.; Li, Y.; Gao, Q.; Ning, Y.; Yu, L.; Cai, Y.; Pan, C.; Zhang, X.; Huang, N.; et al. Identification and fine mapping of qPBR10-1, a novel locus controlling panicle blast resistance in Pigm-containing P/TGMS line. Mol. Breed. 2021, 41, 75. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Yan, B.X.; Shou, J.Y.; Tang, J.; Wang, X.; Zhai, K.R.; Liu, J.Y.; Li, Q.; Luo, M.Z.; Deng, Y.W.; et al. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos. Trans. R. Soc. London Biol. Sci. 2019, 4, 374. [Google Scholar] [CrossRef]
- Peng, Z.R.; Li, L.; Wu, S.H.; Chen, X.L.; Shi, Y.F.; He, Q.; Shu, F.; Zhang, W.H.; Sun, P.Y.; Deng, H.F.; et al. Frequencies and variations of Magnaporthe oryzae avirulence genes in Hunan province, China. Plant Dis. 2021, 105, 3829–3834. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Chen, Y.; Pan, C.H.; Xiao, N.; Yu, L.; Li, Y.H.; Zhang, X.X.; Pan, X.B.; Chen, X.J.; Liang, C.Z.; et al. Development and evaluation of near-isogenic lines with different blast resistance alleles at the Piz locus in japonica rice from the lower region of the Yangtze River, China. Plant Dis. 2017, 101, 1283–1291. [Google Scholar] [CrossRef]
- He, N.Q.; Yang, D.W.; Zheng, X.H.; Huang, F.H.; Cheng, C.P.; Ye, N. Improving blast resistance of R20 by molecular marker-assisted selection of Pigm-1 gene. J. Nucl. Agric. Sci. 2022, 36, 245–250. [Google Scholar]
- Yang, D.W.; He, N.Q.; Huang, F.H. Pyramiding Pigm-1 and Xa23 genes in rice (Oryza sativa L.) by marker-assisted selection. J. Northwest A F Univ. (Nat. Sci. Ed.) 2023, 51, 37–45. [Google Scholar]
- Peng, P.; Jiang, H.Y.; Luo, L.H.; Ye, C.R.; Xiao, Y.H. Pyramiding of multiple genes to improve rice blast resistance of photo-thermo sensitive male sterile line, without yield penalty in hybrid rice production. Plants 2023, 12, 1389. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.P.; Wang, G.; Qin, R.; Gong, C.Q.; Zhou, D.; Li, D.K.; Luo, B.J.; Jin, J.H.; Deng, Q.M.; Wang, S.Q.; et al. Improvement of quality and disease resistance for a heavy-panicle hybrid restorer line, R600, in rice (Oryza sativa L.) by gene pyramiding breeding. Curr. Issues Mol. Biol. 2024, 46, 10762–10778. [Google Scholar] [CrossRef]
- Liang, D.; Yang, D.Y.; Li, T.; Zhu, Z.; Yan, B.X.; He, Y.; Li, X.Y.; Zhai, K.R.; Liu, J.Y.; Kawano, Y.; et al. A PRA-Rab trafficking machinery modulates NLR immune receptor plasma membrane microdomain anchoring and blast resistance in rice. Sci. Bull. 2024, 9, S2095-9273(24)00911-3. [Google Scholar] [CrossRef]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.J.; Zhu, X.B.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef]
- Zhao, D.D.; Chung, H.; Jang, Y.H.; Farooq, M.; Choi, S.Y.; Du, X.X.; Kim, K.M. Analysis of rice blast fungus genetic diversity and identification of a novel blast resistance OsDRq12 gene. Phytopathology 2024, 114, 1917–1925. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.W.; Li, S.P.; Lu, L.; Zheng, Z.C.; Tang, D.Z.; Cui, H.T. Transcriptome analysis of rice response to blast fungus identified core genes involved in immunity. Plant Cell Environ. 2021, 44, 3103–3121. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Sun, X.C.; Zhang, S.L.; Liang, W.H.; Zhou, L.H.; Zhao, Q.Y.; Yao, S.; Zhao, L.; Zhao, C.F.; Zhu, Z.; et al. Development and application of Pigm-specific molecular markers for broad-spectrum resistance gene in rice blast. Chin. J. Rice Sci. 2020, 34, 28–36. [Google Scholar]
- Beena, R.; Kirubakaran, S.; Nithya, N.; Manickavelu, A.; Sah, R.P.; Abida, P.S.; Sreekumar, J.; Jaslam, P.M.; Rejeth, R.; Jayalekshmy, V.G.; et al. Association mapping of drought tolerance and agronomic traits in rice (Oryza sativa L.) landraces. BMC Plant Biol. 2021, 21, 484. [Google Scholar] [CrossRef] [PubMed]

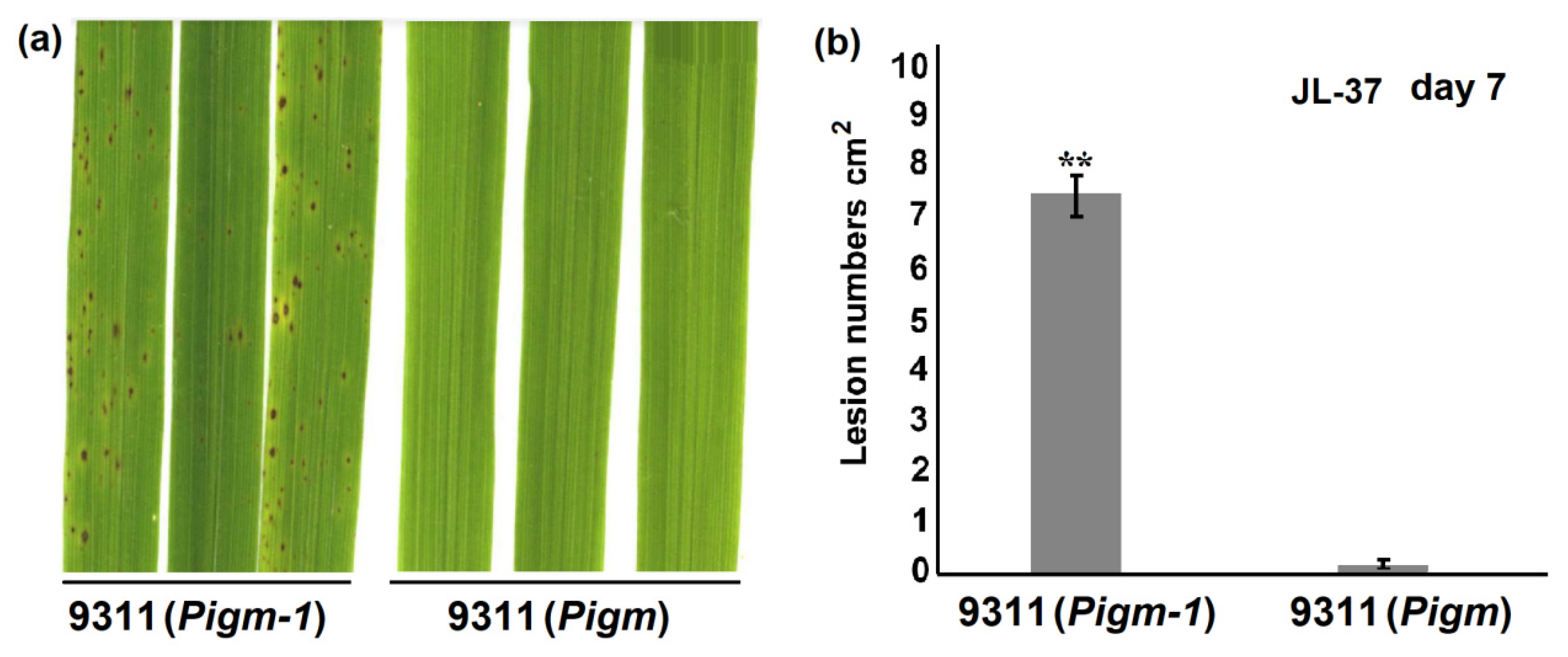


| Traits | 9311 (Pigm-1) | 9311 (Pigm) | 9311 |
|---|---|---|---|
| Plant height (cm) | 115.4 ± 2.24 * | 107.3 ± 2.14 | 106.8 ± 2.11 |
| Panicle length (cm) | 23.1 ± 0.34 * | 20.7 ± 0.31 | 19.4 ± 0.30 |
| Number of effective panicle | 7.68 ± 1.06 | 7.91 ± 1.13 | 7.84 ± 1.19 |
| Spikelets per panicle | 197.6 ± 6.46 ** | 163.7 ± 5.96 | 160.8 ± 5.16 |
| Seed setting rate (%) | 91.06 ± 2.23 | 90.15 ± 1.92 | 88.85 ± 1.62 |
| 1000-grain weight (g) | 32.13± 0.91 | 30.98± 0.81 | 31.16± 0.81 |
| Grain length (mm) | 9.75 ± 0.18 | 9.68 ± 0.21 | 9.71 ± 0.21 |
| Grain width (mm) | 3.26 ± 0.10 | 3.23 ± 0.10 | 3.24 ± 0.11 |
| Name | 9311 (Pigm-1) | 9311 (Pigm) | 9311 | Name | 9311 (Pigm-1) | 9311 (Pigm) | 9311 |
|---|---|---|---|---|---|---|---|
| JL-1 | R | R | S | SH-41 | R | R | S |
| JL-2 | R | R | S | SH-42 | R | R | S |
| JL-3 | R | R | S | SH-43 | R | R | S |
| JL-4 | R | R | S | SH-44 | R | R | S |
| JL-5 | R | R | S | SH-45 | R | R | S |
| JL-6 | R | R | S | SH-46 | R | R | S |
| JL-7 | R | R | S | SH-47 | R | R | S |
| JL-8 | R | R | S | SH-48 | R | R | S |
| JL-9 | R | R | S | SH-49 | R | R | S |
| JL-10 | R | R | S | SH-50 | R | R | S |
| JL-11 | R | R | R | SH-51 | R | R | S |
| JL-12 | R | R | R | SH-52 | R | R | S |
| JL-13 | R | R | S | SH-53 | R | R | S |
| JL-14 | R | R | S | SH-54 | R | R | S |
| JL-15 | R | R | S | SH-55 | R | R | S |
| JL-16 | R | R | S | SH-56 | R | R | S |
| JL-17 | R | R | S | SH-57 | R | R | S |
| JL-18 | R | R | S | SH-58 | R | R | S |
| JL-19 | R | R | S | SH-59 | R | R | S |
| JL-20 | R | R | S | SH-60 | R | R | S |
| JL-21 | R | R | S | SH-61 | R | R | S |
| JL-22 | R | R | S | SH-62 | R | R | S |
| JL-23 | R | R | S | SH-63 | R | R | S |
| JL-24 | R | R | S | SH-64 | R | R | S |
| JL-25 | R | R | S | SH-65 | R | R | S |
| JL-26 | R | R | S | SH-66 | R | R | S |
| JL-27 | R | R | S | SH-67 | R | R | S |
| JL-28 | R | R | S | SH-68 | R | R | R |
| JL-29 | R | R | S | SH-69 | R | R | S |
| JL-30 | R | R | S | SH-70 | R | R | S |
| JL-31 | R | R | S | SH-71 | R | R | S |
| JL-32 | R | R | S | SH-72 | R | R | S |
| JL-33 | R | R | S | SH-73 | R | R | S |
| JL-34 | R | R | S | SH-74 | R | R | S |
| JL-35 | R | R | S | SH-75 | R | R | S |
| JL-36 | R | R | S | SH-76 | R | R | S |
| JL-37 | S | R | S | SH-77 | R | R | S |
| JL-38 | R | R | S | SH-78 | R | R | S |
| JL-39 | R | R | S | SH-79 | R | R | S |
| JL-40 | R | R | S | SH-80 | R | R | S |
| JL-41 | R | R | S | SH-81 | R | R | S |
| JL-42 | R | R | S | SH-82 | R | R | S |
| JL-43 | R | R | S | SH-83 | R | R | S |
| JL-44 | R | R | S | SH-84 | R | R | S |
| JL-45 | R | R | S | SH-85 | R | R | S |
| JL-46 | R | R | S | SH-86 | R | R | S |
| JL-47 | R | R | S | SH-87 | R | R | S |
| JL-48 | R | R | S | SH-88 | R | R | S |
| JL-49 | R | R | S | SH-89 | R | R | S |
| JL-50 | R | R | S | JY-1 | R | R | S |
| JL-51 | R | R | S | JY-2 | R | R | S |
| JL-52 | R | R | S | JY-3 | R | R | S |
| JL-53 | R | R | S | JY-4 | R | R | S |
| JL-54 | R | R | S | JY-5 | R | R | S |
| JL-55 | R | R | S | JY-6 | R | R | S |
| JL-56 | R | R | S | JY-7 | R | R | R |
| JL-57 | R | R | S | JY-8 | R | R | S |
| JL-58 | R | R | S | JY-9 | R | R | S |
| JL-59 | R | R | S | JY-10 | R | R | S |
| JL-60 | R | R | S | JY-11 | R | R | S |
| SH-1 | R | R | S | JY-12 | R | R | S |
| SH-2 | R | R | S | JY-13 | R | R | S |
| SH-3 | R | R | S | JY-14 | R | R | S |
| SH-4 | R | R | S | JY-15 | R | R | S |
| SH-5 | R | R | S | JY-16 | R | R | S |
| SH-6 | R | R | S | JY-17 | R | R | S |
| SH-7 | R | R | S | JY-18 | R | R | S |
| SH-8 | R | R | S | JY-19 | R | R | S |
| SH-9 | R | R | S | JY-20 | R | R | S |
| SH-10 | R | R | S | JY-21 | R | R | S |
| SH-11 | R | R | S | JY-22 | R | R | S |
| SH-12 | R | R | S | JY-23 | R | R | S |
| SH-13 | R | R | S | JY-24 | R | R | S |
| SH-14 | R | R | S | JY-25 | R | R | S |
| SH-15 | R | R | S | JY-26 | R | R | S |
| SH-16 | R | R | S | JY-27 | R | R | S |
| SH-17 | R | R | R | JY-28 | R | R | S |
| SH-18 | R | R | S | JY-29 | R | R | S |
| SH-19 | R | R | S | JY-30 | R | R | S |
| SH-20 | R | R | S | JY-31 | R | R | S |
| SH-21 | R | R | S | JY-32 | R | R | S |
| SH-22 | R | R | S | JY-33 | R | R | S |
| SH-23 | R | R | S | JY-34 | R | R | S |
| SH-24 | R | R | S | JY-35 | R | R | S |
| SH-25 | R | R | S | JY-36 | R | R | S |
| SH-26 | R | R | R | JY-37 | R | R | S |
| SH-27 | R | R | S | JY-38 | R | R | S |
| SH-28 | R | R | S | JY-39 | R | R | S |
| SH-29 | R | R | S | JY-40 | R | R | S |
| SH-30 | R | R | S | JY-41 | R | R | S |
| SH-31 | R | R | S | JY-42 | R | R | S |
| SH-32 | R | R | S | JY-43 | R | R | S |
| SH-33 | R | R | S | JY-44 | R | R | S |
| SH-34 | R | R | S | JY-45 | R | R | S |
| SH-35 | R | R | S | JY-46 | R | R | S |
| SH-36 | R | R | S | ||||
| SH-37 | R | R | S | ||||
| SH-38 | R | R | S | ||||
| SH-39 | R | R | S | ||||
| SH-40 | R | R | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; He, N.; Cheng, Z.; Lin, S.; Huang, F.; Wang, W.; Li, Q.Q.; Yang, D. Resistance Spectrum Analysis and Breeding Utilization of Rice Blast Resistance Gene Pigm-1. Plants 2025, 14, 535. https://doi.org/10.3390/plants14040535
Jin Y, He N, Cheng Z, Lin S, Huang F, Wang W, Li QQ, Yang D. Resistance Spectrum Analysis and Breeding Utilization of Rice Blast Resistance Gene Pigm-1. Plants. 2025; 14(4):535. https://doi.org/10.3390/plants14040535
Chicago/Turabian StyleJin, Yidan, Niqing He, Zhaoping Cheng, Shaojun Lin, Fenghuang Huang, Wenxiao Wang, Qingshun Q. Li, and Dewei Yang. 2025. "Resistance Spectrum Analysis and Breeding Utilization of Rice Blast Resistance Gene Pigm-1" Plants 14, no. 4: 535. https://doi.org/10.3390/plants14040535
APA StyleJin, Y., He, N., Cheng, Z., Lin, S., Huang, F., Wang, W., Li, Q. Q., & Yang, D. (2025). Resistance Spectrum Analysis and Breeding Utilization of Rice Blast Resistance Gene Pigm-1. Plants, 14(4), 535. https://doi.org/10.3390/plants14040535






