Agave macroacantha Transcriptome Reveals Candidate CNGC Genes Responsive to Cold Stress in Agave
Abstract
1. Introduction
2. Results
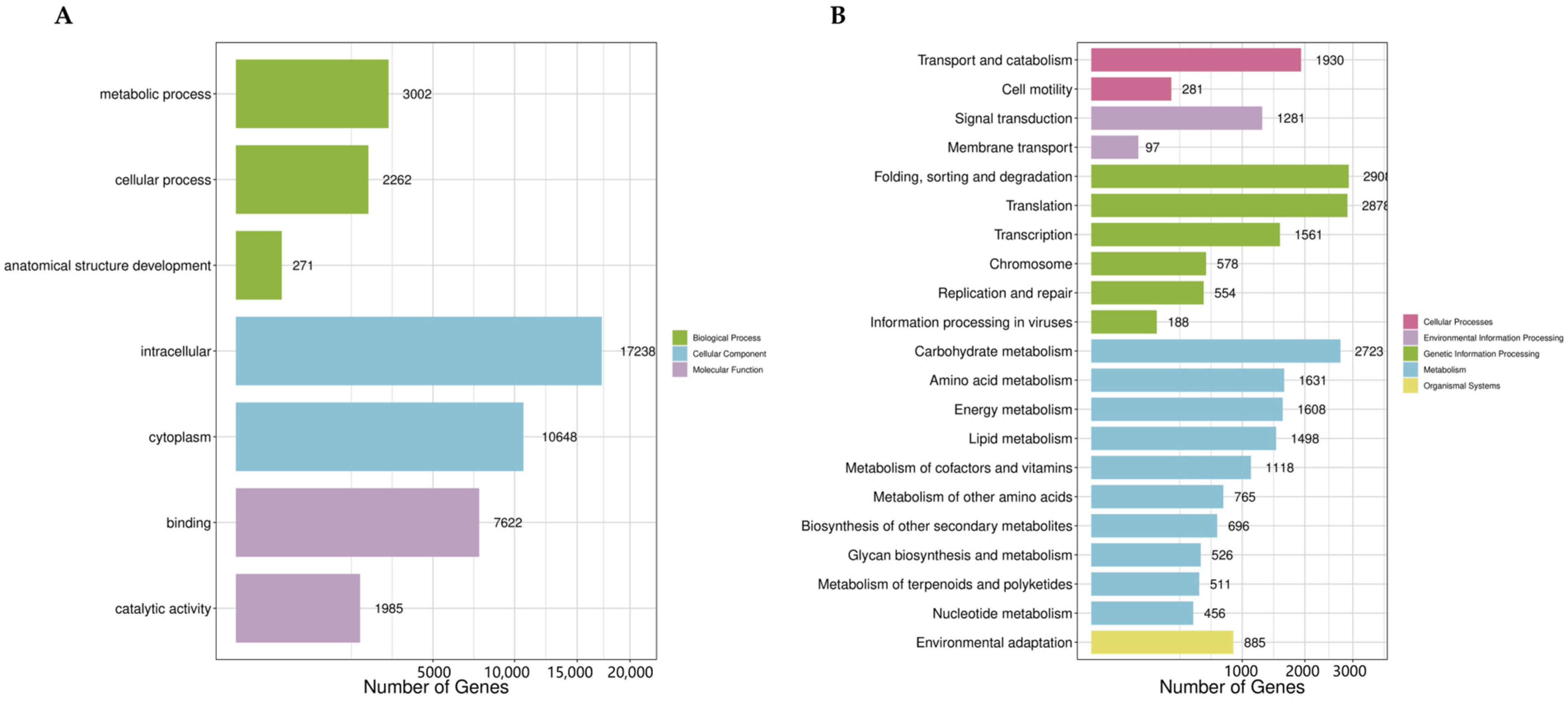
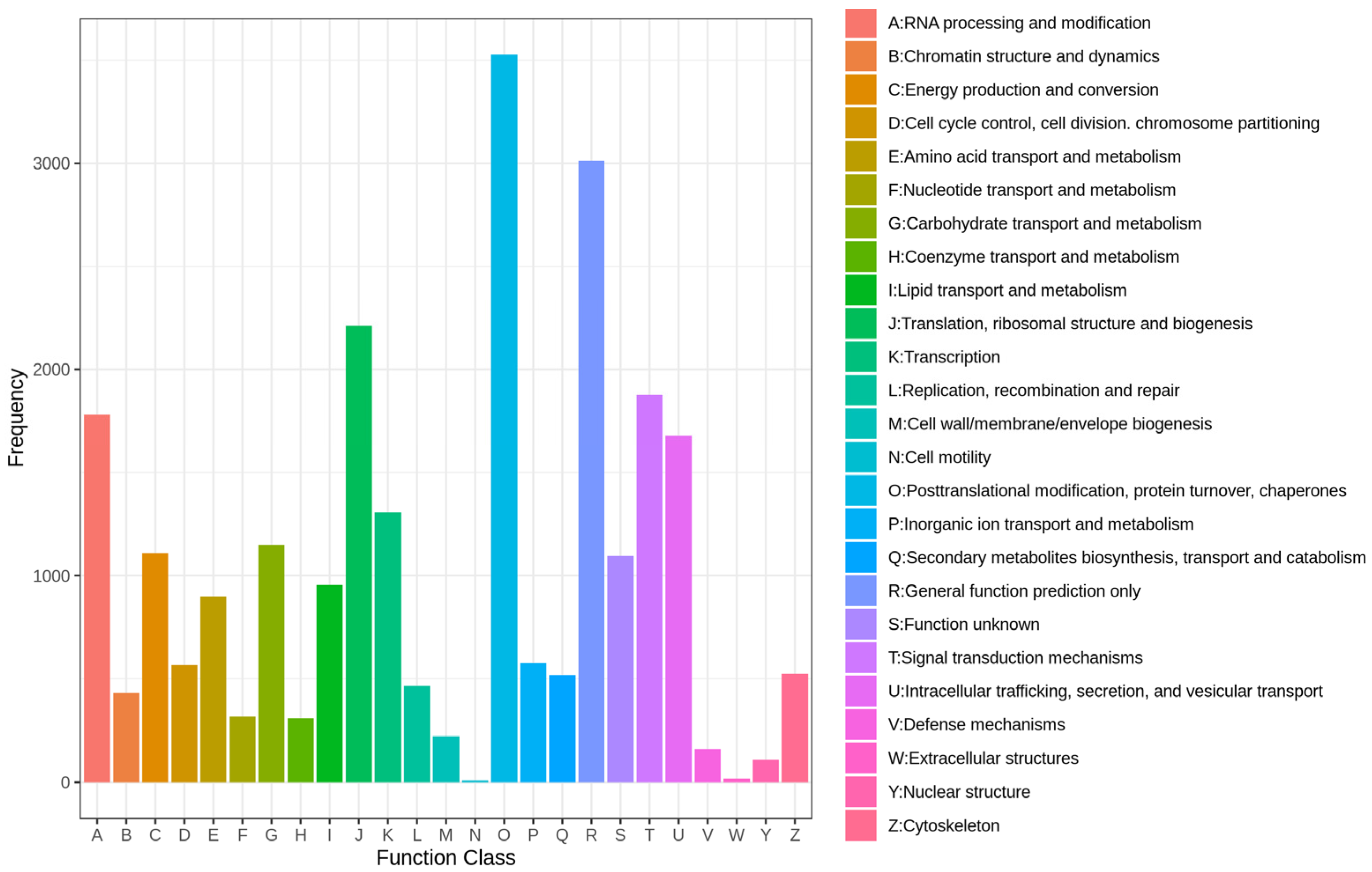
2.1. Tanscriptome Analysis of A. macroacantha
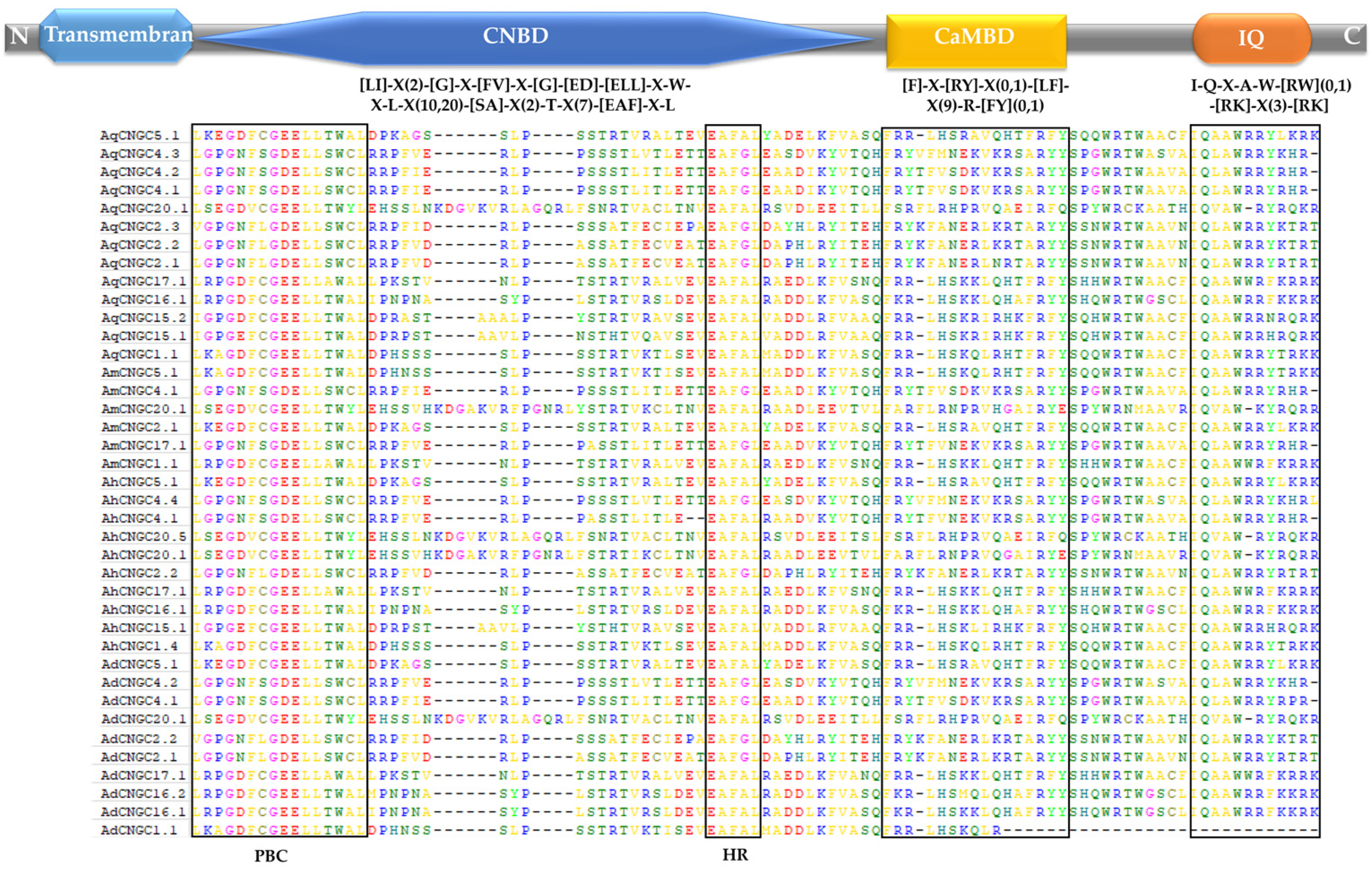
2.2. Identification and Phylogenetic Analysis of CNGC Genes in Agave Species
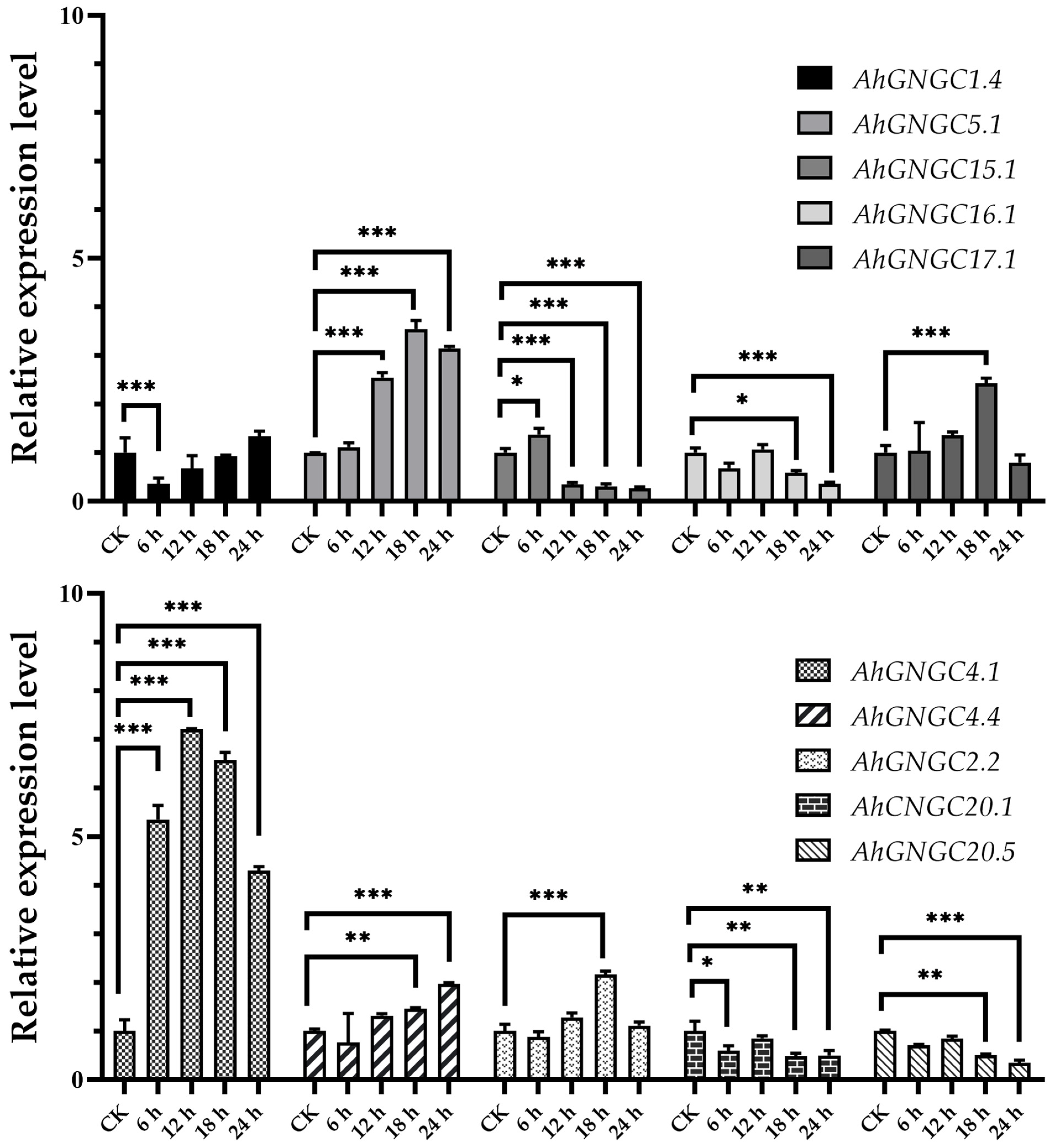
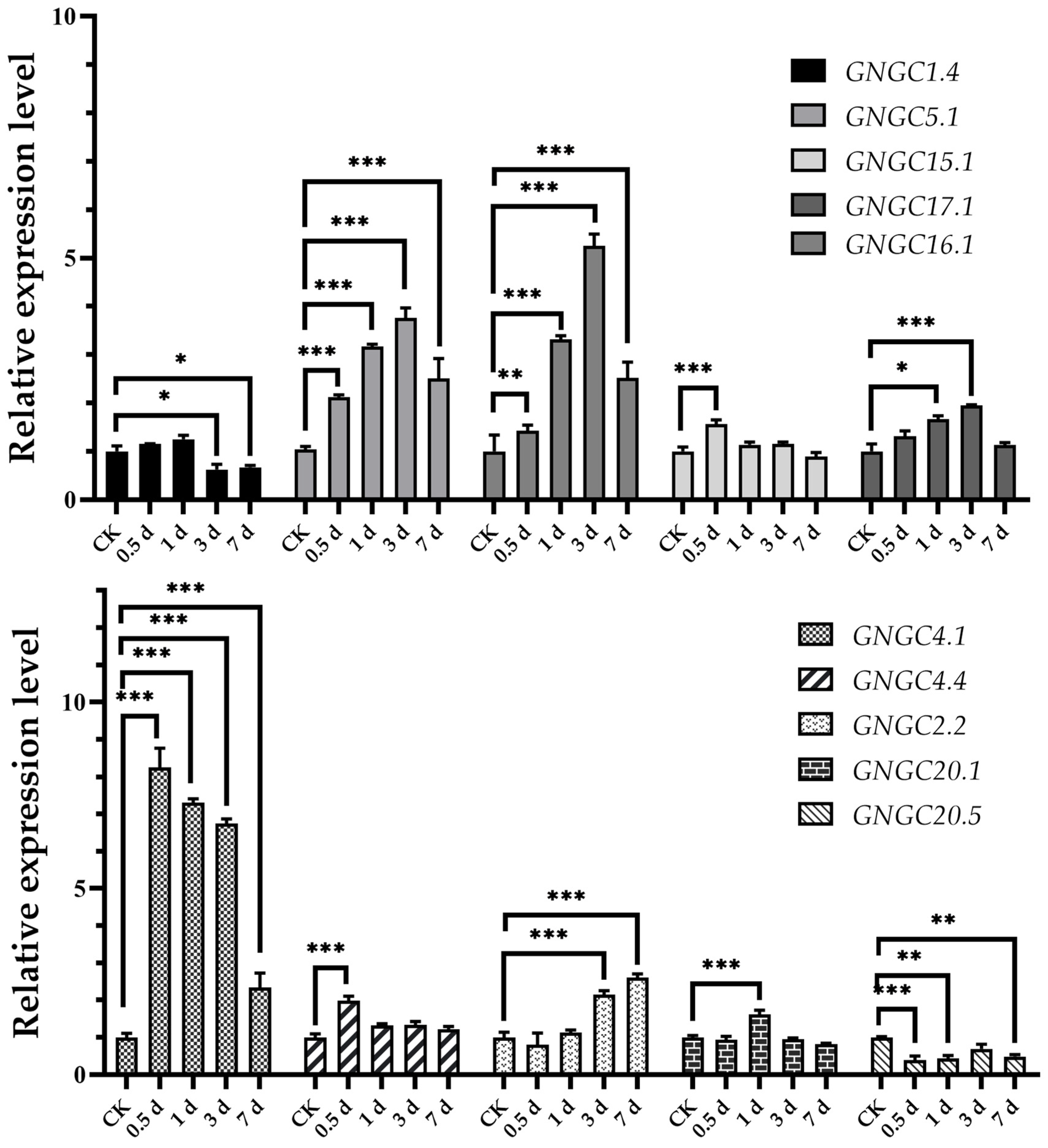
2.3. Expression Profiles of AhCNGCs
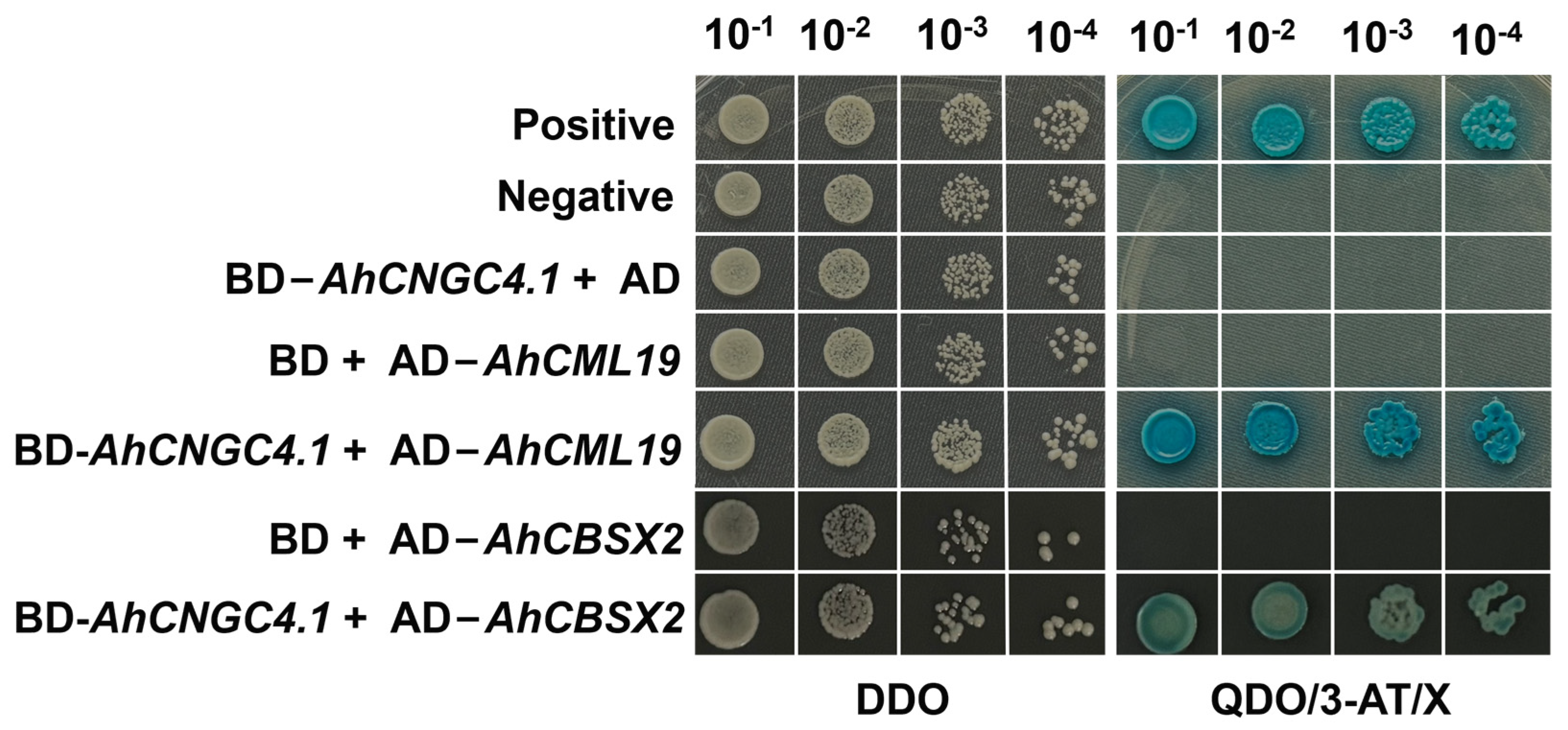
2.4. Yeast Two-Hybrid Screening for AhCNGC4.1 Interacting Proteins
3. Discussion
3.1. Characterization of A. macroacantha Transcriptome
3.2. Candidate CNGC Genes in Development of Agave
3.3. CNGC Genes in Abiotic Resistance
3.4. CNGC Genes and Interaction Protein Screening
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. Transcriptome Sequencing, De Novo Assembly, and Annotation
4.3. Identification and Phylogeny of CNGC Genes
4.4. Quantitative PCR Experiments
4.5. Yeast Two-Hybrid Screening (Y2H)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alducin-Martínez, C.; Ruiz Mondragón, K.Y.; Jiménez-Barrón, O.; Aguirre-Planter, E.; Gasca-Pineda, J.; Eguiarte, L.E.; Medellin, R.A. Uses, Knowledge and Extinction Risk Faced by Agave Species in Mexico. Plants 2022, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Chávez, J.; Villamiel, M.; Santos-Zea, L.; Ramírez-Jiménez, A.K. Agave By-Products: An Overview of Their Nutraceutical Value, Current Applications, and Processing Methods. Polysaccharides 2021, 2, 720–743. [Google Scholar] [CrossRef]
- Xu, B.; Tan, S.; Qin, X.; Huang, X.; Xi, J.; Chen, H.; Qin, J.; Chen, T.; Yi, K. The complete chloroplast genome of Agave amaniensis (Asparagales:Asparagaceae:Agavoideae). Mitochondrial DNA Part B 2022, 7, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Simonet, A.M.; Durán, A.G.; Macías, F.A. Dereplication of Bioactive Agave saponin Fractions: The Hidden Saponins. J. Agric. Food Chem. 2024, 72, 13740–13756. [Google Scholar] [CrossRef]
- Sarwar, M.B.; Ahmad, Z.; Rashid, B.; Hassan, S.; Gregersen, P.L.; Leyva, M.D.; Nagy, I.; Asp, T.; Husnain, T. De novo assembly of Agave sisalana transcriptome in response to drought stress provides insight into the tolerance mechanisms. Sci. Rep. 2019, 9, 396. [Google Scholar] [CrossRef]
- Betancourt Yánez, P.; Mujica, Y. Efecto de la fertilización cálcica y el sistema de siembra en sisal en un sector semiárido del estado Lara. Agron. Trop. 2013, 63, 85–91. [Google Scholar]
- Demidchik, V.; Shabala, S.; Isayenkov, S.; Cuin, T.A.; Pottosin, I. Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 2018, 220, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Gao, H.H.; Li, S.X.; Han, Z.; Li, B. Calcium signaling networks mediate nitrate sensing and responses in Arabidopsis. Plant Signal. Behav. 2021, 16, 7. [Google Scholar] [CrossRef]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium Signals: The Lead Currency of Plant Information Processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Chen, M.; Ni, L.; Chen, J.; Sun, M.; Qin, C.; Zhang, G.; Zhang, A.; Jiang, M. Rice calcium/calmodulin-dependent protein kinase directly phosphorylates a mitogen-activated protein kinase kinase to regulate abscisic acid responses. Plant Cell 2021, 33, 1790–1812. [Google Scholar] [CrossRef]
- Zhang, X.P.; Ma, C.X.; Sun, L.R.; Hao, F.S. Roles and mechanisms of Ca2+ in regulating primary root growth of plants. Plant Signal. Behav. 2020, 15, 1748283. [Google Scholar] [CrossRef]
- Liu, K.-h.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Chung, H.S.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311. [Google Scholar] [CrossRef] [PubMed]
- Sago, Y. Effects of Light Intensity and Growth Rate on Tipburn Development and Leaf Calcium Concentration in Butterhead Lettuce. Hortscience 2016, 51, 1087–1091. [Google Scholar] [CrossRef]
- Xu, G.; Moeder, W.; Yoshioka, K.; Shan, L. A tale of many families: Calcium channels in plant immunity. Plant Cell 2022, 34, 1551–1567. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Steinhorst, L.; Kudla, J. How plants perceive salt. Nature 2019, 572, 318–320. [Google Scholar] [CrossRef]
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The Arabidopsis Calcium-Dependent Protein Kinases (CDPKs) and Their Roles in Plant Growth Regulation and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 1900. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Roessner, N.; Hoang, M.T.T.; Alejandro, S.; Peiter, E. Transport, functions, and interaction of calcium and manganese in plant organellar compartments. Plant Physiol. 2021, 187, 1940–1972. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, U.M.; Manohar, M.; Gaur, V.S. Calcium transport from source to sink: Understanding the mechanism(s) of acquisition, translocation, and accumulation for crop biofortification. Acta Physiol. Plant. 2015, 37, 1722. [Google Scholar] [CrossRef]
- Bai, R.; Bai, C.; Han, X.; Liu, Y.; Yong, J.W.H. The significance of calcium-sensing receptor in sustaining photosynthesis and ameliorating stress responses in plants. Front. Plant Sci. 2022, 13, 1019505. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.M.; Ludwig, A.; Furch, A.C.U.; Mithoefer, A.; Scholz, S.; Reichelt, M.; Oelmueller, R. The Beneficial Root-Colonizing Fungus Mortierella hyalina Promotes the Aerial Growth of Arabidopsis and Activates Calcium-Dependent Responses That Restrict Alternaria brassicae-Induced Disease Development in Roots. Mol. Plant Microbe Interact. 2019, 32, 351–363. [Google Scholar] [CrossRef]
- Gfeller, A.; Baerenfaller, K.; Loscos, J.; Chetelat, A.; Baginsky, S.; Farmer, E.E. Jasmonate Controls Polypeptide Patterning in Undamaged Tissue in Wounded Arabidopsis Leaves. Plant Physiol. 2011, 156, 1797–1807. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Choi, W.-G.; Gilroy, S.; Morris, R.J. A ROS-Assisted Calcium Wave Dependent on the AtRBOHD NADPH Oxidase and TPC1 Cation Channel Propagates the Systemic Response to Salt Stress. Plant Physiol. 2016, 171, 1771–1784. [Google Scholar] [CrossRef]
- Wu, X.; Wan, Y.; Bian, Y.; Ren, Y.; Xu, X.; Zhou, F.; Ding, H. A critical review on plant annexin: Structure, function, and mechanism. Plant Physiol. Biochem. 2022, 190, 81–89. [Google Scholar] [CrossRef]
- Nakayama, M.; Tsunooka-Ohta, M.; Kojima-Aikawa, K. Annexin Lectins: Ca2+-Dependent Heparin-Binding Activity, Phosphatidylserine-Binding Activity, and Anticoagulant Activity. Methods Mol. Biol. 2020, 2132, 661–668. [Google Scholar] [CrossRef]
- Duszyn, M.; Swiezawska, B.; Szmidt-Jaworska, A.; Jaworski, K. Cyclic nucleotide gated channels (CNGCs) in plant signalling-Current knowledge and perspectives. J. Plant Physiol. 2019, 241, 153035. [Google Scholar] [CrossRef]
- Jarratt-Barnham, E.; Wang, L.; Ning, Y.; Davies, J.M. The Complex Story of Plant Cyclic Nucleotide-Gated Channels. Int. J. Mol. Sci. 2021, 22, 874. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Moeder, W.; Yoshioka, K. Opening the Gates: Insights into Cyclic Nucleotide-Gated Channel-Mediated Signaling. Trends Plant Sci. 2016, 21, 903–906. [Google Scholar] [CrossRef]
- Saponaro, A.; Pauleta, S.R.; Cantini, F.; Matzapetakis, M.; Hammann, C.; Donadoni, C.; Hu, L.; Thiel, G.; Banci, L.; Santoro, B.; et al. Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function. Proc. Natl. Acad. Sci. USA 2014, 111, 14577–14582. [Google Scholar] [CrossRef]
- Puljung, M.C.; DeBerg, H.A.; Zagotta, W.N.; Stoll, S. Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proc. Natl. Acad. Sci. USA 2014, 111, 9816–9821. [Google Scholar] [CrossRef]
- Nawaz, Z.; Kakar, K.U.; Saand, M.A.; Shu, Q.-Y. Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genom. 2014, 15, 853. [Google Scholar] [CrossRef] [PubMed]
- James, Z.M.; Zagotta, W.N. Structural insights into the mechanisms of CNBD channel function. J. Gen. Physiol. 2018, 150, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Zagotta, W.N.; Olivier, N.B.; Black, K.D.; Young, E.C.; Olson, R.; Gouaux, E. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 2003, 425, 200–205. [Google Scholar] [CrossRef]
- Dai, G.; Aman, T.K.; DiMaio, F.; Zagotta, W.N. Electromechanical coupling mechanism for activation and inactivation of an HCN channel. Nat. Commun. 2021, 12, 2802. [Google Scholar] [CrossRef] [PubMed]
- Beck, E.M.; Parnell, E.; Cowley, A.; Porter, A.; Gillespie, J.; Robinson, J.; Robinson, L.; Pannifer, A.D.; Hamon, V.; Jones, P.; et al. Identification of A Novel Class of Benzofuran Oxoacetic Acid-Derived Ligands that Selectively Activate Cellular EPAC1. Cells 2019, 8, 1425. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; DeFalco, T.A.; Karia, P.; Snedden, W.A.; Moeder, W.; Yoshioka, K.; Dietrich, P. Calmodulin as a Ca2+-Sensing Subunit of Arabidopsis Cyclic Nucleotide-Gated Channel Complexes. Plant Cell Physiol. 2017, 58, 1208–1221. [Google Scholar] [CrossRef] [PubMed]
- Tunc-Ozdemir, M.; Tang, C.; Ishka, M.R.; Brown, E.; Groves, N.R.; Myers, C.T.; Rato, C.; Poulsen, L.R.; McDowell, S.; Miller, G.; et al. A Cyclic Nucleotide-Gated Channel (CNGC16) in Pollen Is Critical for Stress Tolerance in Pollen Reproductive Development. Plant Physiol. 2013, 161, 1010–1020. [Google Scholar] [CrossRef]
- Kugler, A.; Kohler, B.; Palme, K.; Wolff, P.; Dietrich, P. Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol. 2009, 9, 140. [Google Scholar] [CrossRef]
- Cui, Y.; Lu, S.; Li, Z.; Cheng, J.; Hu, P.; Zhu, T.; Wang, X.; Jin, M.; Wang, X.; Li, L. CYCLIC NUCLEOTIDE-GATED ION CHANNELs 14 and 16 Promote Tolerance to Heat and Chilling in Rice. Plant Physiol. 2020, 183, 1794–1808. [Google Scholar] [CrossRef]
- Shi, J.; Du, X. Transcriptome analysis reveals the regulation of cyclic nucleotide-gated ion channels in response to exogenous abscisic acid and calcium treatment under drought stress in tomato. Front. Genet. 2023, 14, 1139087. [Google Scholar] [CrossRef]
- Guo, J.; Islam, M.A.; Lin, H.; Ji, C.; Duan, Y.; Liu, P.; Zeng, Q.; Day, B.; Kang, Z.; Guo, J. Genome-Wide Identification of Cyclic Nucleotide-Gated Ion Channel Gene Family in Wheat and Functional Analyses of TaCNGC14 and TaCNGC16. Front. Plant Sci. 2018, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Zia, K.; Rao, M.J.; Sadaqat, M.; Azeem, F.; Fatima, K.; Ul Qamar, M.T.; Alshammari, A.; Alharbi, M. Pangenome-wide analysis of cyclic nucleotide-gated channel (CNGC) gene family in Citrus spp. Revealed their intraspecies diversity and potential roles in abiotic stress tolerance. Front. Genet. 2022, 13, 27. [Google Scholar] [CrossRef]
- Lu, Z.Y.; Yin, G.; Chai, M.; Sun, L.; Wei, H.L.; Chen, J.; Yang, Y.F.; Fu, X.K.; Li, S.Y. Systematic analysis of CNGCs in cotton and the positive role of GhCNGC32 and GhCNGC35 in salt tolerance. BMC Genom. 2022, 23, 13. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Liu, Z.; Dai, L.; Zhang, M.; Wang, L.; Zhao, J.; Liu, M. Genome-wide identification of CNGC genes in Chinese jujube (Ziziphus jujuba Mill.) and ZjCNGC2 mediated signalling cascades in response to cold stress. BMC Genom. 2020, 21, 191. [Google Scholar] [CrossRef]
- Kakar, K.U.; Nawaz, Z.; Kakar, K.; Ali, E.; Almoneafy, A.A.; Ullah, R.; Ren, X.L.; Shu, Q.Y. Comprehensive genomic analysis of the CNGC gene family in Brassica oleracea: Novel insights into synteny, structures, and transcript profiles. BMC Genom. 2017, 18, 18. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Guo, Y.; Li, J.; Liu, L.; Liu, J.; Yuan, L.; Wei, C.; Ma, J.; Zhang, Y.; Ahammed, G.J.; et al. Cyclic nucleotide-gated ion channel 20 regulates melatonin-induced calcium signaling and cold tolerance in watermelon. Plant Physiol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Peng, L.; Chen, M.; Ye, X.; Li, Y.; Li, Z.; Wen, Q.; Zhu, H. Genome-Wide Identification of the Cyclic Nucleotide-Gated Ion Channel Gene Family and Expression Profiles Under Low-Temperature Stress in Luffa cylindrica L. Int. J. Mol. Sci. 2024, 25, 11330. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Ren, Y.L.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.B.; Zhu, S.S.; Wan, H.; Yang, Y.; et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant. 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Mallano, A.I.; Li, F.; Li, P.; Wu, Q.; Wang, Y.; Li, Y.; Ahmad, N.; Tong, W.; Li, Y.; et al. Characterization of CsWRKY29 and CsWRKY37 transcription factors and their functional roles in cold tolerance of tea plant. Beverage Plant Res. 2022, 2, 1–13. [Google Scholar] [CrossRef]
- Bhat, K.A.; Mahajan, R.; Pakhtoon, M.M.; Urwat, U.; Bashir, Z.; Shah, A.A.; Agrawal, A.; Bhat, B.; Sofi, P.A.; Masi, A.; et al. Low Temperature Stress Tolerance: An Insight into the Omics Approaches for Legume Crops. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xin, H.; Fan, P.; Zhang, J.; Liu, Y.; Dong, Y.; Wang, Z.; Yang, Y.; Zhang, Q.; Ming, R.; et al. The genome of Shanputao (Vitis amurensis) provides a new insight into cold tolerance of grapevine. Plant J. 2021, 105, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genom. 2019, 20, 227. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Xiao, M.; Xi, J.G.; He, C.P.; Zheng, J.L.; Chen, H.L.; Gao, J.M.; Zhang, S.Q.; Wu, W.H.; Liang, Y.Q.; et al. De Novo Transcriptome Assembly of Agave H11648 by Illumina Sequencing and Identification of Cellulose Synthase Genes in Agave Species. Genes 2019, 10, 103. [Google Scholar] [CrossRef]
- Wang, X.; Hu, X.; Lin, C.; Liu, Q.; Li, Y.; Du, D.; Mkapa, D.; Zhang, W.; Huang, X.; Yi, K. Agave schidigera Transcriptome Reveals Stress-Responsive Phenylalanine ammonia-lyase Genes in Agave. Agronomy 2024, 14, 2520. [Google Scholar] [CrossRef]
- Moeder, W.; Phan, V.; Yoshioka, K. Ca2+ to the rescue—Ca2+ channels and signaling in plant immunity. Plant Sci. 2019, 279, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hameed, A.; Liao, W.; Prasad, K.V.S.K.; Reddy, A.S.N. CAMTAs, a family of calmodulin-binding transcription factors, are versatile regulators of biotic and abiotic stress responses in plants. Crit. Rev. Plant Sci. 2024, 43, 171–210. [Google Scholar] [CrossRef]
- Liu, X.; Li, W.; Li, X.; Chen, Y. The functional roles of calcium-dependent protein kinases in plant growth and stress response. Chin. Sci. Bull. 2023, 69, 1082–1095. [Google Scholar] [CrossRef]
- Lu, S.; Zhu, T.Q.; Luo, L.L.; Ouyang, N.N.; Hua, J.; Zou, B.H. Divergent Roles of CNGC2 and CNGC4 in the Regulation of Disease Resistance, Plant Growth and Heat Tolerance in Arabidopsis. Agronomy 2022, 12, 2176. [Google Scholar] [CrossRef]
- Peng, Y.; Ming, Y.; Jiang, B.; Zhang, X.; Fu, D.; Lin, Q.; Zhang, X.; Wang, Y.; Shi, Y.; Gong, Z.; et al. Differential phosphorylation of Ca2+-permeable channel CNGC20 modulates calcium-mediated freezing tolerance in Arabidopsis. Plant Cell 2024, 36, 4356–4371. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Yang, J.; Yang, X.; Chen, S.; Xie, Z.; Zhang, M.; Huang, Y.; Zhang, J.; Huang, X. Genome-Wide Analysis and Expression of Cyclic Nucleotide–Gated Ion Channel (CNGC) Family Genes under Cold Stress in Mango (Mangifera indica). Plants 2023, 12, 592. [Google Scholar] [CrossRef]
- Jiang, Z.; Du, L.; Shen, L.; He, J.; Xia, X.; Zhang, L.; Yang, X. Genome-Wide Exploration and Expression Analysis of the CNGC Gene Family in Eggplant (Solanum melongena L.) under Cold Stress, with Functional Characterization of SmCNGC1a. Int. J. Mol. Sci. 2023, 24, 13049. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, N.; Sacks, M.; Snir, P.; Rosenberg, R. Prevention of Flower Abortion and Leaf Damages in Anemone coronaria by Ca Supplements. Isr. J. Plant Sci. 2018, 65, 97–108. [Google Scholar] [CrossRef]
- Heidari, S.; Azizi, M.; Soltani, F.; Hadian, J. Foliar application of Ca(NO3)2 and KNO3 affects growth, essential oil content, and oil composition of French tarragon. Ind. Crop. Prod. 2014, 62, 526–532. [Google Scholar] [CrossRef]
- Cheng, M.; Guo, Y.; Liu, Q.; Nan, S.; Xue, Y.; Wei, C.; Zhang, Y.; Luan, F.; Zhang, X.; Li, H. H2O2 and Ca2+ Signaling Crosstalk Counteracts ABA to Induce Seed Germination. Antioxidants 2022, 11, 1594. [Google Scholar] [CrossRef]
- Chang, J.; Zhang, R.; Fu, Z.; Wang, Y.; Lei, J.; Cheng, J.; Ren, C.; Xu, K.; Gu, C.; Song, Y.; et al. OsCNGC7 modulates calcium dynamics and accelerates leaf senescence in rice. Plant Physiol. Biochem. 2024, 216, 109193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cui, Y.; An, L.; Li, J.; Yao, Y.; Bai, Y.; Li, X.; Yao, X.; Wu, K. Genome-wide identification of the CNGC gene family and negative regulation of drought tolerance by HvCNGC3 and HvCNGC16 in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2024, 210, 108593. [Google Scholar] [CrossRef]
- Lee, S.; Lee, H.Y.; Kang, H.J.; Seo, Y.E.; Lee, J.H.; Choi, D. Oomycete effector AVRblb2 targets cyclic nucleotide-gated channels through calcium sensors to suppress pattern-triggered immunity. New Phytol. 2023, 241, 1277–1291. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.; Park, J.H.; Wi, S.D.; Kang, C.H.; Chi, Y.H.; Chae, H.B.; Paeng, S.K.; Ji, M.G.; Kim, W.-Y.; Kim, M.G.; et al. Author Correction: Redox-dependent structural switch and CBF activation confer freezing tolerance in plants. Nat. Plants 2022, 8, 1493. [Google Scholar] [CrossRef]
- Gross, S.M.; Martin, J.A.; Simpson, J.; Abraham-Juarez, M.J.; Wang, Z.; Visel, A. De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genom. 2013, 14, 563. [Google Scholar] [CrossRef]


| Gene ID | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| AhCNGC1.4 | CTTCAACCAGTACGCCCCTT | ACCAAAAGTTGACCGCGACA |
| AhCNGC2.2 | ATGCCCCATCGACACTCAAA | CGTCTCATGTAGCTGGTGGG |
| AhCNGC4.1 | TTCGGGACTGGTTTGTGACC | AGCCGTTGGAGTTAAGGGTG |
| AhCNGC4.4 | TGAACCATCTCGCACTCGTC | CACAGGTGGCCTCATACTCG |
| AhCNGC5.1 | CCTCTTCTTCCCTTCGCCTG | CCAGTCAGTTCAGACGGCTT |
| AhCNGC15.1 | CCATAGGTTTCTCGCGTCGT | CCATAGGTTTCTCGCGTCGT |
| AhCNGC16.1 | TGATGGAAACTGCATGGGCT | TGATGGAAACTGCATGGGCT |
| AhCNGC17.1 | CCCTACATCCCCTGGACTCA | TGGCGAGGGTTTGACTTGTT |
| AhCNGC20.1 | AATCCTGCCAATGAGGCGTT | TATTGGCTGGGCATGTCGTT |
| AhCNGC20.5 | GCTCTTCATCTTACCCCGCA | GCCAACGAAGGACAGCTACT |
| AhPP2A | CCTCCTCCTCCTTCGGTTTG | GCCATGAATGTCACCGCAGA |
| Primers ID | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| AhCNGC4.1-BD | AACCGCTCTTCCGATCTGATGGCC- ACCGATCACT | CGCTAACTAGTGTCGACTCACAT- TAAGAATTCA |
| AhCML19-AD | GCCATGGAGGCCAGTGAATTCATGATG- CTCACAGAGA | ATTCATCTGCAGCTCGAGCTCTCAGA- GCATCATGACC |
| AhCBSX3-AD | GCCATGGAGGCCAGTGAATTCATGCA AGGTGCAATTC | ATTCATCTGCAGCTCGAGCTCCTAGT- AACCGCCTTGT |
| AD | TAATACGACTCACTATAGGG | AGATGGTGCACGATGCACAG |
| BD | TAATACGACTCACTATAGGG | TAAGAGTCACTTTAAAATTTGTATAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Hu, X.; Mkapa, D.S.; Xie, L.; Guo, P.; Tan, S.; Zhang, W.; Chen, H.; Huang, X.; Yi, K. Agave macroacantha Transcriptome Reveals Candidate CNGC Genes Responsive to Cold Stress in Agave. Plants 2025, 14, 513. https://doi.org/10.3390/plants14040513
Li Y, Hu X, Mkapa DS, Xie L, Guo P, Tan S, Zhang W, Chen H, Huang X, Yi K. Agave macroacantha Transcriptome Reveals Candidate CNGC Genes Responsive to Cold Stress in Agave. Plants. 2025; 14(4):513. https://doi.org/10.3390/plants14040513
Chicago/Turabian StyleLi, Yubo, Xiaoli Hu, Dietram Samson Mkapa, Li Xie, Pingan Guo, Shibei Tan, Weiyi Zhang, Helong Chen, Xing Huang, and Kexian Yi. 2025. "Agave macroacantha Transcriptome Reveals Candidate CNGC Genes Responsive to Cold Stress in Agave" Plants 14, no. 4: 513. https://doi.org/10.3390/plants14040513
APA StyleLi, Y., Hu, X., Mkapa, D. S., Xie, L., Guo, P., Tan, S., Zhang, W., Chen, H., Huang, X., & Yi, K. (2025). Agave macroacantha Transcriptome Reveals Candidate CNGC Genes Responsive to Cold Stress in Agave. Plants, 14(4), 513. https://doi.org/10.3390/plants14040513







