Comprehensive Omics Analysis Reveals Cold-Induced Metabolic Reprogramming and Alternative Splicing in Dendrobium officinale
Abstract
1. Introduction
2. Results
2.1. The Core Pathway Involved in the Cold Response of D. officinale
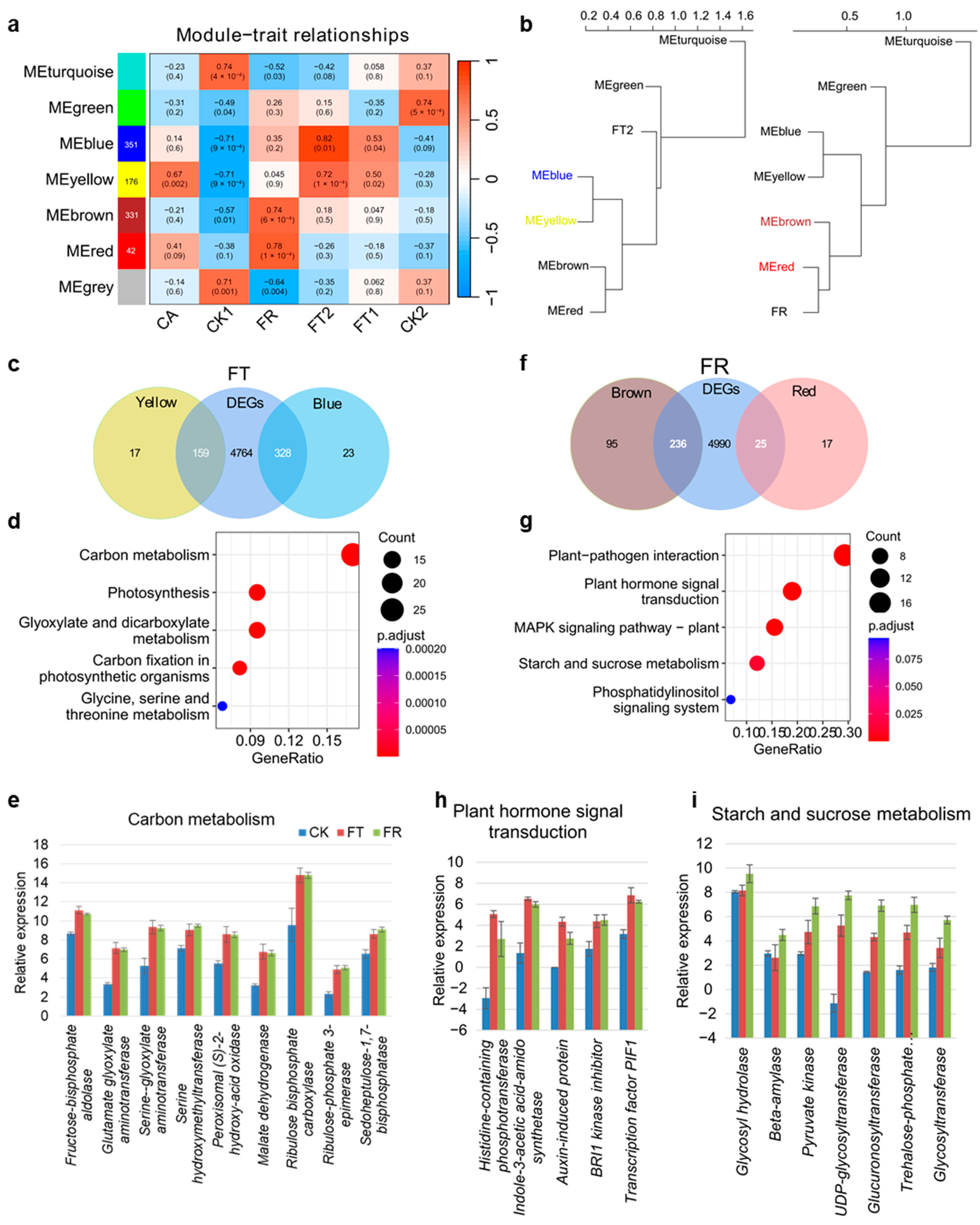
2.2. Temperature-Specific Response in D. officinale
2.3. Metabolite-Specific Accumulation Under Cold Treatment in D. officinale
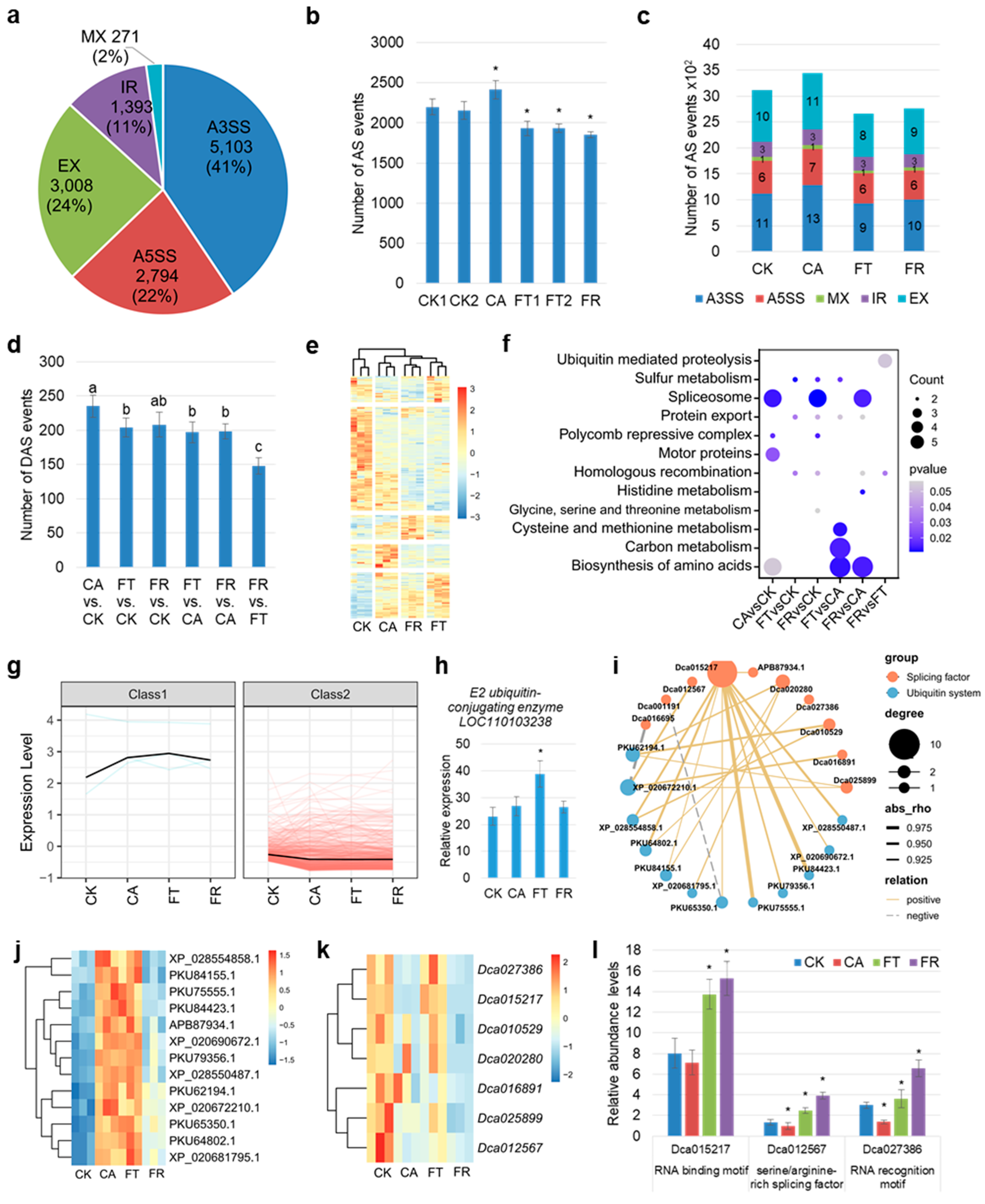
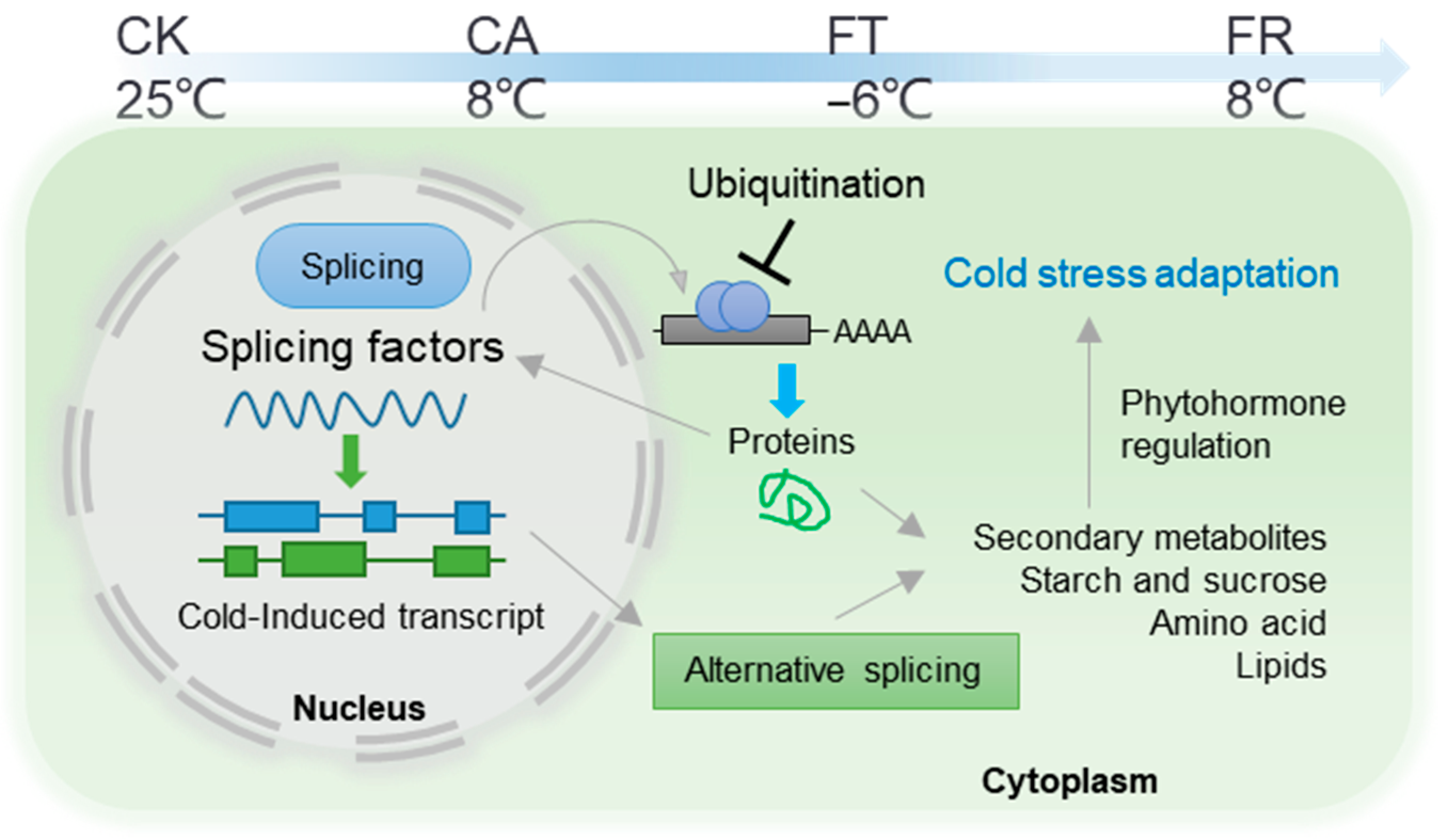
2.4. Identification of Key Splicing Factors in Response to Cold in D. officinale
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatments
4.2. Multi-Omics Analysis
4.3. Alternative Splicing Analysis
4.4. Real-Time Quantitative PCR
4.5. Gene Suppression Using Candidate Antisense Oligonucleotides
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steponkus, P.L. Role of the Plasma Membrane in Freezing Injury and Cold Acclimation. Annu. Rev. Plant Physiol. 1984, 35, 543–584. [Google Scholar] [CrossRef]
- Pearce, R.S. Plant Freezing and Damage. Ann. Bot. 2001, 87, 417–424. [Google Scholar] [CrossRef]
- Miura, K.; Furumoto, T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef] [PubMed]
- Porto, D.D.; Bruneau, M.; Perini, P.; Anzanello, R.; Renou, J.P.; dos Santos, H.P.; Fialho, F.B.; Revers, L.F. Transcription profiling of the chilling requirement for bud break in apples: A putative role for FLC-like genes. J. Exp. Bot. 2015, 66, 2659–2672. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, C.; Ren, J.; Dong, J.; Shi, X.; Zhao, X.; Wang, X.; Wang, J.; Zhong, C.; Zhao, S.; et al. An Advanced Lipid Metabolism System Revealed by Transcriptomic and Lipidomic Analyses Plays a Central Role in Peanut Cold Tolerance. Front. Plant Sci. 2020, 11, 1110. [Google Scholar] [CrossRef]
- Shen, Z.J.; Qin, Y.Y.; Luo, M.R.; Li, Z.; Ma, D.N.; Wang, W.H.; Zheng, H.L. Proteome analysis reveals a systematic response of cold-acclimated seedlings of an exotic mangrove plant Sonneratia apetala to chilling stress. J. Proteom. 2021, 248, 104349. [Google Scholar] [CrossRef]
- Lee, J.G.; Yi, G.; Seo, J.; Kang, B.C.; Choi, J.H.; Lee, E.J. Jasmonic acid and ERF family genes are involved in chilling sensitivity and seed browning of pepper fruit after harvest. Sci. Rep. 2020, 10, 17949. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Shen, X.; Li, H.; Liu, X.; Niu, C.; et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Dikaya, V.; El Arbi, N.; Rojas-Murcia, N.; Nardeli, S.M.; Goretti, D.; Schmid, M. Insights into the role of alternative splicing in plant temperature response. J. Exp. Bot. 2021, 72, 7384–7403. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhong, J.; Zhang, L.; Wang, Y.; Song, P.; Liu, W.; Li, X.; Han, D. Overexpression of a Fragaria vesca MYB Transcription Factor Gene (FvMYB82) Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 10538. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liang, X.; Cai, W.; Wang, H.; Liu, X.; Cheng, L.; Song, P.; Luo, G.; Han, D. Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 13418. [Google Scholar] [CrossRef] [PubMed]
- Marquez, Y.; Brown, J.W.S.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Luo, Y.; Sun, J.; Qin, X.; Gan, P.; Zhou, Z.; Qian, Y.; Zhao, R.; Zhao, Z.; Cai, W.; et al. Pan-transcriptomic analysis reveals alternative splicing control of cold tolerance in rice. Plant Cell 2024, 36, 2117–2139. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xiao, W.; Hui, L.; Yang, T.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q. The Genome of Dendrobium officinale Illuminates the Biology of the Important Traditional Chinese Orchid Herb. Mol. Plant 2015, 8, 922–934. [Google Scholar]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef]
- Zhan, X.; Qian, Y.; Mao, B. Metabolic Profiling of Terpene Diversity and the Response of Prenylsynthase-Terpene Synthase Genes during Biotic and Abiotic Stresses in Dendrobium catenatum. Int. J. Mol. Sci. 2022, 23, 6398. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Qi, J.; Zhou, B.; Mao, B. Metabolomic and transcriptomic analyses reveal the regulation of pigmentation in the purple variety of Dendrobium officinale. Sci. Rep. 2020, 10, 17700. [Google Scholar] [CrossRef]
- Zhan, X.; Qi, J.; Shen, Q.; He, B.; Mao, B. Regulation of phenylpropanoid metabolism during moderate freezing and post-freezing recovery in Dendrobium officinale. J. Plant Interact. 2022, 17, 290–300. [Google Scholar] [CrossRef]
- Wu, Z.G.; Jiang, W.; Chen, S.L.; Mantri, N.; Tao, Z.M.; Jiang, C.X. Insights from the Cold Transcriptome and Metabolome of Dendrobium officinale: Global Reprogramming of Metabolic and Gene Regulation Networks during Cold Acclimation. Front. Plant Sci. 2016, 7, 1653. [Google Scholar] [CrossRef]
- Zhan, X.; Qian, Y.; Mao, B. Identification of Two GDSL-Type Esterase/Lipase Genes Related to Tissue-Specific Lipolysis in Dendrobium catenatum by Multi-Omics Analysis. Life 2022, 12, 1563. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Qiu, H.L.; Huang, Y.; Zhang, L.; Si, J.P. Genome-wide identification and expression profiling of SET DOMAIN GROUP family in Dendrobium catenatum. BMC Plant Biol. 2020, 20, 40. [Google Scholar] [CrossRef]
- Wu, Z.; Guo, Z.; Wang, K.; Wang, R.; Fang, C. Comparative Metabolomic Analysis Reveals the Role of OsHPL1 in the Cold-Induced Metabolic Changes in Rice. Plants 2023, 12, 2032. [Google Scholar] [CrossRef]
- Sun, C.X.; Gao, X.X.; Li, M.Q.; Fu, J.Q.; Zhang, Y.L. Plastic responses in the metabolome and functional traits of maize plants to temperature variations. Plant Biol. 2016, 18, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Chen, K.; Arora, R. Short versus prolonged freezing differentially impacts freeze—Thaw injury in spinach leaves: Mechanistic insights through metabolite profiling. Physiol. Plant. 2020, 168, 777–789. [Google Scholar] [CrossRef]
- Ma, L.; Cheng, K.; Li, J.; Deng, Z.; Zhang, C.; Zhu, H. Roles of Plant Glycine-Rich RNA-Binding Proteins in Development and Stress Responses. Int. J. Mol. Sci. 2021, 22, 5849. [Google Scholar] [CrossRef]
- Xiaochuang, C.; Chu, Z.; Lianfeng, Z.; Junhua, Z.; Hussain, S.; Lianghuan, W.; Qianyu, J. Glycine increases cold tolerance in rice via the regulation of N uptake, physiological characteristics, and photosynthesis. Plant Physiol. Bioch 2017, 112, 251–260. [Google Scholar] [CrossRef]
- Obata, T.; Witt, S.; Lisec, J.; Palacios-Rojas, N.; Florez-Sarasa, I.; Yousfi, S.; Araus, J.L.; Cairns, J.E.; Fernie, A.R. Metabolite Profiles of Maize Leaves in Drought, Heat, and Combined Stress Field Trials Reveal the Relationship between Metabolism and Grain Yield. Plant Physiol. 2015, 169, 2665–2683. [Google Scholar] [CrossRef]
- Vyse, K.; Penzlin, J.; Sergeant, K.; Hincha, D.K.; Arora, R.; Zuther, E. Repair of sub-lethal freezing damage in leaves of Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Couée, I.; Sulmon, C.; Gouesbet, G.; El Amrani, A. Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 2006, 57, 449–459. [Google Scholar] [CrossRef]
- Guo, J.; Beemster, G.T.S.; Liu, F.; Wang, Z.; Li, X. Abscisic Acid Regulates Carbohydrate Metabolism, Redox Homeostasis and Hormonal Regulation to Enhance Cold Tolerance in Spring Barley. Int. J. Mol. Sci. 2023, 24, 11348. [Google Scholar] [CrossRef]
- Cai, P.; Lan, Y.; Gong, F.; Li, C.; Xia, F.; Li, Y.; Fang, C. Comparative physiology and transcriptome response patterns in cold-tolerant and cold-sensitive varieties of Solanum melongena. BMC Plant Biol. 2024, 24, 256. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, B.O.; Laxalt, A.M.; ter Riet, B.; van Schooten, B.; Merquiol, E.; Testerink, C.; Haring, M.A.; Bartels, D.; Munnik, T. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 2009, 50, 78–89. [Google Scholar] [CrossRef]
- Jan, S.; Rustgi, S.; Barmukh, R.; Shikari, A.B.; Leske, B.; Bekuma, A.; Sharma, D.; Ma, W.; Kumar, U.; Kumar, U.; et al. Advances and opportunities in unraveling cold-tolerance mechanisms in the world’s primary staple food crops. Plant Genome 2024, 17, e20402. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef]
- Zhao, X.; Tan, L.; Wang, S.; Shen, Y.; Guo, L.; Ye, X.; Liu, S.; Feng, Y.; Wu, W. The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa. Int. J. Mol. Sci. 2021, 22, 11369. [Google Scholar] [CrossRef]
- Albuquerque-Martins, R.; Szakonyi, D.; Rowe, J.; Jones, A.M.; Duque, P. ABA signaling prevents phosphodegradation of the SR45 splicing factor to alleviate inhibition of early seedling development in Arabidopsis. Plant Commun. 2023, 4, 100495. [Google Scholar] [CrossRef]
- Gao, C.; Tang, D.; Wang, W. The Role of Ubiquitination in Plant Immunity: Fine-Tuning Immune Signaling and Beyond. Plant Cell Physiol. 2022, 63, 1405–1413. [Google Scholar] [CrossRef]
- Lan, W.; Qiu, Y.; Xu, Y.; Liu, Y.; Miao, Y. Ubiquitination and Ubiquitin-Like Modifications as Mediators of Alternative Pre-mRNA Splicing in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 869870. [Google Scholar] [CrossRef]
- McKay, S.L.; Johnson, T.L. A bird’s-eye view of post-translational modifications in the spliceosome and their roles in spliceosome dynamics. Mol. Biosyst. 2010, 6, 2093–2102. [Google Scholar] [CrossRef][Green Version]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Mulvaney, K.M.; Blomquist, C.; Acharya, N.; Li, R.; Ranaghan, M.J.; O’Keefe, M.; Rodriguez, D.J.; Young, M.J.; Kesar, D.; Pal, D.; et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol. Cell 2021, 81, 3481–3495.e3487. [Google Scholar] [CrossRef]
- Wilkinson, C.R.; Dittmar, G.A.; Ohi, M.D.; Uetz, P.; Jones, N.; Finley, D. Ubiquitin-like protein Hub1 is required for pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr. Biol. 2004, 14, 2283–2288. [Google Scholar] [CrossRef]
- Zhan, X.; Qian, Y.; Mao, B. Dendrobium Multi-Omics Reveal Lipid Remodeling in Response to Freezing. Metabolites 2022, 12, 1216. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhan, X.; Shen, Q.; Wang, X.; Hong, Y. The sulfoquinovosyltransferase-like enzyme SQD2.2 is involved in flavonoid glycosylation, regulating sugar metabolism and seed setting in rice. Sci. Rep. 2017, 7, 4685. [Google Scholar] [CrossRef]
- Zhao, M.; Jin, J.; Gao, T.; Zhang, N.; Jing, T.; Wang, J.; Ban, Q.; Schwab, W.; Song, C. Glucosyltransferase CsUGT78A14 Regulates Flavonols Accumulation and Reactive Oxygen Species Scavenging in Response to Cold Stress in Camellia sinensis. Front. Plant Sci. 2019, 10, 1675. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, X.; Li, Z.; Pang, M.; Yao, G.; Mao, B. Comprehensive Omics Analysis Reveals Cold-Induced Metabolic Reprogramming and Alternative Splicing in Dendrobium officinale. Plants 2025, 14, 412. https://doi.org/10.3390/plants14030412
Zhan X, Li Z, Pang M, Yao G, Mao B. Comprehensive Omics Analysis Reveals Cold-Induced Metabolic Reprogramming and Alternative Splicing in Dendrobium officinale. Plants. 2025; 14(3):412. https://doi.org/10.3390/plants14030412
Chicago/Turabian StyleZhan, Xinqiao, Zhangqun Li, Minxia Pang, Guoxiang Yao, and Bizeng Mao. 2025. "Comprehensive Omics Analysis Reveals Cold-Induced Metabolic Reprogramming and Alternative Splicing in Dendrobium officinale" Plants 14, no. 3: 412. https://doi.org/10.3390/plants14030412
APA StyleZhan, X., Li, Z., Pang, M., Yao, G., & Mao, B. (2025). Comprehensive Omics Analysis Reveals Cold-Induced Metabolic Reprogramming and Alternative Splicing in Dendrobium officinale. Plants, 14(3), 412. https://doi.org/10.3390/plants14030412






