Genetic Analysis and Fingerprint Construction for Thick-Skinned Melon (Cucumis melo subsp. melo) Based on InDel Markers
Abstract
1. Introduction
2. Results
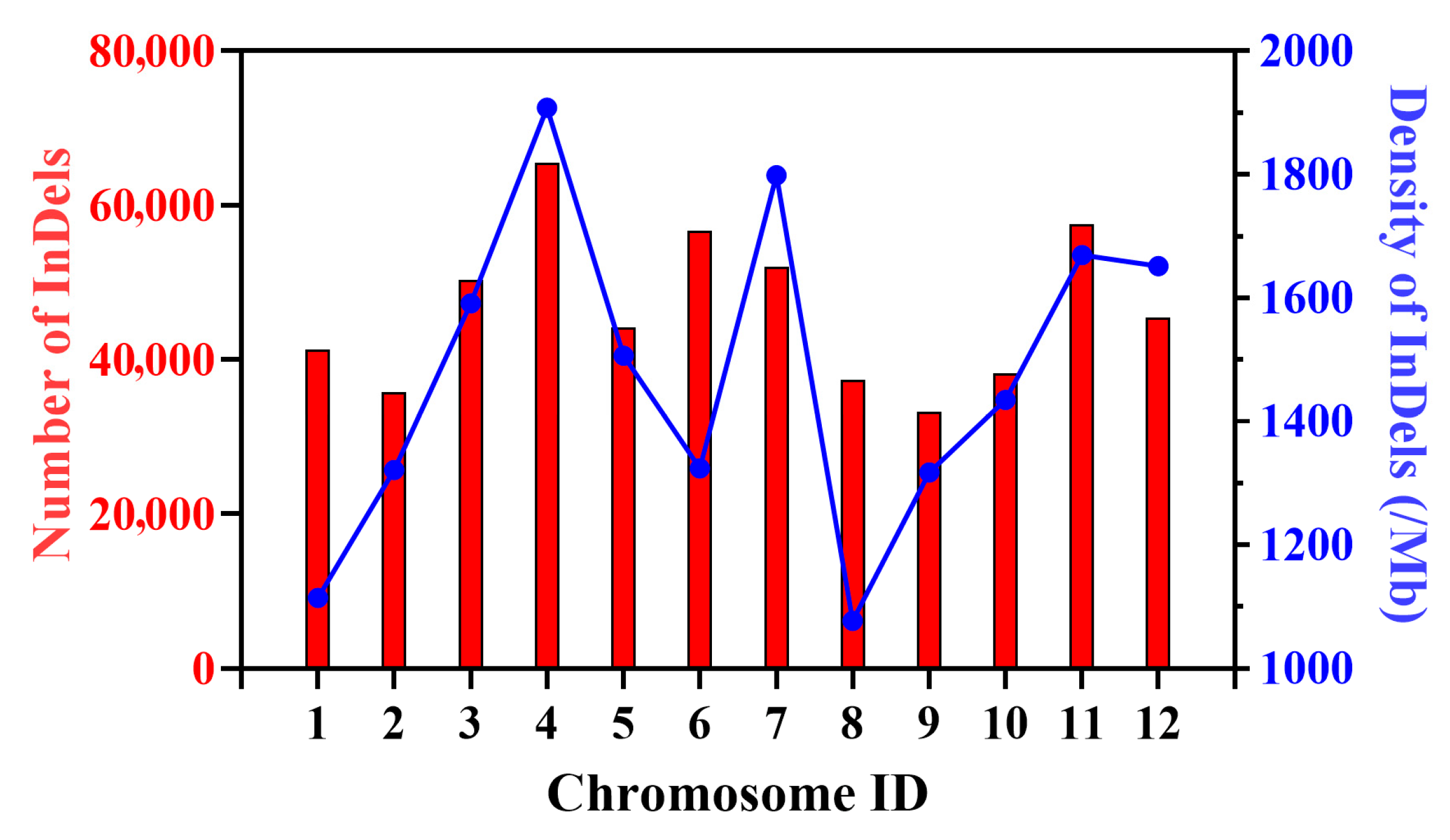
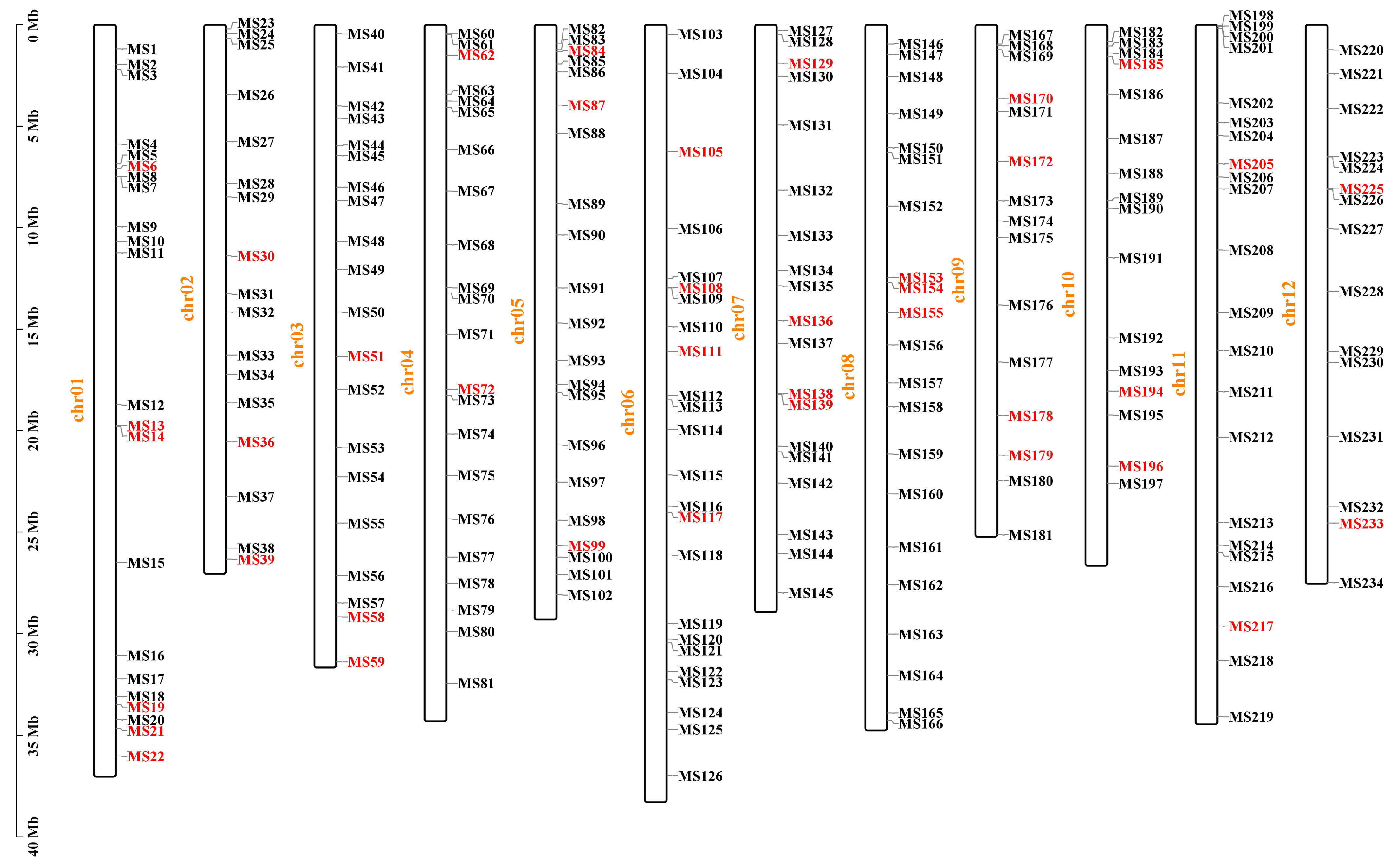
2.1. The Chromosomal Distribution Characteristics of InDel Variations
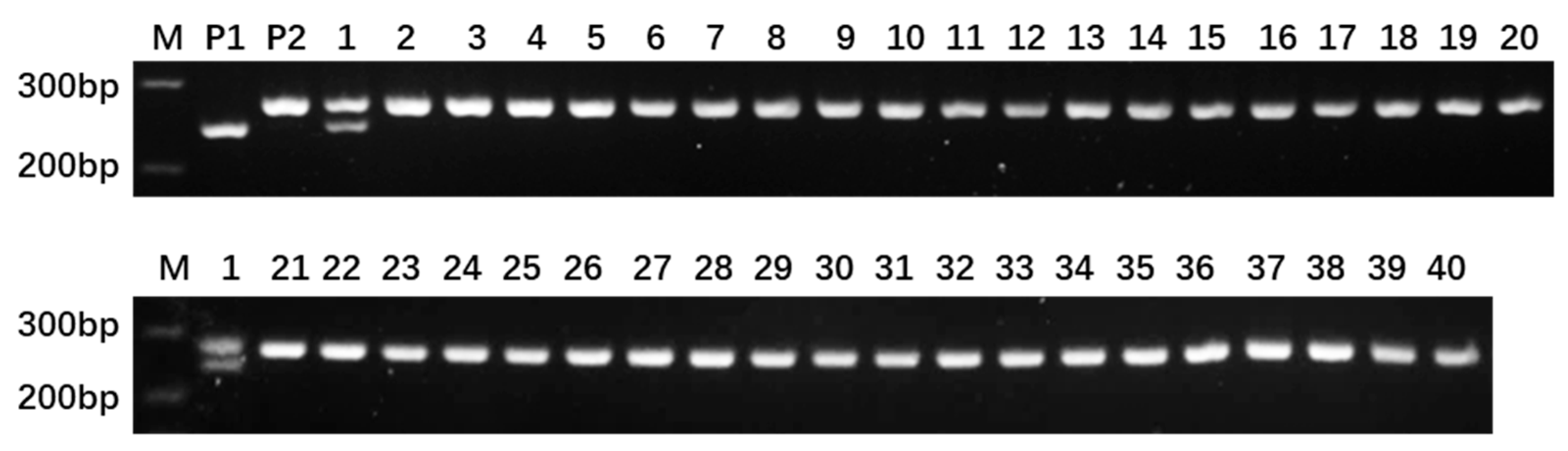
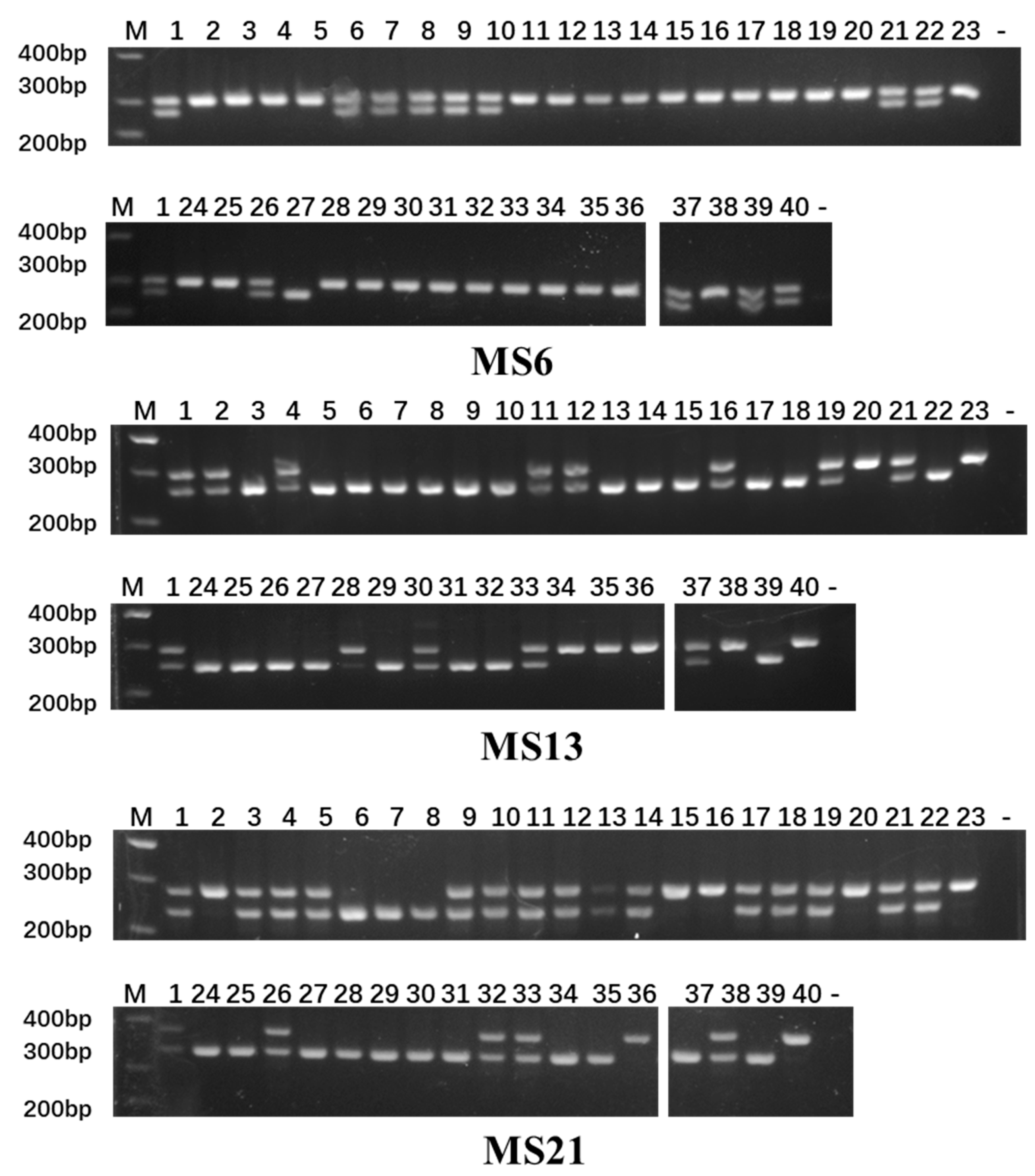
2.2. Development and Screening of InDel Markers
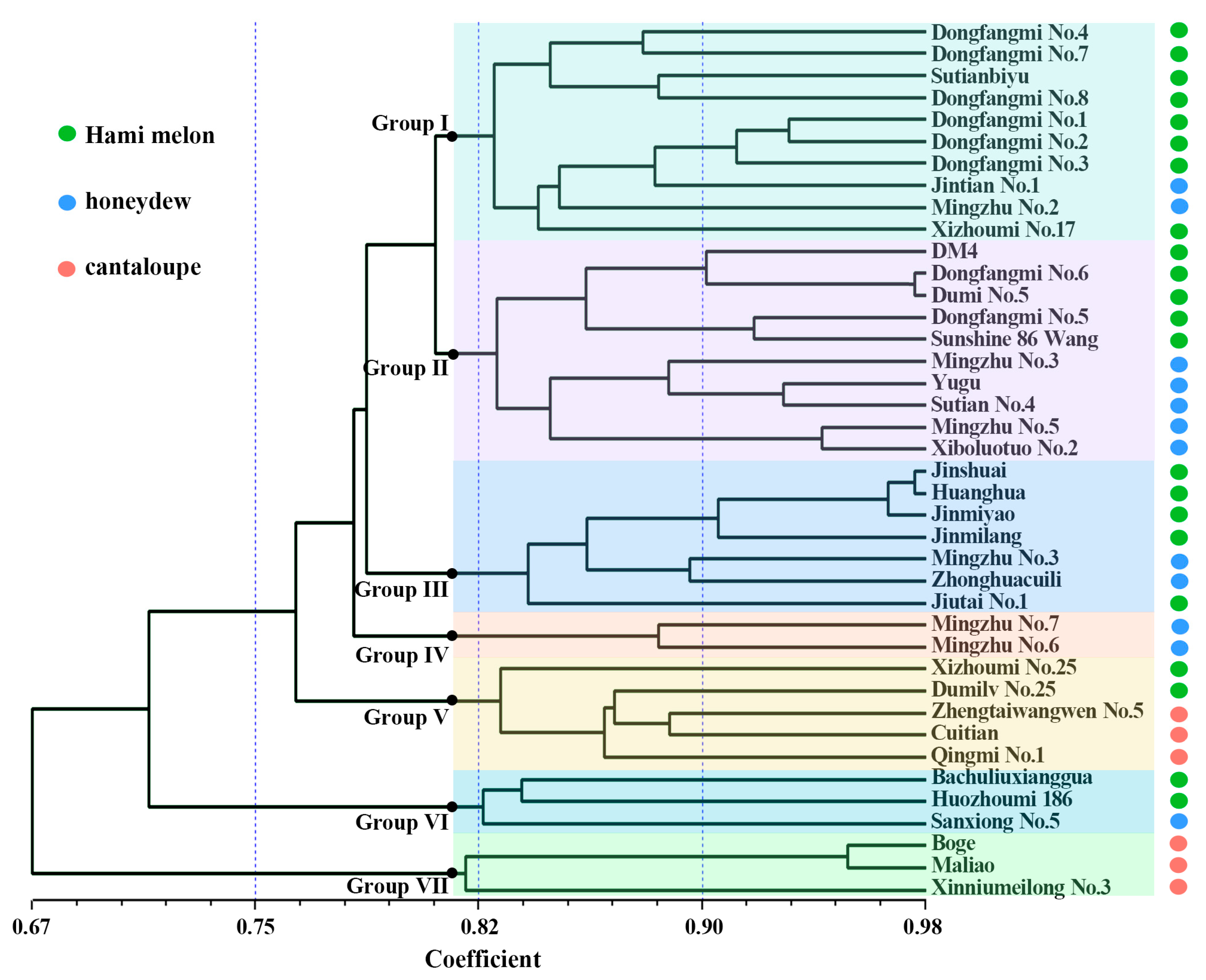
2.3. Genetic Diversity Analysis
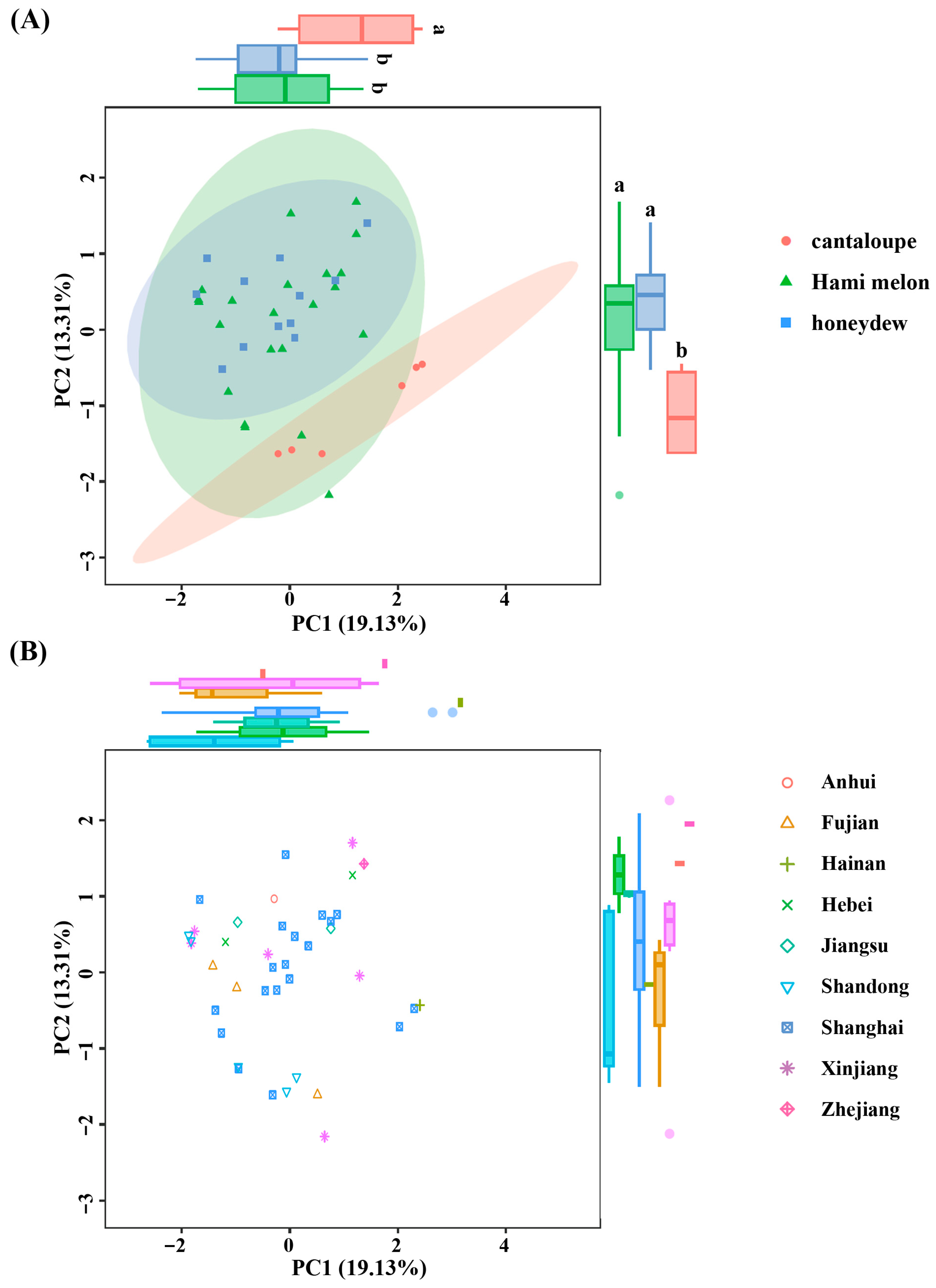
2.4. Genetic Differentiation
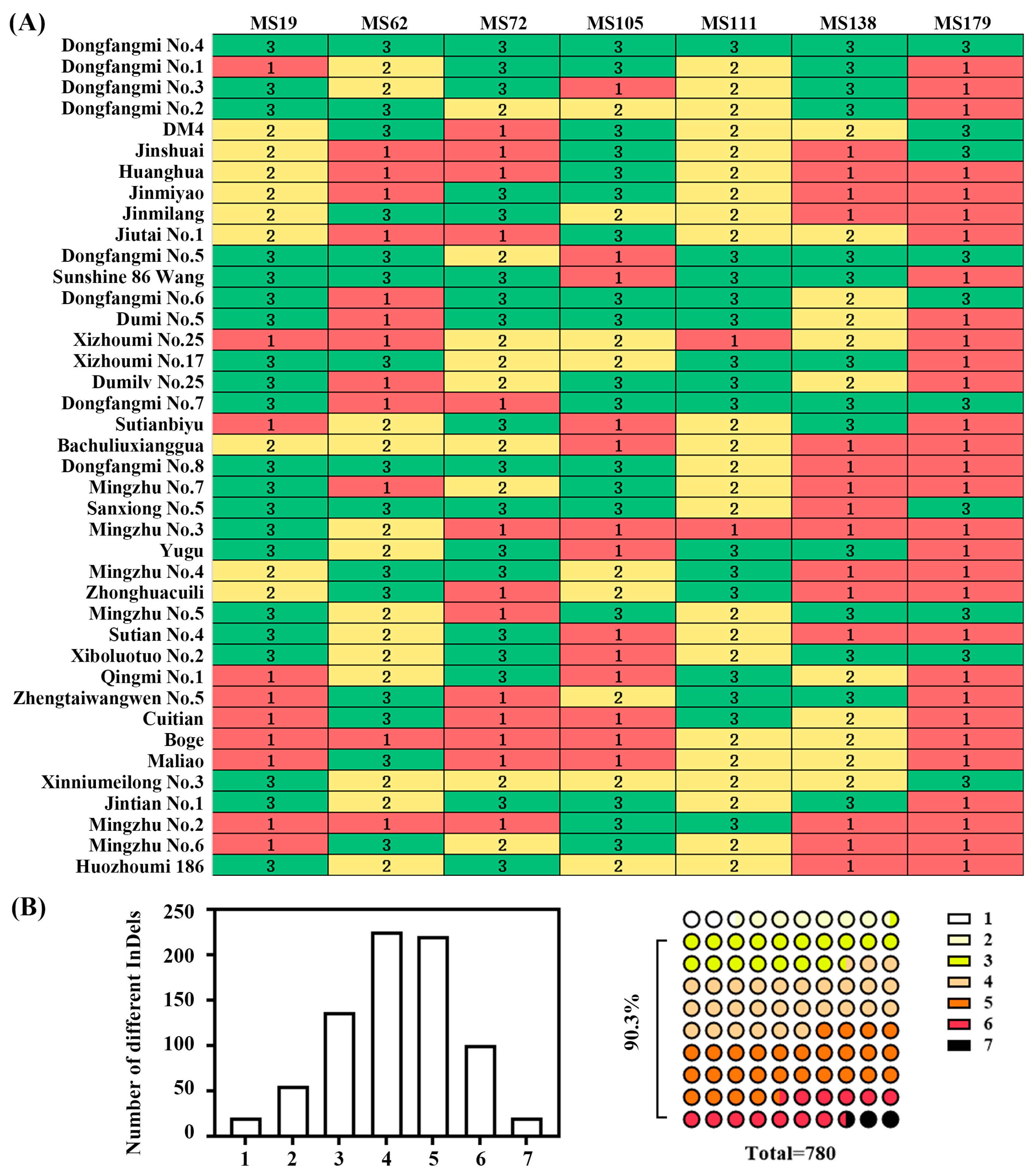
2.5. Construction of DNA Fingerprint Profiles
3. Discussion
3.1. Distribution Characteristics of InDels and Their Value in Marker Development
3.2. The History of Breeding Exchanges Reflected by Population Structure
3.3. Prospects for the Identification Application of Core Marker Combinations
4. Materials and Methods
4.1. Plant Materials
| Number | Cultivar | Origin | Type | Peel Color | Pulp Color | Fruit Shape | Surface |
|---|---|---|---|---|---|---|---|
| 1 | Dongfangmi No.4 | Shanghai | Hami melon | yellow | orange | oval | net |
| 2 | Dongfangmi No.1 | Shanghai | Hami melon | white | orange | oval | smooth |
| 3 | Dongfangmi No.3 | Shanghai | Hami melon | white | orange | oval | smooth |
| 4 | Dongfangmi No.2 | Shanghai | Hami melon | yellow | orange | oval | net |
| 5 | DM4 | Shanghai | Hami melon | yellow | orange | oval | net |
| 6 | Jinshuai | Xinjiang | Hami melon | yellow | orange | oval | net |
| 7 | Huanghua | Shandong | Hami melon | yellow | orange | oval | net |
| 8 | Jinmiyao | Xinjiang | Hami melon | yellow | orange | oval | net |
| 9 | Jinmilang | Hebei | Hami melon | yellow | orange | oval | net |
| 10 | Jiutai No.1 | Fujian | Hami melon | yellow | orange | oval | net |
| 11 | Dongfangmi No.5 | Shanghai | Hami melon | chartreuse | orange | oval | net |
| 12 | Sunshine 86 Wang | Xinjiang | Hami melon | chartreuse | orange | oval | net |
| 13 | Dongfangmi No.6 | Shanghai | Hami melon | green | orange | oval | net |
| 14 | Dumi No.5 | Shandong | Hami melon | green | orange | oval | net |
| 15 | Xizhoumi No.25 | Xinjiang | Hami melon | Celadon green | orange | oval | net |
| 16 | Xizhoumi No.17 | Xinjiang | Hami melon | Celadon green | orange | oval | net |
| 17 | Dumilv No.25 | Shandong | Hami melon | Celadon green | green | oval | net |
| 18 | Dongfangmi No.7 | Shanghai | Hami melon | Celadon green | green | oval | net |
| 19 | Sutianbiyu | Jiangsu | Hami melon | green | green | oval | net |
| 20 | Bachuliuxianggua | Xinjiang | Hami melon | green | green | oval | net |
| 21 | Dongfangmi No.8 | Shanghai | Hami melon | black-green | white | fusiform | smooth |
| 22 | Mingzhu No.7 | Shanghai | honeydew | yellow | green-white | circle | smooth |
| 23 | Sanxiong No.5 | Zhejiang | honeydew | yellow | green-white | circle | smooth |
| 24 | Mingzhu No.3 | Shanghai | honeydew | white | green | circle | smooth |
| 25 | Yugu | Fujian | honeydew | white | green | circle | smooth |
| 26 | Mingzhu No.4 | Shanghai | honeydew | white | white | oval | smooth |
| 27 | Zhonghuacuili | Shandong | honeydew | white | white | oval | smooth |
| 28 | Mingzhu No.5 | Shanghai | honeydew | white | white | circle | smooth |
| 29 | Sutian No.4 | Jiangsu | honeydew | white | orange | oval | smooth |
| 30 | Xiboluotuo No.2 | Shanghai | honeydew | white | white | circle | smooth |
| 31 | Qingmi No.1 | Shanghai | cantaloupe | Celadon green | green | circle | net |
| 32 | Zhengtaiwangwen No.5 | Shandong | cantaloupe | Celadon green | green | circle | net |
| 33 | Cuitian | Fujian | cantaloupe | Celadon green | green | circle | net |
| 34 | Boge | Shanghai | cantaloupe | Celadon green | green | circle | net |
| 35 | Maliao | Shanghai | cantaloupe | Celadon green | green | circle | net |
| 36 | Xinniumeilong No.3 | Hainan | cantaloupe | Celadon green | green | circle | net |
| 37 | Jintian No.1 | Anhui | honeydew | yellow | orange | oval | smooth |
| 38 | Mingzhu No.2 | Shanghai | honeydew | white | white | circle | net |
| 39 | Mingzhu No.6 | Shanghai | honeydew | yellow | white | circle | smooth |
| 40 | Huozhoumi 186 | Hebei | Hami melon | yellow | orange | oval | net |
4.2. Melon Material Planting and Sampling
4.3. Genome-Wide Resequencing and Variants Calling
- (1)
- Homozygous loci in both parental lines;
- (2)
- Genotypic differences between parental lines;
- (3)
- Absence of missing genotypes in bi-parents;
- (4)
- Heterozygous genotypes in offspring;
- (5)
- Genotype quality (GQ) > q20.
4.4. InDel Primer Synthesis and Screening
- (1)
- clear, single-band amplification in both parents and F1;
- (2)
- visible polymorphism (size difference) between parents;
- (3)
- consistent Mendelian inheritance in the F1;
- (4)
- absence of non-specific or stutter bands.
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Endl, J.; Achigan-Dako, E.G.; Pandey, A.K.; Monforte, A.J.; Pico, B.; Schaefer, H. Repeated domestication of melon (Cucumis melo) in Africa and Asia and a new close relative from India. Am. J. Bot. 2018, 105, 1662–1671. [Google Scholar] [CrossRef] [PubMed]
- Imoh, O.N.; Shigita, G.; Sugiyama, M.; Dung, T.P.; Tanaka, K.; Takahashi, M.; Nishimura, K.; Monden, Y.; Nishida, H.; Goda, M.; et al. Molecular polymorphisms of the nuclear and chloroplast genomes among African melon germplasms reveal abundant and unique genetic diversity, especially in Sudan. Ann. Bot. 2025, 135, 1329–1344. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, P.; Zhu, Q.; Zhu, Z.; Liu, H.; Wang, X.; Weng, Y.; Gao, M.; Luan, F. Resequencing of 297 melon accessions reveals the genomic history of improvement and loci related to fruit traits in melon. Plant Biotechnol. J. 2020, 18, 2545–2558. [Google Scholar] [CrossRef]
- Shahwar, D.; Khan, Z.; Park, Y. Molecular marker-assisted mapping, candidate gene identification, and breeding in melon (Cucumis melo L.): A Review. Int. J. Mol. Sci. 2023, 24, 15490. [Google Scholar] [CrossRef]
- Gao, P.; Ma, H.; Luan, F.; Song, H. DNA fingerprinting of Chinese melon provides evidentiary support of seed quality appraisal. PLoS ONE 2012, 7, e52431. [Google Scholar] [CrossRef]
- Hu, J.; Yao, J.; Lu, J.; Liu, W.; Zhao, Z.; Li, Y.; Jiang, L.; Zha, L. The complete chloroplast genome sequences of nine melon varieties (Cucumis melo L.): Lights into comparative analysis and phylogenetic relationships. Front. Genet. 2024, 15, 1417266. [Google Scholar] [CrossRef]
- Jeffrey, C. A review of the Cucurbitaceae. Bot. J. Linn. Soc. 1980, 81, 233–247. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Lv, Y.; Zhang, X.; Xia, C.; Zhao, H.; Wen, C. Genetic diversity analysis and variety identification using SSR and SNP markers in melon. BMC Plant Biol. 2023, 23, 39. [Google Scholar] [CrossRef]
- Li, X.; Song, D.; Li, M.; Li, D.; You, M.; Peng, Y.; Yan, J.; Bai, S. An initial exploration of core collection construction and DNA fingerprinting in Elymus sibiricus L. using SNP markers. Front. Plant Sci. 2025, 16, 1534085. [Google Scholar] [CrossRef]
- Sadeghpour, N.; Asadi-Gharneh, H.A.; Nasr-Esfahani, M.; Khankahdani, H.H.; Golabadi, M. Assessing genetic diversity and population structure of Iranian melons (Cucumis melo) collection using primer pair markers in association with resistance to Fusarium wilt. Funct. Plant Biol. 2023, 50, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Flores-León, A.; Peréz Moro, C.; Martí, R.; Beltran, J.; Roselló, S.; Cebolla-Cornejo, J.; Picó, B. Spanish melon landraces: Revealing useful diversity by genomic, morphological, and metabolomic analysis. Int. J. Mol. Sci. 2022, 23, 7162. [Google Scholar] [CrossRef]
- Yang, J.; Meng, P.; Mi, H.; Wang, X.; Yang, J.; Fu, S.; Gong, W.; Bao, R.; Deng, W.; Wu, H.; et al. The development of ideal insertion and deletion (InDel) markers and initial indel map variation in cucumber using re-sequenced data. BMC Genom. 2025, 26, 391. [Google Scholar] [CrossRef]
- Moghaddam, S.M.; Song, Q.; Mamidi, S.; Schmutz, J.; Lee, R.; Cregan, P.; Osorno, J.M.; Mcclean, P.E. Developing market class specific InDel markers from next generation sequence data in Phaseolus vulgaris L. Front. Plant Sci. 2014, 5, 185. [Google Scholar] [CrossRef]
- Wu, D.; Wu, H.; Wang, C.; Tseng, H.; Hwu, K. Genome-wide InDel marker system for application in rice breeding and mapping studies. Euphytica 2013, 192, 131–143. [Google Scholar] [CrossRef]
- Li, H.; Ikram, M.; Xia, Y.; Li, R.; Yuan, Q.; Zhao, W.; Siddique, K.H.M.; Guo, P. Genome-wide identification and development of InDel markers in tobacco (Nicotiana tabacum L.) using RAD-seq. Physiol. Mol. Biol. Plants 2022, 28, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Yuan, Y.; Real, N.; Tang, M.; Ren, J.; Wei, J.; Liu, B.; Zhang, X. Fine mapping and identification of candidate genes associated with powdery mildew resistance in melon (Cucumis melo L.). Hortic. Res. 2024, 11, uhae222. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Dong, X.; Zhang, X.; Cao, J.; Liu, M.; Zhou, X.; Long, H.; Cao, H.; Lin, H.; Zhang, L. Genome assembly and resequencing shed light on evolution, population selection, and sex identification in Vernicia montana. Hortic. Res. 2024, 11, uhae141. [Google Scholar] [CrossRef]
- Lyu, X.; Xia, Y.; Wang, C.; Zhang, K.; Deng, G.; Shen, Q.; Gao, W.; Zhang, M.; Liao, N.; Ling, J.; et al. Pan-genome analysis sheds light on structural variation-based dissection of agronomic traits in melon crops. Plant Physiol. 2023, 193, 1330–1348. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, T.; Meng, X.; Song, J.; Zhang, C.; Gao, P. Genetic Mapping and QTL Analysis of Fruit Traits in Melon (Cucumis melo L.). Curr. Issues Mol. Biol. 2023, 45, 3419–3433. [Google Scholar] [CrossRef]
- Chattopadhyay, T.; Sangam, S.; Akhtar, S. Rapid genotyping in tomato by VPCR using agarose gel-resolvable InDel markers. 3 Biotech 2023, 13, 85. [Google Scholar] [CrossRef]
- Sathapondecha, P.; Suksri, P.; Nuanpirom, J.; Nakkanong, K.; Nualsri, C.; Whankaew, S. Development of gene-based InDel markers on putative drought stress-responsive genes and genetic diversity of durian (Durio zibethinus). Biochem. Genet. 2024, 62, 4396–4407. [Google Scholar] [CrossRef] [PubMed]
- Oren, E.; Dafna, A.; Tzuri, G.; Halperin, I.; Isaacson, T.; Elkabetz, M.; Meir, A.; Saar, U.; Ohali, S.; La, T.; et al. Pan-genome and multi-parental framework for high-resolution trait dissection in melon (Cucumis melo). Plant J. 2022, 112, 1525–1542. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wu, W.; Su, L.; Lv, H.; Cheng, Z.; Yang, W.; Nong, L.; Liu, T.; Chen, Y.; Wang, P.; et al. Development and application of InDel markers linked to fruit-shape and peel-colour Genes in wax gourd. Genes 2022, 13, 1567. [Google Scholar] [CrossRef]
- Jiang, X.L.; An, M.; Zheng, S.S.; Deng, M.; Su, Z.H. Geographical isolation and environmental heterogeneity contribute to the spatial genetic patterns of Quercus kerrii (Fagaceae). Heredity 2018, 120, 219–233. [Google Scholar] [CrossRef]
- Ahatović Hajro, A.; Hasanović, M.; Murtić, S.; Kalajdžić, A.; Pojskić, N.; Durmić-Pašić, A. Serpentine environment prevails over geographic distribution in shaping the genetic diversity of Medicago lupulina L. Mol. Genet. Genom. 2024, 299, 28. [Google Scholar] [CrossRef]
- Tanaka, K.; Sugiyama, M.; Shigita, G.; Murakami, R.; Duong, T.T.; Aierken, Y.; Artemyeva, A.M.; Mamypbelov, Z.; Ishikawa, R.; Nishida, H.; et al. Melon diversity on the Silk Road by molecular phylogenetic analysis in Kazakhstan melons. Breed. Sci. 2023, 73, 219–229. [Google Scholar] [CrossRef]
- Shigita, G.; Dung, T.P.; Pervin, M.N.; Duong, T.T.; Imoh, O.N.; Monden, Y.; Nishida, H.; Tanaka, K.; Sugiyama, M.; Kawazu, Y.; et al. Elucidation of genetic variation and population structure of melon genetic resources in the NARO Genebank, and construction of the World Melon Core Collection. Breed. Sci. 2023, 73, 269–277. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.; Dong, Y.; Xu, Y.; Xu, K.; Zhang, Q.; Yao, Z.; Yu, Q.; Zhang, H.; Zhang, Z. A wild melon reference genome provides novel insights into the domestication of a key gene responsible for melon fruit acidity. Theor. Appl. Genet. 2024, 137, 144. [Google Scholar] [CrossRef]
- Aboul-Maaty, N.A.-F.; Oraby, H.A.-S. Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull. Natl. Res. Cent. 2019, 43, 25. [Google Scholar] [CrossRef]
- Takahiro, N.; Kaoru, D.; Takashi, M.; Yukio, N. Development of Indel markers for the selection of Satsuma mandarin (Citrus unshiu Marc.) hybrids that can be used for low-cost genotyping with agarose gels. Euphytica 2020, 216, 115. [Google Scholar] [CrossRef]
- Xing, X.; Xu, H.; Dong, Y.; Cui, H.; Sun, M.; Wang, H.; Liu, Y.; Meng, L.; Zheng, C. Genetic analysis and fingerprint construction for Isatis indigotica fort. using SSR markers. Curr. Issues Mol. Biol. 2025, 47, 146. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, version 2.1; Exeter Publishing Setauket: New York, NY, USA, 2000.
| Number | Position | Primer-F | Primer-R |
|---|---|---|---|
| MS6 | chr01_7074391 | TCCTTCTTTTTATCTTCTGCATGGC | CGGGTACAAGGCAACAGCTA |
| MS13 | chr01_19733999 | ACACATCAATATGATACATGGTCAAGT | TTCTCATTGGGGCCTTGTCC |
| MS14 | chr01_19795754 | GTTCTGTTTGTTGGGTCCACG | AGCTACCTCCATTGCGGAAG |
| MS19 | chr01_33481852 | TAGCGGGACAGAGTGTTTGG | GGGGCCCAAAGGATCAAAGA |
| MS21 | chr01_34675645 | TGGATTTACGATAAGAGGGCCT | TCCCCAAGTTGCAAGCTTGT |
| MS22 | chr01_36018052 | CGTGACATCCAAATTGATATTCGGT | CTCGACCTCAAGCCTCTGTC |
| MS30 | chr02_11391369 | CCAAGCTACCTAACACTTCCAAC | TGGAATTGTTGAGATGGGGGA |
| MS36 | chr02_20541361 | ACCAATGGCTCTATTTTATCTCCTCT | TTTTGCGGCTCTTTGTGCAA |
| MS39 | chr02_26332867 | TTGTCTTCGGATTCGGAAACCT | AGCATATCAGTGTCTATCGATGTCT |
| MS51 | chr03_16330935 | AGATTCGTGCTCAGGAAGTCA | ACAATGAAATCTCGTGCAAATTGC |
| MS58 | chr03_29176371 | AGGTTTTCTTACCAGGAAGCTACT | AGCACCCCATTGATTAAAAGCTG |
| MS59 | chr03_31365283 | CAAAGCATCGGATGGCTGAG | CGTACATTTTGTATCAACCAATGGC |
| MS62 | chr04_1488958 | CGACCAATAGTAGCTACCAACGA | TCGTAACTAAGCATTAATGAAAGCTGT |
| MS72 | chr04_17954161 | TGGTGCAAGGGATGAGAGTG | AATAGCGCTCCACGTGATGT |
| MS84 | chr05_1320764 | ACTTCACCGTCCTCCACAAG | ACCCAATTTTGTTCTCGAGTTTTCA |
| MS87 | chr05_3963301 | TGCAATGACCTTCAACGCTTC | TGGTGCAGCTTCGACTGATT |
| MS99 | chr05_25658229 | ACCTCATCAACTGTGAGGCC | AGGATGGAGCACTGATGAAGAA |
| MS105 | chr06_6246570 | GGTCCATTCAAGGCCTAGCA | TCCAGAAATCTCGTGTGAACAGA |
| MS108 | chr06_12951796 | CCCCTACCGTGCTCAAACTT | GTCCTTCGTTTCATTGGCGG |
| MS111 | chr06_16084050 | GGGATAGGCTCGGCTTGATT | CCGTGGTTTTAGTCGGACCA |
| MS117 | chr06_24013568 | AATCCTCCATTGGGCCAGTC | ACTGGACGTCCTAAAAAGCGA |
| MS129 | chr07_1885541 | CCAGCCGTGAAAGTAAAATAAAACG | GCACACATGCCATGGATAACA |
| MS136 | chr07_14576627 | CCCTTGACCAAAGTTCGTGG | AGAATTGAGTTGTGGGACCCA |
| MS138 | chr07_18178680 | TGATTGAGGGAGGAAGAGGGA | TCTGTCGTCCTGTGGTTGTG |
| MS139 | chr07_18211629 | AGGCGTTCAAAAGAAGGGAAC | TGTGGTCCTTTCACCCCTTG |
| MS153 | chr08_12448261 | TGGCGAAGATGAAGTTGGGT | TGGGGCGTTACATTCTTCGAT |
| MS154 | chr08_12700987 | TGTCCAACACATGCAAACGG | TCCCTGTGTTCGACCCTAGAT |
| MS155 | chr08_14175099 | AGAAGGTGTGGAGAAGCACC | TGTTCGCCGTCTCTTGTTCT |
| MS170 | chr09_3616045 | CAATTTCGCAGGGAGAGGGA | GCAGTGCCATGGCTTCAAAT |
| MS172 | chr09_6721132 | GCAACCAATAAGTTCCTGCCA | AGCATCAAACATCATTGACTAGGG |
| MS178 | chr09_19253495 | GTTGGCGACCAAACAACTCC | ATGTGTGTGTGTGTGAGCCT |
| MS179 | chr09_21200612 | GTTGGATGATGGAGGAGAAGCT | AGTGGGAAGGAAAGTTGCACT |
| MS185 | chr10_1552627 | TCAATAGTGCCACGTGGGG | ACGCAAAGTTGAAACCGACC |
| MS194 | chr10_18050725 | GAGTGGCAGTACTCAAGCTCA | TGGCAGTGAAGGGAAGAGAAG |
| MS196 | chr10_21741047 | TGGATGGAGAAGCCCTCTTTG | AGCTCACGACACTCAAGTTGT |
| MS205 | chr11_6850271 | TGAACTGTAGTCATAAGGTGTACTCAA | TCTTGTTGGACTTGAGCACTCA |
| MS217 | chr11_29625033 | AGCATCACTTGGTCTAGCTTCT | CCTGTGTATGGGGAAGTGCA |
| MS225 | chr12_8076169 | CAACTCCAACGTTTCGTCCG | GTCCTCGGCTCGGATCAATT |
| MS233 | chr12_24555285 | GGCCCTGTCTCTCTGTTACC | AAGGAAAAGGTTGGAAGCGC |
| Order | Na | Ne | He | Ho | I | PIC | MAF |
|---|---|---|---|---|---|---|---|
| MS6 | 2.000 | 1.406 | 0.289 | 0.300 | 0.464 | 0.454 | 0.175 |
| MS13 | 2.000 | 1.406 | 0.289 | 0.300 | 0.464 | 0.454 | 0.175 |
| MS14 | 2.000 | 1.663 | 0.399 | 0.300 | 0.588 | 0.564 | 0.275 |
| MS19 | 2.000 | 1.999 | 0.500 | 0.525 | 0.693 | 0.611 | 0.488 |
| MS21 | 2.000 | 1.941 | 0.485 | 0.475 | 0.678 | 0.621 | 0.413 |
| MS22 | 2.000 | 1.995 | 0.499 | 0.500 | 0.692 | 0.624 | 0.475 |
| MS30 | 2.000 | 1.663 | 0.399 | 0.400 | 0.588 | 0.558 | 0.275 |
| MS36 | 2.000 | 1.956 | 0.489 | 0.650 | 0.682 | 0.505 | 0.425 |
| MS39 | 2.000 | 1.941 | 0.485 | 0.575 | 0.678 | 0.564 | 0.413 |
| MS51 | 2.000 | 1.311 | 0.237 | 0.175 | 0.400 | 0.366 | 0.138 |
| MS58 | 2.000 | 1.051 | 0.049 | 0.050 | 0.117 | 0.095 | 0.025 |
| MS59 | 2.000 | 1.600 | 0.375 | 0.300 | 0.562 | 0.540 | 0.250 |
| MS62 | 2.000 | 1.999 | 0.500 | 0.375 | 0.693 | 0.664 | 0.488 |
| MS72 | 2.000 | 1.980 | 0.495 | 0.450 | 0.688 | 0.641 | 0.450 |
| MS84 | 2.000 | 1.280 | 0.219 | 0.200 | 0.377 | 0.359 | 0.125 |
| MS87 | 2.000 | 1.835 | 0.455 | 0.300 | 0.647 | 0.620 | 0.350 |
| MS99 | 2.000 | 1.438 | 0.305 | 0.275 | 0.483 | 0.466 | 0.188 |
| MS105 | 2.000 | 1.980 | 0.495 | 0.450 | 0.688 | 0.641 | 0.450 |
| MS108 | 2.000 | 1.025 | 0.025 | 0.025 | 0.067 | 0.048 | 0.013 |
| MS111 | 2.000 | 1.568 | 0.362 | 0.375 | 0.548 | 0.526 | 0.238 |
| MS117 | 2.000 | 1.859 | 0.462 | 0.475 | 0.655 | 0.599 | 0.363 |
| MS129 | 2.000 | 1.632 | 0.387 | 0.275 | 0.576 | 0.549 | 0.263 |
| MS136 | 2.000 | 1.923 | 0.480 | 0.450 | 0.673 | 0.626 | 0.400 |
| MS138 | 2.000 | 1.980 | 0.495 | 0.350 | 0.688 | 0.661 | 0.450 |
| MS139 | 2.000 | 1.882 | 0.469 | 0.100 | 0.662 | 0.304 | 0.125 |
| MS153 | 2.000 | 1.250 | 0.200 | 0.075 | 0.352 | 0.266 | 0.113 |
| MS154 | 2.000 | 1.220 | 0.180 | 0.150 | 0.325 | 0.296 | 0.100 |
| MS155 | 2.000 | 1.753 | 0.430 | 0.275 | 0.621 | 0.591 | 0.313 |
| MS170 | 2.000 | 1.438 | 0.305 | 0.325 | 0.483 | 0.471 | 0.188 |
| MS172 | 2.000 | 1.250 | 0.200 | 0.225 | 0.352 | 0.349 | 0.113 |
| MS178 | 2.000 | 1.406 | 0.289 | 0.200 | 0.464 | 0.429 | 0.125 |
| MS179 | 2.000 | 1.280 | 0.219 | 0.250 | 0.377 | 0.375 | 0.175 |
| MS185 | 2.000 | 1.406 | 0.289 | 0.150 | 0.464 | 0.405 | 0.175 |
| MS194 | 2.000 | 1.406 | 0.289 | 0.250 | 0.464 | 0.445 | 0.125 |
| MS196 | 2.000 | 1.904 | 0.475 | 0.425 | 0.668 | 0.628 | 0.388 |
| MS205 | 2.000 | 1.471 | 0.320 | 0.250 | 0.500 | 0.476 | 0.200 |
| MS217 | 2.000 | 1.989 | 0.497 | 0.525 | 0.690 | 0.609 | 0.463 |
| MS225 | 2.000 | 1.374 | 0.272 | 0.275 | 0.444 | 0.139 | 0.038 |
| MS233 | 2.000 | 1.250 | 0.200 | 0.175 | 0.352 | 0.595 | 0.475 |
| Mean | 2.000 | 1.608 | 0.354 | 0.313 | 0.528 | 0.480 | 0.267 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, D.; Liao, J.; Zhang, K.; Zhang, J.; Qu, J.; Ma, G.; Li, J. Genetic Analysis and Fingerprint Construction for Thick-Skinned Melon (Cucumis melo subsp. melo) Based on InDel Markers. Plants 2025, 14, 3782. https://doi.org/10.3390/plants14243782
Ren D, Liao J, Zhang K, Zhang J, Qu J, Ma G, Li J. Genetic Analysis and Fingerprint Construction for Thick-Skinned Melon (Cucumis melo subsp. melo) Based on InDel Markers. Plants. 2025; 14(24):3782. https://doi.org/10.3390/plants14243782
Chicago/Turabian StyleRen, Dandan, Jinglei Liao, Keyan Zhang, Jiaying Zhang, Jingtao Qu, Guobin Ma, and Jufen Li. 2025. "Genetic Analysis and Fingerprint Construction for Thick-Skinned Melon (Cucumis melo subsp. melo) Based on InDel Markers" Plants 14, no. 24: 3782. https://doi.org/10.3390/plants14243782
APA StyleRen, D., Liao, J., Zhang, K., Zhang, J., Qu, J., Ma, G., & Li, J. (2025). Genetic Analysis and Fingerprint Construction for Thick-Skinned Melon (Cucumis melo subsp. melo) Based on InDel Markers. Plants, 14(24), 3782. https://doi.org/10.3390/plants14243782





