Lipid Metabolism and Actin Cytoskeleton Regulation Underlie Yield and Disease Resistance in Two Coffea canephora Breeding Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Analytical Design and Rationale
2.2. Experimental Populations and Data
2.3. Phenotypic and Genotypic Data Acquisition
- Production of coffee beans: Measured as the volume of mature fruit (“cherries”) harvested, expressed in 60 kg bags per hectare.
- Green bean yield: Measured as the weight (g) of processed, dried beans relative to the fresh harvest weight.
- Leaf rust incidence: Assessed visually using a 1–9 scale based on sporulation intensity, where 1 indicates absence of symptoms (resistant) and 9 indicates severe sporulation (highly susceptible). Scoring was performed during the period of high natural infection pressure to maximize discrimination between genotypes.
2.4. Single-SNP Association Analysis
2.5. Machine Learning for SNP Importance Analysis
2.5.1. Model Rationale and Implementation
2.5.2. Model Parameters and Variable Importance
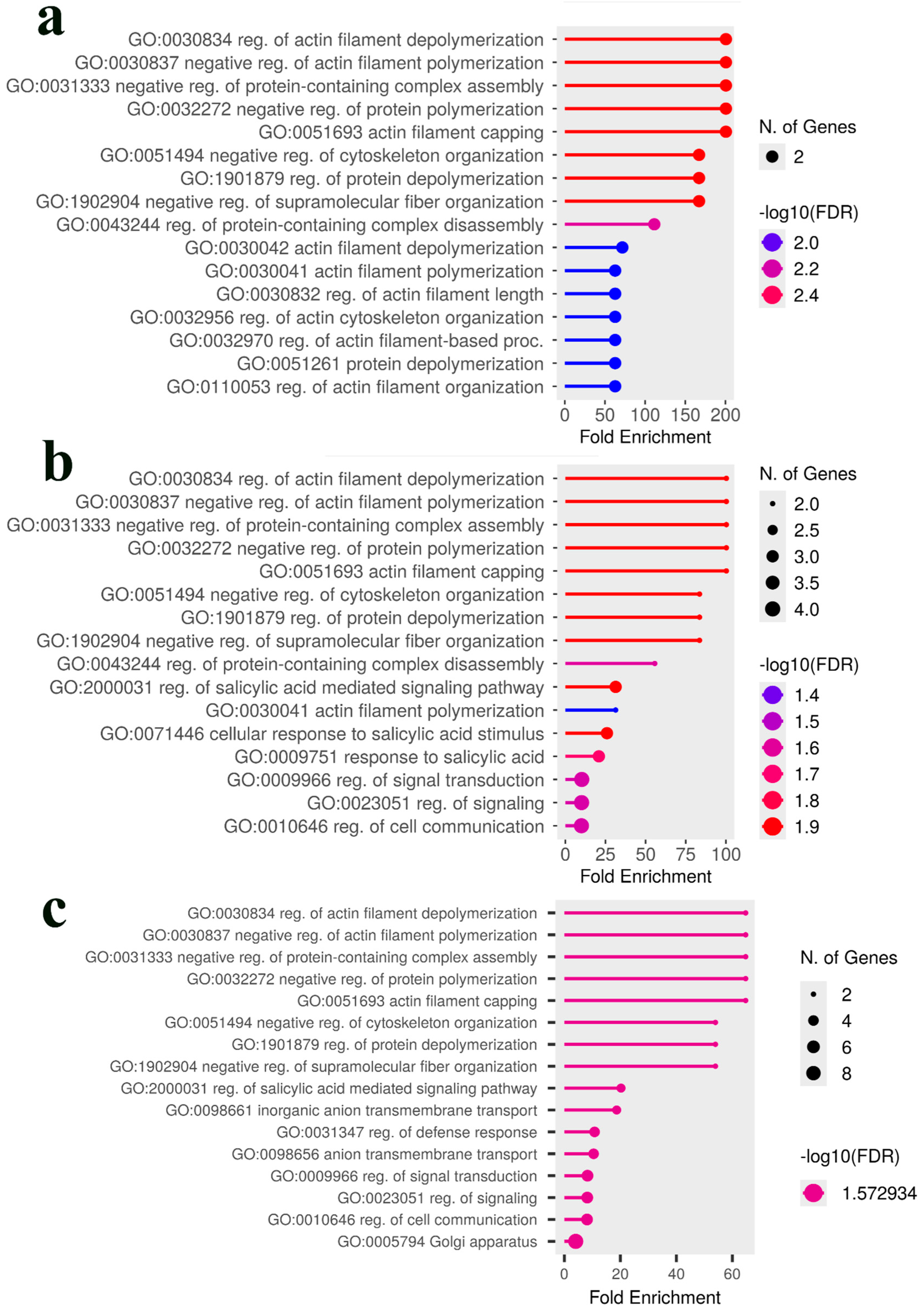
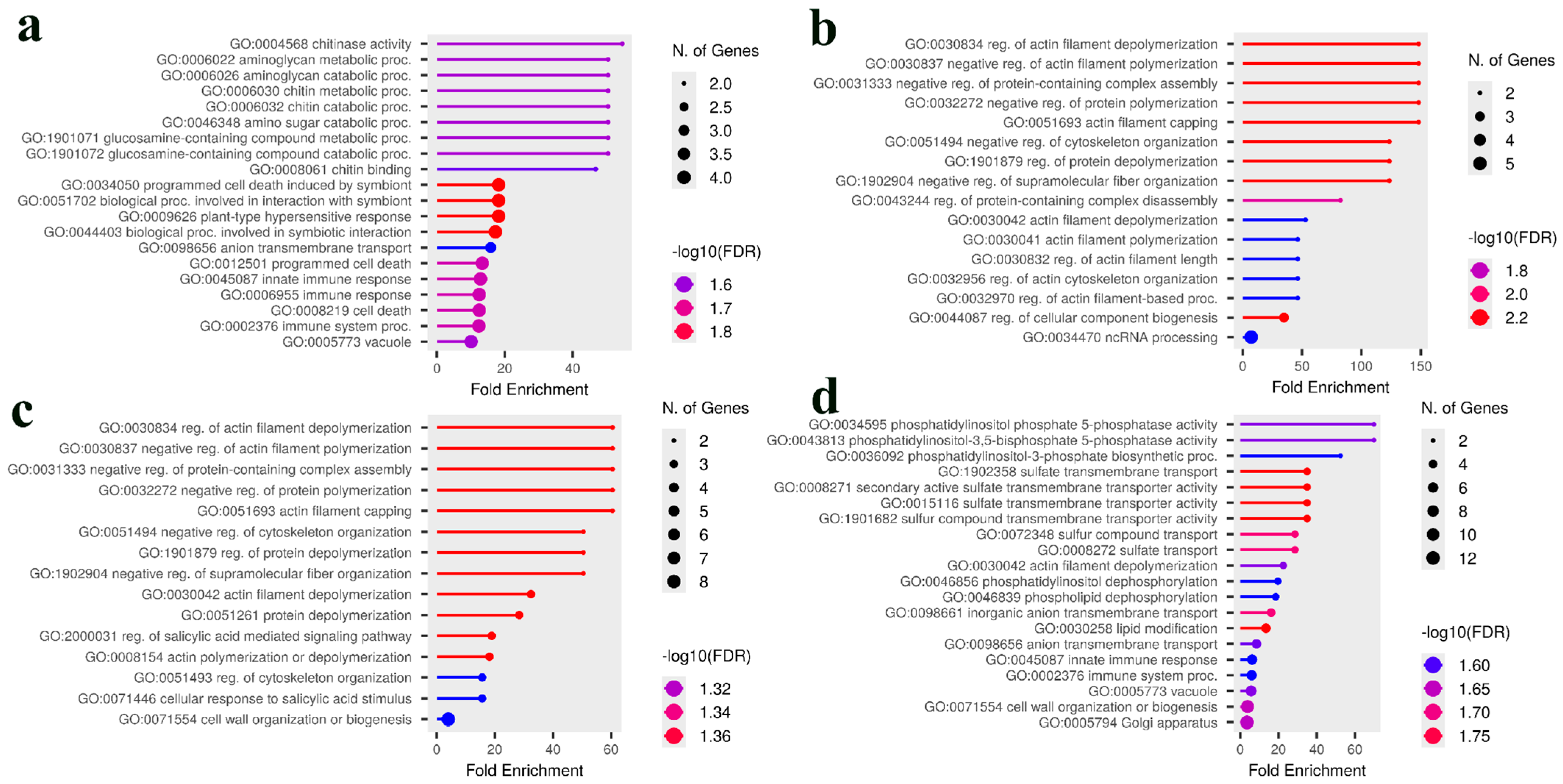
2.6. Candidate Gene Identification and Gene Ontology Enrichment Analysis
2.7. Reproducibility and Software Specifications
3. Results
3.1. Single-SNP Association Analysis of Agronomic Traits
3.2. Bootstrap Forest Analysis of SNP Importance for Agronomic Traits
3.3. Comparative Analysis and Identification of Consensus Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lashermes, P.; Andrade, A.C.; Etienne, H. Genomics of Coffee One of the World’s Largest Traded Commodities. Genom. Trop. Crop Plants 2008, 1, 203–226. [Google Scholar]
- Millet, C.P.; Delahaie, B.; Georget, F.; Allinne, C.; Solano-Sánchez, W.; Zhang, D.; Jeune, W.; Toniutti, L.; Poncet, V. Guadeloupe and Haiti’s Coffee Genetic Resources Reflect the Crop’s Regional and Global History. Plants People Planet 2025, 7, 245–262. [Google Scholar] [CrossRef]
- Campuzano-Duque, L.F.; Herrera, J.C.; Ged, C.; Blair, M.W. Bases for the Establishment of Robusta Coffee (Coffea canephora) as a New Crop for Colombia. Agronomy 2021, 11, 2550. [Google Scholar] [CrossRef]
- Capucho, A.; Zambolim, L.; Lopes, U.; Milagres, N. Chemical Control of Coffee Leaf Rust in Coffea canephora Cv. Conilon. Australas. Plant Pathol. 2013, 42, 667–673. [Google Scholar] [CrossRef]
- Mishra, M.K. Genetic Resources and Breeding of Coffee (Coffea spp.). In Advances in Plant Breeding Strategies: Nut and Beverage Crops: Volume 4; Springer: Berlin/Heidelberg, Germany, 2020; pp. 475–515. [Google Scholar]
- Merrick, L.F.; Herr, A.W.; Sandhu, K.S.; Lozada, D.N.; Carter, A.H. Optimizing Plant Breeding Programs for Genomic Selection. Agronomy 2022, 12, 714. [Google Scholar] [CrossRef]
- Chaves, S.F.; Dias, L.A.; Alves, R.S.; Ferreira, F.M.; Araújo, M.S.; Resende, M.D.; Takahashi, E.K.; Souza, J.E.; Leite, F.P.; Fernandes, S.B. Realized Genetic Gain with Reciprocal Recurrent Selection in a Eucalyptus Breeding Program. Tree Genet. Genomes 2024, 20, 47. [Google Scholar] [CrossRef]
- Vieira, R.A.; Nogueira, A.P.O.; Fritsche-Neto, R. Optimizing the Selection of Quantitative Traits in Plant Breeding Using Simulation. Front. Plant Sci. 2025, 16, 1495662. [Google Scholar] [CrossRef]
- Alemu, A.; Åstrand, J.; Montesinos-Lopez, O.A.; Y Sanchez, J.I.; Fernandez-Gonzalez, J.; Tadesse, W.; Vetukuri, R.R.; Carlsson, A.S.; Ceplitis, A.; Crossa, J. Genomic Selection in Plant Breeding: Key Factors Shaping Two Decades of Progress. Mol. Plant 2024, 17, 552–578. [Google Scholar] [CrossRef]
- Cabrera-Bosquet, L.; Crossa, J.; von Zitzewitz, J.; Serret, M.D.; Luis Araus, J. High-throughput Phenotyping and Genomic Selection: The Frontiers of Crop Breeding Converge F. J. Integr. Plant Biol. 2012, 54, 312–320. [Google Scholar] [CrossRef]
- Alkimim, E.R.; Caixeta, E.T.; Sousa, T.V.; Resende, M.D.V.; da Silva, F.L.; Sakiyama, N.S.; Zambolim, L. Selective Efficiency of Genome-Wide Selection in Coffea canephora Breeding. Tree Genet. Genomes 2020, 16, 41. [Google Scholar] [CrossRef]
- Paixão, P.T.M.; Nascimento, A.C.C.; Nascimento, M.; Azevedo, C.F.; Oliveira, G.F.; da Silva, F.L.; Caixeta, E.T. Factor Analysis Applied in Genomic Selection Studies in the Breeding of Coffea canephora. Euphytica 2022, 218, 42. [Google Scholar] [CrossRef] [PubMed]
- Ferrão, L.F.V.; Ferrão, R.G.; Ferrão, M.A.G.; Fonseca, A.; Carbonetto, P.; Stephens, M.; Garcia, A.A.F. Accurate Genomic Prediction of Coffea canephora in Multiple Environments Using Whole-Genome Statistical Models. Heredity 2019, 122, 261–275. [Google Scholar] [CrossRef]
- Ferrão, M.A.G.; Da Fonseca, A.F.; Volpi, P.S.; de Souza, L.C.; Comério, M.; Filho, A.C.V.; Riva-Souza, E.M.; Munoz, P.R.; Ferrão, R.G.; Ferrão, L.F.V. Genomic-assisted Breeding for Climate-smart Coffee. Plant Genome 2024, 17, e20321. [Google Scholar] [CrossRef]
- Morgante, M.; Salamini, F. From Plant Genomics to Breeding Practice. Curr. Opin. Biotechnol. 2003, 14, 214–219. [Google Scholar] [CrossRef]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The Molecular Genetics of Crop Domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef]
- Sun, L.; Lai, M.; Ghouri, F.; Nawaz, M.A.; Ali, F.; Baloch, F.S.; Nadeem, M.A.; Aasim, M.; Shahid, M.Q. Modern Plant Breeding Techniques in Crop Improvement and Genetic Diversity: From Molecular Markers and Gene Editing to Artificial Intelligence—A Critical Review. Plants 2024, 13, 2676. [Google Scholar] [CrossRef]
- Klimberg, R. Fundamentals of Predictive Analytics with JMP; SAS Institute: Cary, NC, USA, 2023; ISBN 1-68580-001-7. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Basu, S.; Kumbier, K.; Brown, J.B.; Yu, B. Iterative Random Forests to Discover Predictive and Stable High-Order Interactions. Proc. Natl. Acad. Sci. USA 2018, 115, 1943–1948. [Google Scholar] [CrossRef]
- Dereeper, A.; Bocs, S.; Rouard, M.; Guignon, V.; Ravel, S.; Tranchant-Dubreuil, C.; Poncet, V.; Garsmeur, O.; Lashermes, P.; Droc, G. The Coffee Genome Hub: A Resource for Coffee Genomes. Nucleic Acids Res. 2015, 43, D1028–D1035. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G. The Coffee Genome Provides Insight into the Convergent Evolution of Caffeine Biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY Transcription Factors in Plant Responses to Stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY Transcription Factors and Plant Defense Responses: Latest Discoveries and Future Prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Duplan, V.; Rivas, S. E3 Ubiquitin-Ligases and Their Target Proteins during the Regulation of Plant Innate Immunity. Front. Plant Sci. 2014, 5, 42. [Google Scholar] [CrossRef]
- Adunola, P.; Ferrão, M.A.G.; Ferrão, R.G.; Da Fonseca, A.F.; Volpi, P.S.; Comério, M.; Verdin Filho, A.C.; Munoz, P.R.; Ferrão, L.F.V. Genomic Selection for Genotype Performance and Environmental Stability in Coffea canephora. G3 Genes Genomes Genet. 2023, 13, jkad062. [Google Scholar] [CrossRef]
- Bernardo, R. Molecular Markers and Selection for Complex Traits in Plants: Learning from the Last 20 Years. Crop Sci. 2008, 48, 1649–1664. [Google Scholar] [CrossRef]
- Heffner, E.L.; Sorrells, M.E.; Jannink, J.-L. Genomic Selection for Crop Improvement. Crop Sci. 2009, 49, 1–12. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler IV, E.S. Structure of Linkage Disequilibrium in Plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Yu, J.; Buckler, E.S. Genetic Association Mapping and Genome Organization of Maize. Curr. Opin. Biotechnol. 2006, 17, 155–160. [Google Scholar] [CrossRef]
- Wright, S.I.; Ness, R.W.; Foxe, J.P.; Barrett, S.C. Genomic Consequences of Outcrossing and Selfing in Plants. Int. J. Plant Sci. 2008, 169, 105–118. [Google Scholar] [CrossRef]
- Casler, M. Agricultural Fitness of Smooth Bromegrass Populations Selected for Divergent Fiber Concentration. Crop Sci. 2005, 45, 36–43. [Google Scholar] [CrossRef]
- Roncallo, P.F.; Larsen, A.O.; Achilli, A.L.; Pierre, C.S.; Gallo, C.A.; Dreisigacker, S.; Echenique, V. Linkage Disequilibrium Patterns, Population Structure and Diversity Analysis in a Worldwide Durum Wheat Collection Including Argentinian Genotypes. BMC Genom. 2021, 22, 233. [Google Scholar] [CrossRef]
- Ladizinsky, G. Founder Effect in Crop-Plant Evolution. Econ. Bot. 1985, 39, 191–199. [Google Scholar] [CrossRef]
- Pelc, S.E.; Couillard, D.M.; Stansell, Z.J.; Farnham, M.W. Genetic Diversity and Population Structure of Collard Landraces and Their Relationship to Other Brassica Oleracea Crops. Plant Genome 2015, 8, plantgenome2015.04.0023. [Google Scholar] [CrossRef]
- Hedrich, R.; Sauer, N.; Neuhaus, H.E. Sugar Transport across the Plant Vacuolar Membrane: Nature and Regulation of Carrier Proteins. Curr. Opin. Plant Biol. 2015, 25, 63–70. [Google Scholar] [CrossRef]
- Takatsuka, H.; Higaki, T.; Ito, M. At the Nexus between Cytoskeleton and Vacuole: How Plant Cytoskeletons Govern the Dynamics of Large Vacuoles. Int. J. Mol. Sci. 2023, 24, 4143. [Google Scholar] [CrossRef]
- Wingenter, K.; Schulz, A.; Wormit, A.; Wic, S.; Trentmann, O.; Hoermiller, I.I.; Heyer, A.G.; Marten, I.; Hedrich, R.; Neuhaus, H.E. Increased Activity of the Vacuolar Monosaccharide Transporter TMT1 Alters Cellular Sugar Partitioning, Sugar Signaling, and Seed Yield in Arabidopsis. Plant Physiol. 2010, 154, 665–677. [Google Scholar] [CrossRef]
- Suzuki, T.; Ashihara, H.; Waller, G.R. Purine and Purine Alkaloid Metabolism in Camellia and Coffea Plants. Phytochemistry 1992, 31, 2575–2584. [Google Scholar] [CrossRef]
- Ashihara, H.; Sano, H.; Crozier, A. Caffeine and Related Purine Alkaloids: Biosynthesis, Catabolism, Function and Genetic Engineering. Phytochemistry 2008, 69, 841–856. [Google Scholar] [CrossRef]
- Kato, M.; Kitao, N.; Ishida, M.; Morimoto, H.; Irino, F.; Mizuno, K. Expression for Caffeine Biosynthesis and Related Enzymes in Camellia Sinensis. Z. Für Naturforschung C 2010, 65, 245–256. [Google Scholar] [CrossRef]
- Fu, X.; Li, G.; Hu, F.; Huang, J.; Lou, Y.; Li, Y.; Li, Y.; He, H.; Lv, Y.; Cheng, J. Comparative Transcriptome Analysis in Peaberry and Regular Bean Coffee to Identify Bean Quality Associated Genes. BMC Genom. Data 2023, 24, 12. [Google Scholar] [CrossRef]
- Wasteneys, G.O.; Galway, M.E. Remodeling the Cytoskeleton for Growth and Form: An Overview with Some New Views. Annu. Rev. Plant Biol. 2003, 54, 691–722. [Google Scholar] [CrossRef]
- Pokotylo, I.; Pejchar, P.; Potocký, M.; Kocourková, D.; Krčková, Z.; Ruelland, E.; Kravets, V.; Martinec, J. The Plant Non-Specific Phospholipase C Gene Family. Novel Competitors in Lipid Signalling. Prog. Lipid Res. 2013, 52, 62–79. [Google Scholar] [CrossRef]
- Cai, G.; Fan, C.; Liu, S.; Yang, Q.; Liu, D.; Wu, J.; Li, J.; Zhou, Y.; Guo, L.; Wang, X. Nonspecific Phospholipase C6 Increases Seed Oil Production in Oilseed Brassicaceae Plants. New Phytol. 2020, 226, 1055–1073. [Google Scholar] [CrossRef]
- Nakamura, Y.; Ngo, A.H. Non-Specific Phospholipase C (NPC): An Emerging Class of Phospholipase C in Plant Growth and Development. J. Plant Res. 2020, 133, 489–497. [Google Scholar] [CrossRef]
- Blatch, G.L.; Lässle, M. The Tetratricopeptide Repeat: A Structural Motif Mediating Protein-protein Interactions. BIOEEJ 1999, 21, 932–939. [Google Scholar] [CrossRef]
- Schapire, A.L.; Valpuesta, V.; Botella, M.A. TPR Proteins in Plant Hormone Signaling. Plant Signal. Behav. 2006, 1, 229–230. [Google Scholar] [CrossRef]
- Schlegel, T.; Mirus, O.; Von Haeseler, A.; Schleiff, E. The Tetratricopeptide Repeats of Receptors Involved in Protein Translocation across Membranes. Mol. Biol. Evol. 2007, 24, 2763–2774. [Google Scholar] [CrossRef]
- Zeytuni, N.; Zarivach, R. Structural and Functional Discussion of the Tetra-Trico-Peptide Repeat, a Protein Interaction Module. Structure 2012, 20, 397–405. [Google Scholar] [CrossRef]
- Zhang, T.; Meng, L.; Kong, W.; Yin, Z.; Wang, Y.; Schneider, J.D.; Chen, S. Quantitative Proteomics Reveals a Role of JAZ7 in Plant Defense Response to Pseudomonas Syringae DC3000. J. Proteom. 2018, 175, 114–126. [Google Scholar] [CrossRef]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of Salicylic Acid for the Induction of Systemic Acquired Resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.A. Systemic Acquired Resistance and Salicylic Acid: Past, Present, and Future. Mol. Plant. Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, G.; Pandharikar, G.; Frendo, P. Salicylic Acid in Plant Symbioses: Beyond Plant Pathogen Interactions. Biology 2022, 11, 861. [Google Scholar] [CrossRef]
- Shariatipour, N.; Yazdani, M.; Carlsson, A.; Bengtsson, T.; Kianian, S.F.; Jalli, M.; Rahmatov, M.; PPP RobOat Consortium. Genetic Dissection of Crown Rust Resistance in Oat and the Identification of Key Adult Plant Resistance Genes. Plant Genome 2025, 18, e70059. [Google Scholar] [CrossRef]
- Mu, X.; Luo, J. Evolutionary Analyses of NIN-like Proteins in Plants and Their Roles in Nitrate Signaling. Cell. Mol. Life Sci. 2019, 76, 3753–3764. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Crute, I.R.; Holub, E.B.; Beynon, J.L. RPP13 Is a Simple Locus in Arabidopsis Thaliana for Alleles That Specify Downy Mildew Resistance to Different Avirulence Determinants in Peronospora Parasitica. Plant J. 2000, 21, 177–188. [Google Scholar] [CrossRef]
- Bachlava, E.; Radwan, O.E.; Abratti, G.; Tang, S.; Gao, W.; Heesacker, A.F.; Bazzalo, M.E.; Zambelli, A.; Leon, A.J.; Knapp, S.J. Downy Mildew (Pl 8 and Pl 14) and Rust (R Adv) Resistance Genes Reside in Close Proximity to Tandemly Duplicated Clusters of Non-TIR-like NBS-LRR-Encoding Genes on Sunflower Chromosomes 1 and 13. Theor. Appl. Genet. 2011, 122, 1211–1221. [Google Scholar] [CrossRef]
- Bish, M.D.; Ramachandran, S.R.; Wright, A.; Lincoln, L.M.; Whitham, S.A.; Graham, M.A.; Pedley, K.F. The Soybean Rpp3 Gene Encodes a TIR-NBS-LRR Protein That Confers Resistance to Phakopsora pachyrhizi. Mol. Plant. Microbe Interact. 2024, 37, 561–570. [Google Scholar] [CrossRef]
- Yuan, B.; Li, C.; Wang, Q.; Yao, Q.; Guo, X.; Zhang, Y.; Wang, Z. Identification and Functional Characterization of the RPP13 Gene Family in Potato (Solanum tuberosum L.) for Disease Resistance. Front. Plant Sci. 2025, 15, 1515060. [Google Scholar] [CrossRef]
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.; Krasileva, K.V. Comparative Analysis of Plant Immune Receptor Architectures Uncovers Host Proteins Likely Targeted by Pathogens. BMC Biol. 2016, 14, 8. [Google Scholar] [CrossRef]
- Chandra, S.; Kazmi, A.Z.; Ahmed, Z.; Roychowdhury, G.; Kumari, V.; Kumar, M.; Mukhopadhyay, K. Genome-Wide Identification and Characterization of NB-ARC Resistant Genes in Wheat (Triticum aestivum L.) and Their Expression during Leaf Rust Infection. Plant Cell Rep. 2017, 36, 1097–1112. [Google Scholar] [CrossRef]
- Dubey, N.; Singh, K. Role of NBS-LRR Proteins in Plant Defense. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; pp. 115–138. [Google Scholar]
- Wang, J.; Chen, T.; Han, M.; Qian, L.; Li, J.; Wu, M.; Han, T.; Cao, J.; Nagalakshmi, U.; Rathjen, J.P. Plant NLR Immune Receptor Tm-22 Activation Requires NB-ARC Domain-Mediated Self-Association of CC Domain. PLoS Pathog. 2020, 16, e1008475. [Google Scholar] [CrossRef]
- Zipfel, C. Early Molecular Events in PAMP-Triggered Immunity. Curr. Opin. Plant Biol. 2009, 12, 414–420. [Google Scholar] [CrossRef]
- García, Y.H.; Zamora, O.R.; Troncoso-Rojas, R.; Tiznado-Hernández, M.E.; Báez-Flores, M.E.; Carvajal-Millan, E.; Rascón-Chu, A. Toward Understanding the Molecular Recognition of Fungal Chitin and Activation of the Plant Defense Mechanism in Horticultural Crops. Molecules 2021, 26, 6513. [Google Scholar] [CrossRef] [PubMed]
- Fan, A.; Wei, L.; Zhang, X.; Liu, J.; Sun, L.; Xiao, J.; Wang, Y.; Wang, H.; Hua, J.; Singh, R.P. Heterologous Expression of the Haynaldia villosa Pattern-Recognition Receptor CERK1-V in Wheat Increases Resistance to Three Fungal Diseases. Crop J. 2022, 10, 1733–1745. [Google Scholar] [CrossRef]
- Wang, L.; He, Y.; Guo, G.; Xia, X.; Dong, Y.; Zhang, Y.; Wang, Y.; Fan, X.; Wu, L.; Zhou, X. Overexpression of Plant Chitin Receptors in Wheat Confers Broad-spectrum Resistance to Fungal Diseases. Plant J. 2024, 120, 1047–1063. [Google Scholar] [CrossRef]
- Daryani, P.; Darzi Ramandi, H.; Dezhsetan, S.; Mirdar Mansuri, R.; Hosseini Salekdeh, G.; Shobbar, Z.-S. Pinpointing Genomic Regions Associated with Root System Architecture in Rice through an Integrative Meta-Analysis Approach. Theor. Appl. Genet. 2022, 135, 81–106. [Google Scholar] [CrossRef]
- Yin, X.; Bose, D.; Kwon, A.; Hanks, S.C.; Jackson, A.U.; Stringham, H.M.; Welch, R.; Oravilahti, A.; Silva, L.F.; Locke, A.E. Integrating Transcriptomics, Metabolomics, and GWAS Helps Reveal Molecular Mechanisms for Metabolite Levels and Disease Risk. Am. J. Hum. Genet. 2022, 109, 1727–1741. [Google Scholar] [CrossRef]
- DeHaan, L.R.; Van Tassel, D.L. Useful Insights from Evolutionary Biology for Developing Perennial Grain Crops. Am. J. Bot. 2014, 101, 1801–1819. [Google Scholar] [CrossRef]
- Álvarez, M.F.; Mosquera, T.; Blair, M.W. The Use of Association Genetics Approaches in Plant Breeding. Plant Breed. Rev. 2014, 38, 17–68. [Google Scholar]
- Silva, L.F. Estudo de Associação Genômica Ampla (GWAS) em Coffea canephora. Master’s Thesis, Universidade Federal de Viçosa, Viçosa, Brazil, 2018. [Google Scholar]
- de Faria Silva, L.; Alkimim, E.R.; Barreiro, P.R.R.M.; Leichtweis, B.G.; Silva, A.C.A.; da Silva, R.A.; Sousa, T.V.; Nascimento, M.; Caixeta, E.T. Genome-Wide Association Study of Plant Architecture and Diseases Resistance in Coffea canephora. Euphytica 2022, 218, 92. [Google Scholar] [CrossRef]
- Paape, T.; Heiniger, B.; Santo Domingo, M.; Clear, M.R.; Lucas, M.M.; Pueyo, J.J. Genome-Wide Association Study Reveals Complex Genetic Architecture of Cadmium and Mercury Accumulation and Tolerance Traits in Medicago truncatula. Front. Plant Sci. 2022, 12, 806949. [Google Scholar] [CrossRef] [PubMed]
- Altaf, M.T.; Liaqat, W.; Jamil, A.; Mohamed, H.I.; Fahad, M.; Jan, M.F.; Baloch, F.S. A Critical Review: Breeding Objectives, Genomic Resources, and Marker-Assisted Methods in Sorghum (Sorghum bicolor L.). J. Soil Sci. Plant Nutr. 2024, 24, 4597–4623. [Google Scholar] [CrossRef]
- Kumar, R.; Das, S.P.; Choudhury, B.U.; Kumar, A.; Prakash, N.R.; Verma, R.; Chakraborti, M.; Devi, A.G.; Bhattacharjee, B.; Das, R. Advances in Genomic Tools for Plant Breeding: Harnessing DNA Molecular Markers, Genomic Selection, and Genome Editing. Biol. Res. 2024, 57, 80. [Google Scholar] [CrossRef] [PubMed]
- Werner, C.R.; Gaynor, R.C.; Gorjanc, G.; Hickey, J.M.; Kox, T.; Abbadi, A.; Leckband, G.; Snowdon, R.J.; Stahl, A. How Population Structure Impacts Genomic Selection Accuracy in Cross-Validation: Implications for Practical Breeding. Front. Plant Sci. 2020, 11, 592977. [Google Scholar] [CrossRef]
- Dekkers, J.C.; Su, H.; Cheng, J. Predicting the Accuracy of Genomic Predictions. Genet. Sel. Evol. 2021, 53, 55. [Google Scholar] [CrossRef]
- Schrauf, M.F.; de Los Campos, G.; Munilla, S. Comparing Genomic Prediction Models by Means of Cross Validation. Front. Plant Sci. 2021, 12, 734512. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, R.J.; Michno, J.-M.; Jeffers, J.; Hoekenga, O.; Dilkes, B.; Baxter, I.; Myers, C.L. Integrating Coexpression Networks with GWAS to Prioritize Causal Genes in Maize. Plant Cell 2018, 30, 2922–2942. [Google Scholar] [CrossRef]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef]
- Thomson, M.J.; Biswas, S.; Tsakirpaloglou, N.; Septiningsih, E.M. Functional Allele Validation by Gene Editing to Leverage the Wealth of Genetic Resources for Crop Improvement. Int. J. Mol. Sci. 2022, 23, 6565. [Google Scholar] [CrossRef] [PubMed]
- Tsakirpaloglou, N.; Septiningsih, E.M.; Thomson, M.J. Guidelines for Performing CRISPR/Cas9 Genome Editing for Gene Validation and Trait Improvement in Crops. Plants 2023, 12, 3564. [Google Scholar] [CrossRef] [PubMed]
- Sahito, J.H.; Zhang, H.; Gishkori, Z.G.N.; Ma, C.; Wang, Z.; Ding, D.; Zhang, X.; Tang, J. Advancements and Prospects of Genome-Wide Association Studies (GWAS) in Maize. Int. J. Mol. Sci. 2024, 25, 1918. [Google Scholar] [CrossRef] [PubMed]
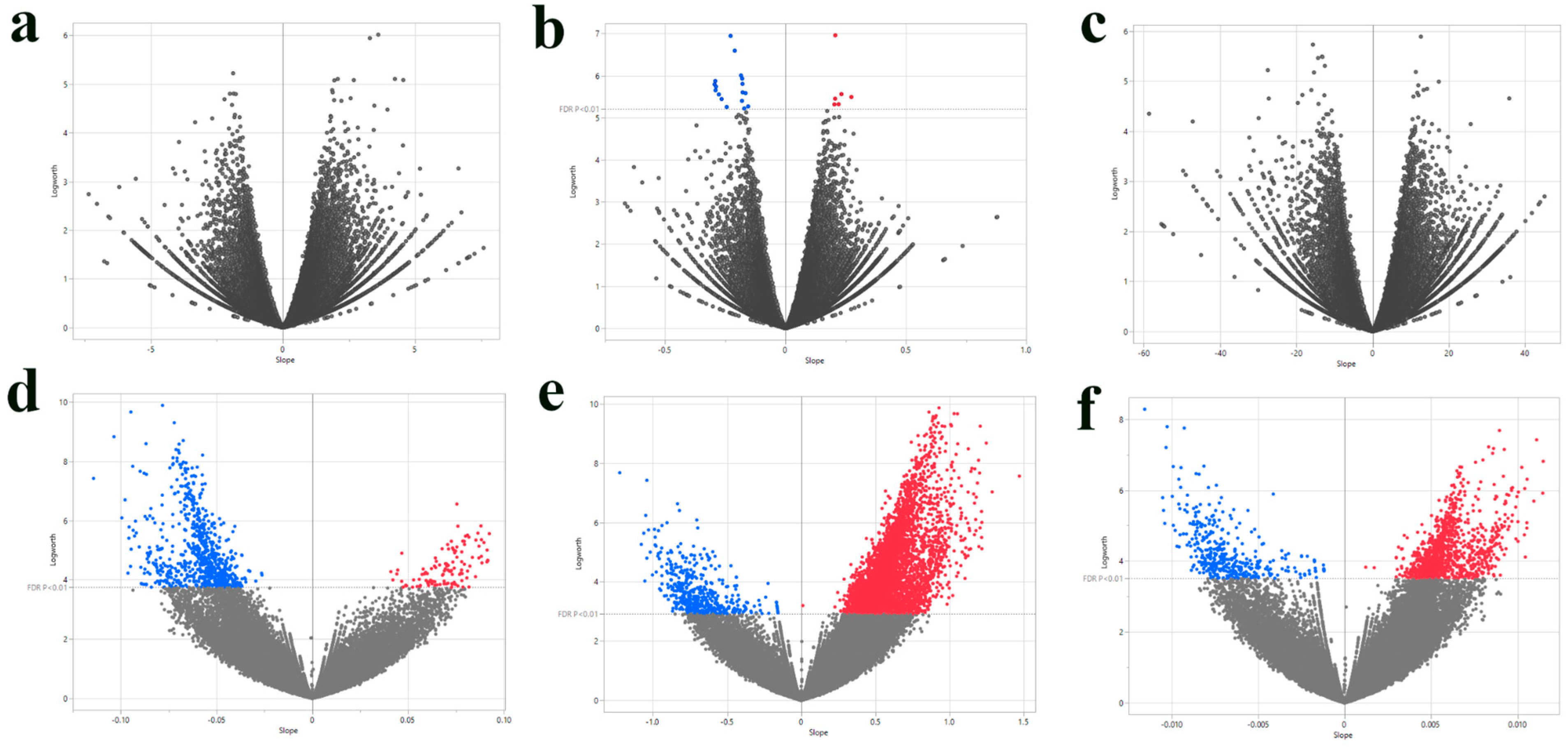
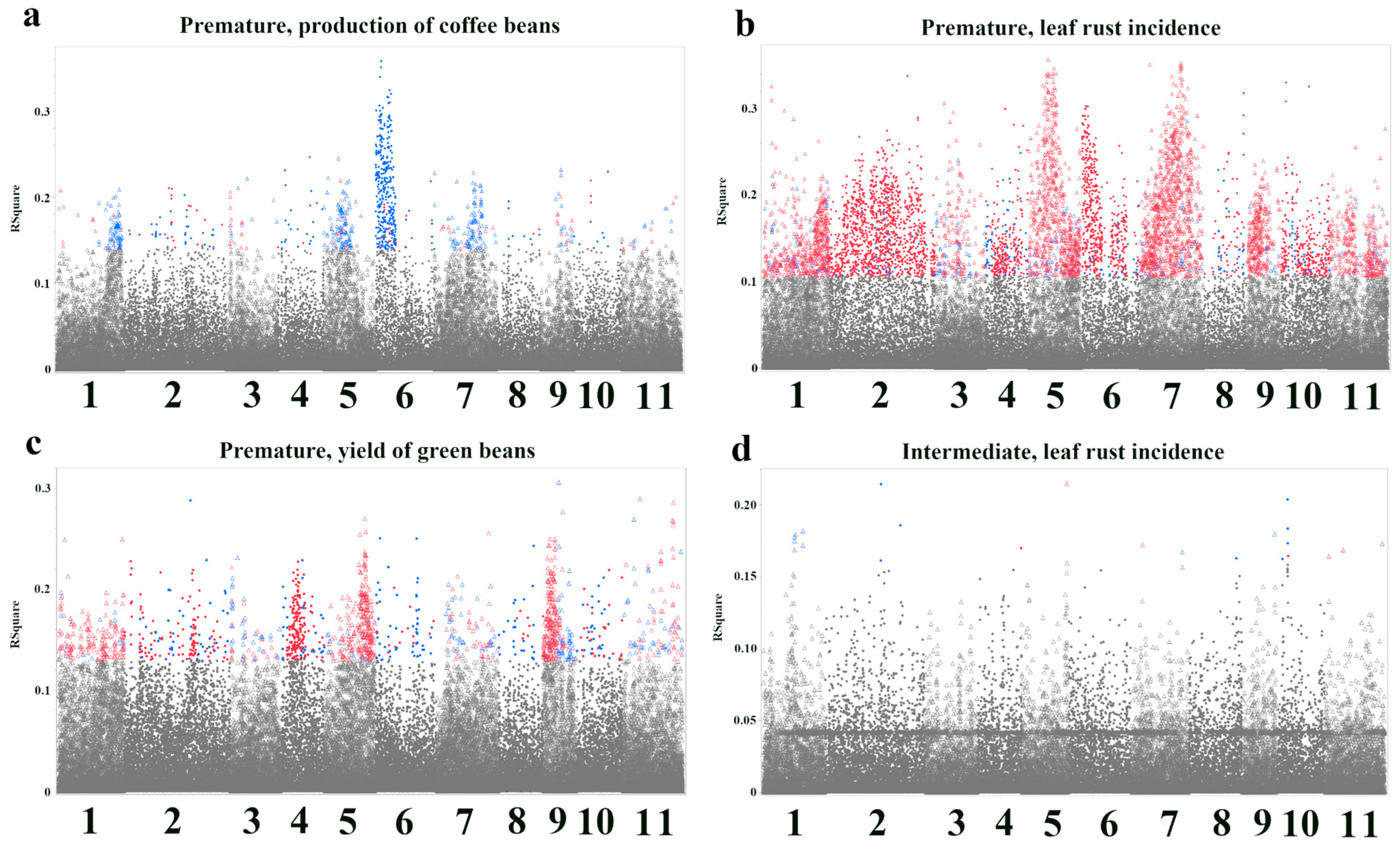

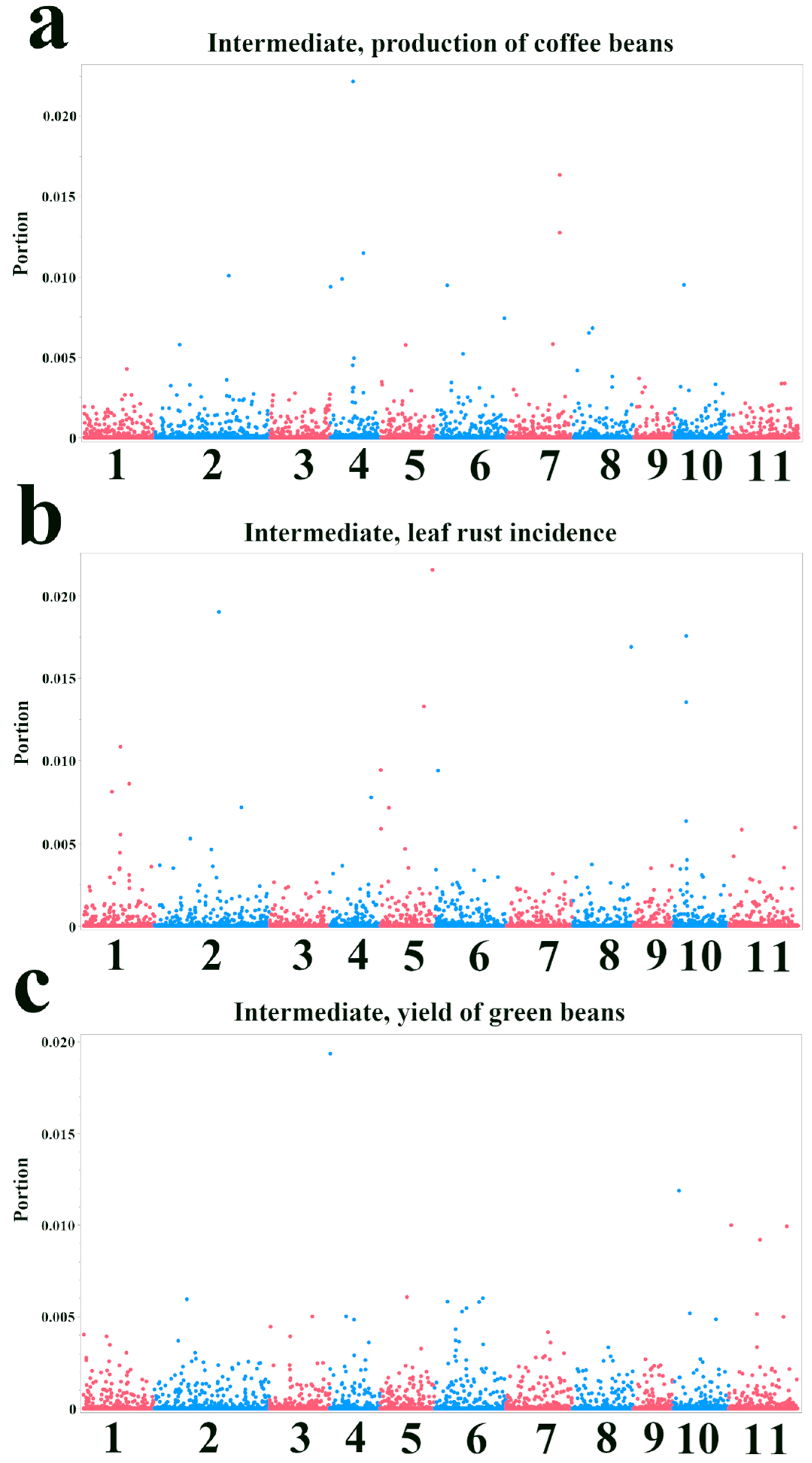
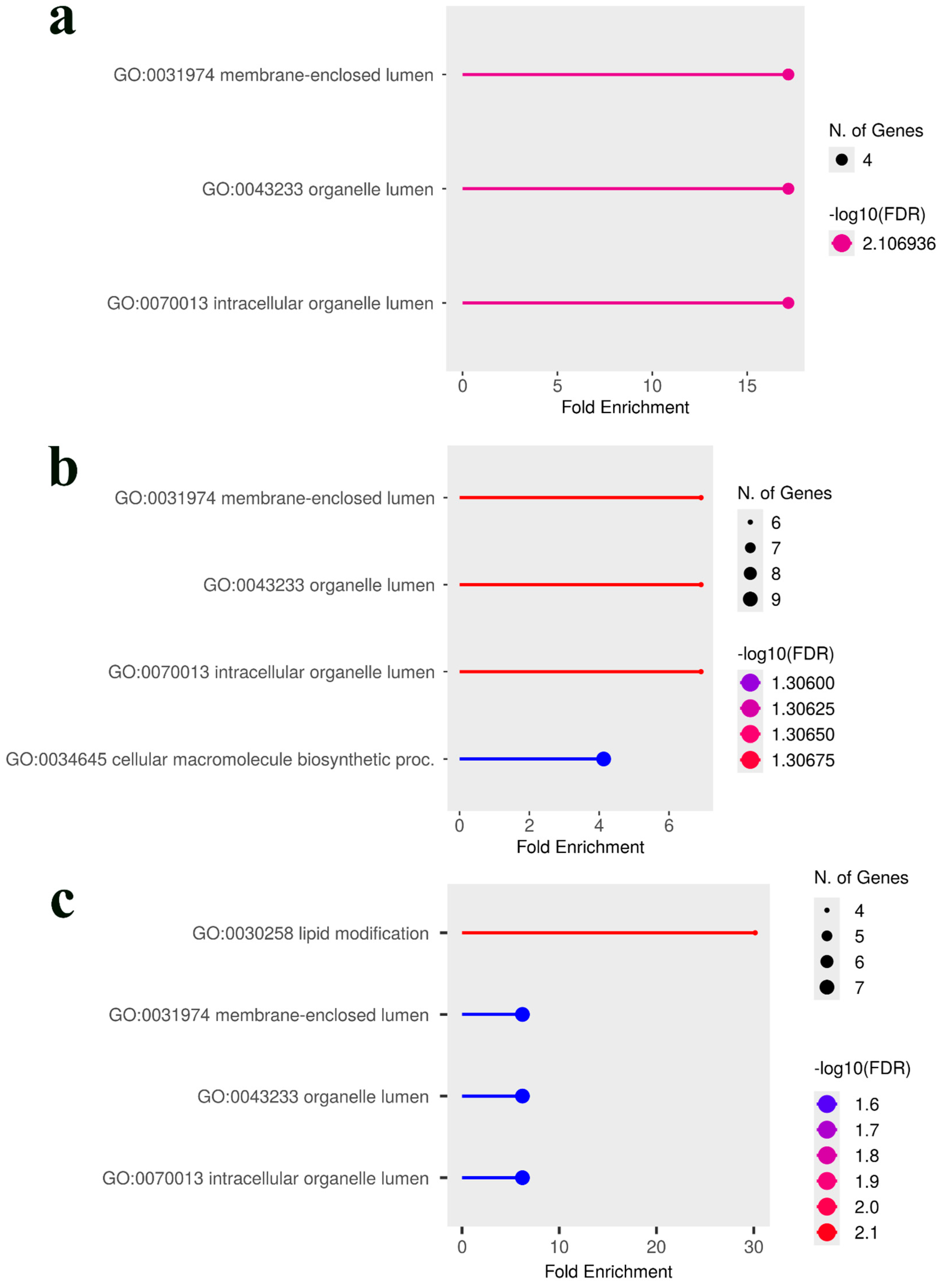

| SNP ID | Nearest Gene and Function | Base Pairs Away | FDR p-Value | Effect Size | R-Square |
|---|---|---|---|---|---|
| Premature, production of coffee beans—Response Screening | |||||
| 6.2295851 | Cc06t02920.1 ADF-H domain-containing protein | 0 | 2.12 × 10−10 | 0.49 | 0.35 |
| 6.2378930 | Cc06t03050.1 IPPc domain-containing protein | 0 | 1.27 × 10−10 | 0.49 | 0.36 |
| 6.2075614 | Cc06t02620.1 E3 ubiquitin-protein ligase | 0 | 0.00000000049 | 0.48 | 0.34 |
| 6.4097514 | Cc06t05200.1 NPL domain-containing protein | −29 | 0.0000000025 | 0.46 | 0.32 |
| 6.4477759 | Cc06t05570.1 IU_nuc | 0 | 0.0000000026 | 0.47 | 0.32 |
| 4.15969587 | Cc04t13140.1 UDP-glycosyltransferase 83A1 | −25,832 | 0.00000027 | 0.41 | 0.25 |
| 10.6890851 | Cc10t07860.1 GH10 domain-containing protein | 0 | 0.0000015 | 0.39 | 0.22 |
| 5.13748592 | Cc05t03040.1 Putative Short-chain dehydrogenase reductase ATA1 | −47,435 | 0.0000015 | 0.39 | 0.22 |
| 2.17352904 2.17352905 | Cc02t19180.1 Stress enhanced protein 1, chloroplastic | 0 | 0.0000026 | 0.38 | 0.21 |
| 2.18145802 | Cc02t20300.1 SCP domain-containing protein | +3008 | 0.0000027 | 0.38 | 0.21 |
| Premature, leaf rust incidence—Response Screening | |||||
| 7.15362801 | Cc07t18070.1 Hexosyltransferase | −1304 | 0.000000021 | 0.54 | 0.29 |
| 11.32751026 | Cc11t16690.1 Urease | 0 | 0.000000038 | 0.53 | 0.28 |
| 4.20658374 | Cc04t14270.1 Putative disease resistance RPP13-like protein 3 | −32,177 | 0.00000023 | 0.5 | 0.25 |
| 3.12306478 | Cc03t09860.1 NB-ARCdomain-containing protein | 0 | 0.00000039 | 0.49 | 0.24 |
| 7.13312636 | Cc07t16380.1 Conserved hypothetical protein | +1562 | 0.00000058 | 0.49 | 0.23 |
| 2.35607328 | Cc02t30380.1 Peroxidase | 0 | 0.0000014 | 0.59 | 0.34 |
| 5.13342765 | Cc05t02930.1 TAF domain-containing protein | +27,160 | 0.0000014 | 0.59 | 0.34 |
| 5.14130887 | Cc05t03220.1 Lycopene beta/epsilon cyclase protein | −3241 | 0.0000014 | 0.59 | 0.34 |
| 5.14494464 5.14494471 5.14494484 | Cc05t03270.1 AT1G05060.1 | 0 | 0.0000014 | 0.58 | 0.34 |
| 5.14625364 | Cc05t03340.1 Chitin elicitor receptor kinase 1 | 0 | 0.0000014 | 0.58 | 0.34 |
| Premature, yield of green beans—Response Screening | |||||
| 11.12510898 | Cc11t03410.1 Protein of unknown function (DUF789) | +10,726 | 0.000000016 | 0.52 | 0.29 |
| 2.24864284 | Cc02t27160.1 Vicianin hydrolase | 0 | 0.000000018 | 0.52 | 0.29 |
| 9.6114101 | Cc09t05750.1 T-complex protein 1 subunit gamma | 0 | 0.0000000052 | 0.54 | 0.31 |
| 11.7875427 | Cc11t02410.1 RING-type domain-containing protein | 0 | 0.000000063 | 0.5 | 0.27 |
| 1.3497181 | Cc01t01920.1 SKP1-like protein 4 | +5743 | 0.00000023 | 0.48 | 0.25 |
| 11.30714537 | Cc11t13980.1 HDAC_interact domain-containing protein | 0 | 0.000000021 | 0.52 | 0.29 |
| 9.8293367 | Cc09t06990.1 Putative caffeine synthase 3 | 0 | 0.000000038 | 0.51 | 0.28 |
| 11.30518300 | Cc11t13720.1 GSDH domain-containing protein | 0 | 0.000000067 | 0.5 | 0.27 |
| 11.30697733 | Cc11t13960.1 TORTIFOLIA1-like protein 4 | 0 | 0.000000071 | 0.5 | 0.27 |
| 5.26247026 | Cc05t12380.1 Transducin/WD40 repeat-like superfamily protein | −3088 | 0.00000006 | 0.5 | 0.27 |
| Intermediate, leaf rust incidence—Response Screening | |||||
| 2.22416916 | Cc02t25100.1 Nitrate regulatory gene 2 protein | 0 | 0.00000011 | 0.56 | 0.21 |
| 10.3678570 | Cc10t04730.1 C2H2-type domain-containing protein | 0 | 0.00000026 | 0.54 | 0.2 |
| 10.3747377 | Cc10t04810.1 WRKY domain-containing protein | 0 | 0.0000012 | 0.52 | 0.18 |
| 1.26450374 1.26450396 | Cc01t08110.1 Putative late blight resistance protein homolog R1B-16 | 0 | 0.0000016 | 0.51 | 0.18 |
| 1.29976260 1.29976261 1.29976262 | Cc01t11280.1 Conserved hypothetical protein | −2741 | 0.0000013 | 0.51 | 0.18 |
| 5.28610373 | Cc05t15840.1 TPR_REGION domain-containing protein | 0 | 0.00000011 | 0.56 | 0.21 |
| 7.3420157 | Cc07t04860.1 AAA domain-containing protein | 0 | 0.0000027 | 0.50 | 0.17 |
| 11.13698759 11.13698762 | Cc11t03510.1 RING-type domain-containing protein | −337,600 | 0.0000035 | 0.50 | 0.17 |
| 4.27729192 | Cc04t17060.1 BHLH domain-containing protein | −85 | 0.0000032 | 0.50 | 0.17 |
| 10.3876532 | Cc10t04930.1 SASA domain-containing protein | −2641 | 0.0000048 | 0.49 | 0.16 |
| SNP ID | Nearest Gene and Function | Base Pairs Away | Importance Score (Portion) |
|---|---|---|---|
| Premature, production of coffee beans—Bootstrap Forest | |||
| 6.4939167 | Cc06t06270.1 Alpha/beta-Hydrolases superfamily protein | 0 | 0.037 |
| 6.2378930 | Cc06t03050.1 IPPc domain-containing protein | +172 | 0.026 |
| 1.2020393 | Cc01t01290.1 Putative 60S ribosomal protein L23a-1 | +6605 | 0.019 |
| 7.15086633 | Cc07t17840.1 NB-ARC domain-containing protein | +19 | 0.015 |
| 6.1079450 | Cc06t01300.1 HEAT repeat-containing protein | 0 | 0.013 |
| Premature, leaf rust incidence—Bootstrap Forest | |||
| 7.12204503 | Cc07t15410.1 Acyl-coenzyme A oxidase | +308 | 0.2 |
| 8.21181446 | Cc08t07800.1 Hydroxyproline-rich glycoprotein family protein | 0 | 0.07 |
| 5.13342765 | Cc05t02930.1 TAF domain-containing protein | +27,160 | 0.06 |
| 1.32477500 | Cc01t14390.1 Increased DNA methylation like | 0 | 0.06 |
| 10.1733570 | Cc10t02280.1 OBG-type G domain-containing protein | 0 | 0.056 |
| Premature, yield of green beans- Bootstrap Forest | |||
| 11.30697733 | Cc11t13960.1 TORTIFOLIA1-like protein 4 | 0 | 0.14 |
| 9.3350785 | Cc09t03950.1 NAD(P)-binding Rossmann-fold superfamily protein | 0 | 0.1 |
| 5.24257788 | Cc05t09820.1 Glucose-1-phosphate adenylyltransferase | −1708 | 0.088 |
| 7.20811138 | Cc07t20130.1 Protein of unknown function (DUF1365) | +26,561 | 0.073 |
| 10.20833232 | Cc10t11950.1 tRNA (guanine(37)-N1)-methyltransferase | 0 | 0.061 |
| SNP ID | Nearest Gene and Function | Base Pairs Away | Importance Score (Portion) |
|---|---|---|---|
| Intermediate, production of coffee beans—Bootstrap Forest | |||
| 4.9597530 | Cc04t10310.1 Non-specific phospholipase C6 | 0 | 0.022 |
| 7.20000085 7.20000113 | Cc07t19900.1 Smr domain-containing protein | −737 | 0.016 |
| 4.15068413 | Cc04t12970.1 Alpha-N-acetylglucosaminidase | +804 | 0.012 |
| 2.26624120 | Cc02t28190.1 BHLH domain-containing protein | +1083 | 0.01 |
| 4.3864735 | Cc04t05180.1 Phytocyanin domain-containing protein | 0 | 0.01 |
| Intermediate, leaf rust incidence—Bootstrap Forest | |||
| 5.28610373 | Cc05t15840.1 TPR_REGION domain-containing protein | 0 | 0.022 |
| 2.22416916 | Cc02t25100.1 Nitrate regulatory gene2 protein | 0 | 0.019 |
| 10.3678570 | Cc10t04730.1 C2H2-type domain-containing protein | 0 | 0.018 |
| 8.30746352 | Cc08t16130.1 Nucleoside diphosphate kinase | 0 | 0.017 |
| 10.3612160 | Cc10t04630.1 BHLH domain-containing protein | 0 | 0.014 |
| Intermediate, yield of green beans—Bootstrap Forest | |||
| 4.234906 | Cc04t00320.1 Conserved hypothetical protein | 0 | 0.019 |
| 10.1467983 | Cc10t01960.1 Regulator of chromosome condensation (RCC1) family protein | 0 | 0.012 |
| 11.1400701 | Cc11t00520.1 Exopolygalacturonase | +2297 | 0.01 |
| 11.30547303 | Cc11t13750.1 Galectin domain-containing protein | −6340 | 0.01 |
| 11.20731087 | Cc11t05390.1 NB-ARC domain-containing protein | 0 | 0.0092 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, E.; Park, S.; Bhatt, J.; Lim, S.; Meinhardt, L.W. Lipid Metabolism and Actin Cytoskeleton Regulation Underlie Yield and Disease Resistance in Two Coffea canephora Breeding Populations. Plants 2025, 14, 3675. https://doi.org/10.3390/plants14233675
Ahn E, Park S, Bhatt J, Lim S, Meinhardt LW. Lipid Metabolism and Actin Cytoskeleton Regulation Underlie Yield and Disease Resistance in Two Coffea canephora Breeding Populations. Plants. 2025; 14(23):3675. https://doi.org/10.3390/plants14233675
Chicago/Turabian StyleAhn, Ezekiel, Sunchung Park, Jishnu Bhatt, Seunghyun Lim, and Lyndel W. Meinhardt. 2025. "Lipid Metabolism and Actin Cytoskeleton Regulation Underlie Yield and Disease Resistance in Two Coffea canephora Breeding Populations" Plants 14, no. 23: 3675. https://doi.org/10.3390/plants14233675
APA StyleAhn, E., Park, S., Bhatt, J., Lim, S., & Meinhardt, L. W. (2025). Lipid Metabolism and Actin Cytoskeleton Regulation Underlie Yield and Disease Resistance in Two Coffea canephora Breeding Populations. Plants, 14(23), 3675. https://doi.org/10.3390/plants14233675






