Transcriptional Analysis of a Tripartite Interaction Between Maize (Zea mays, L.) Roots Inoculated with the Pathogenic Fungus Fusarium verticillioides and Its Bacterial Control Agent Bacillus cereus sensu lato Strain B25
Abstract
1. Introduction
2. Results
2.1. B25 Mediated the Biocontrol of Fv and Enhanced Maize Plant Growth in the Tripartite Interaction
2.2. RNA Sequencing of the Tripartite Assay
2.3. Global Transcriptome Profiles of Maize Roots from the Tripartite Assay
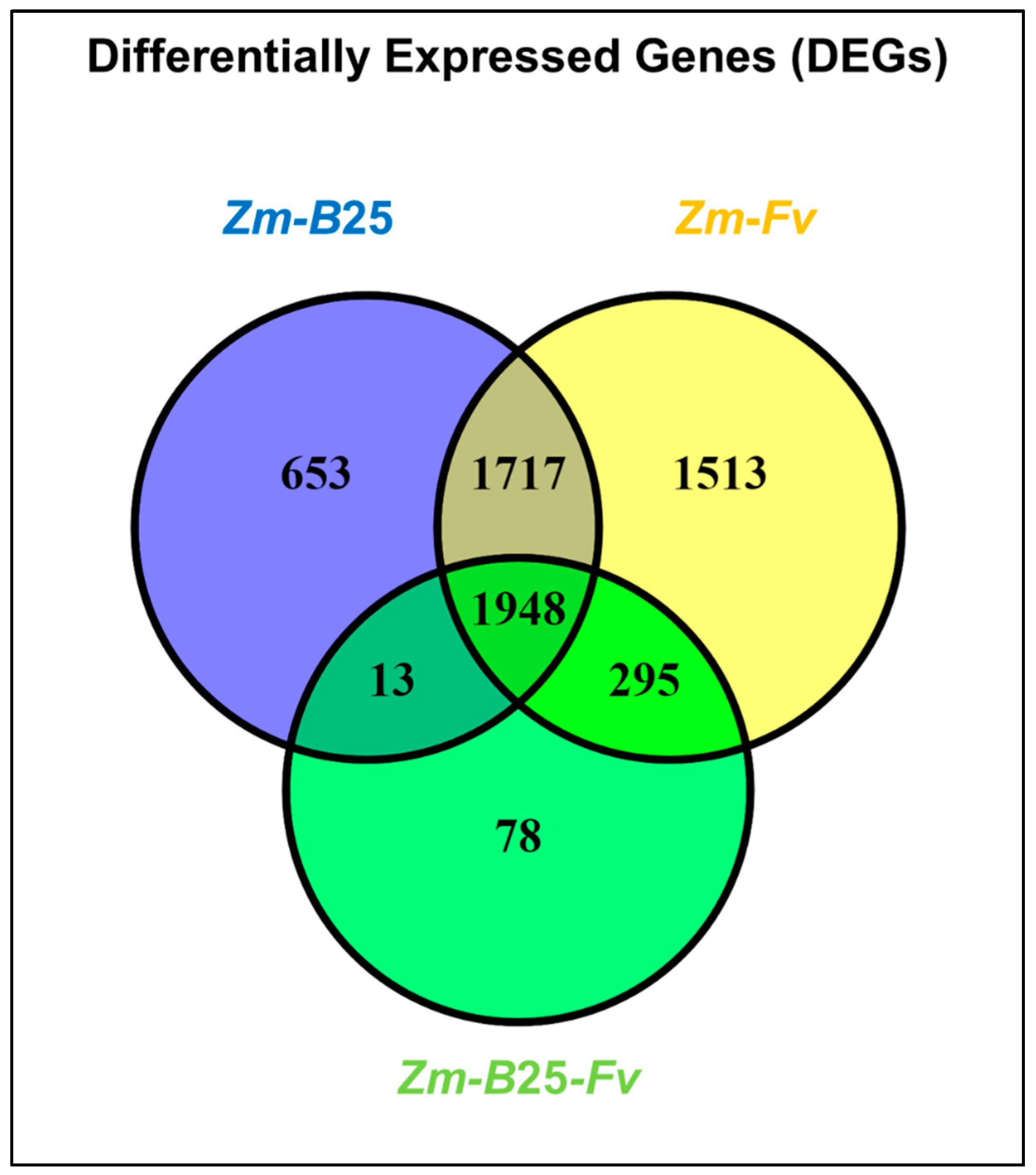
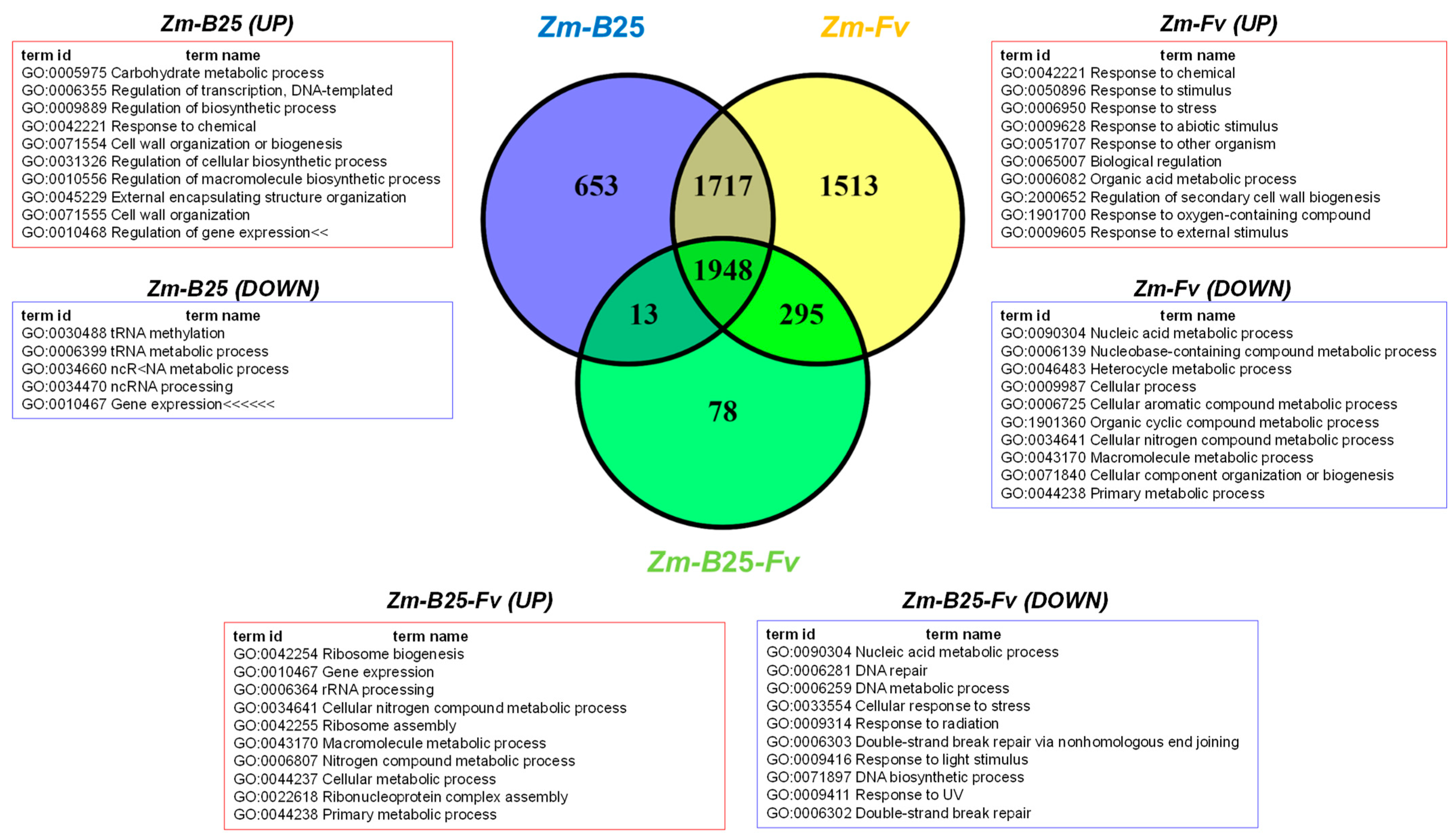
2.4. Differentially Expressed Genes Within and Among Maize Interactions
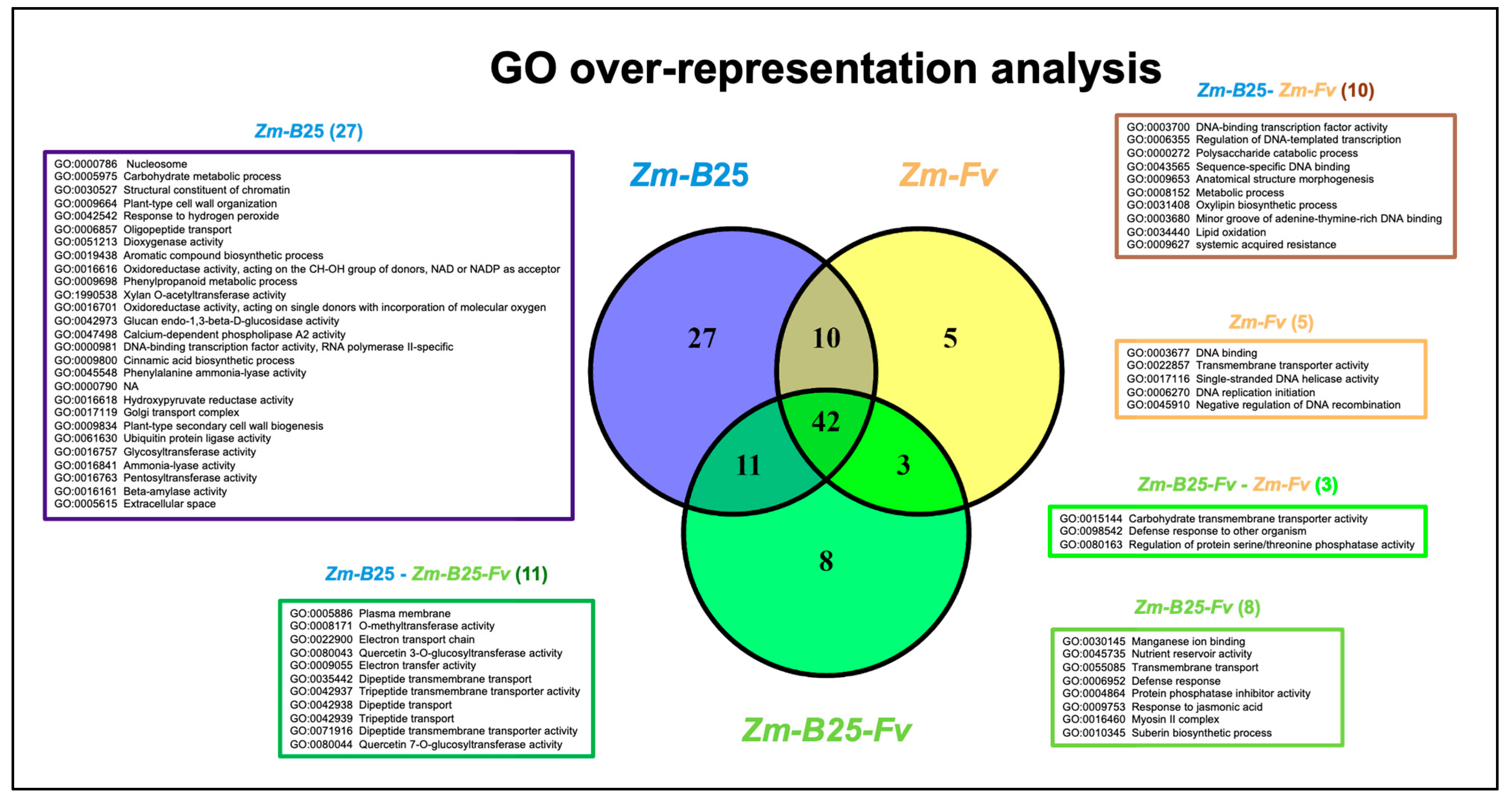
2.5. Overrepresentation Analysis: A General Approach
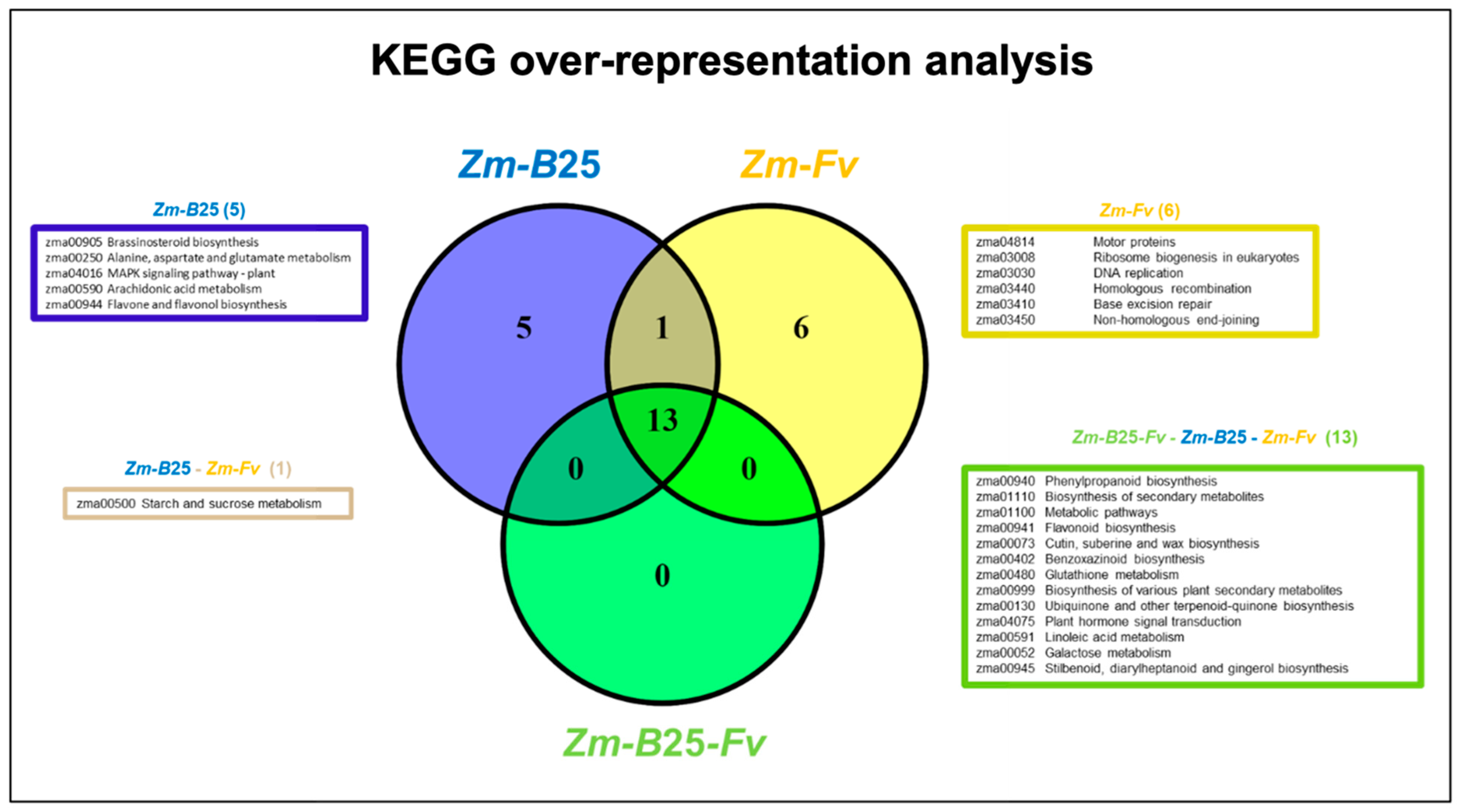
2.6. KEGG Analysis
2.7. Functional Gene Association Networks: Enriched Biological Processes Using A. thaliana Orthologous Genes
2.8. Gene Interaction Networks Based on Co-Expression
2.9. Validation of RNA Sequencing by qRT-PCR
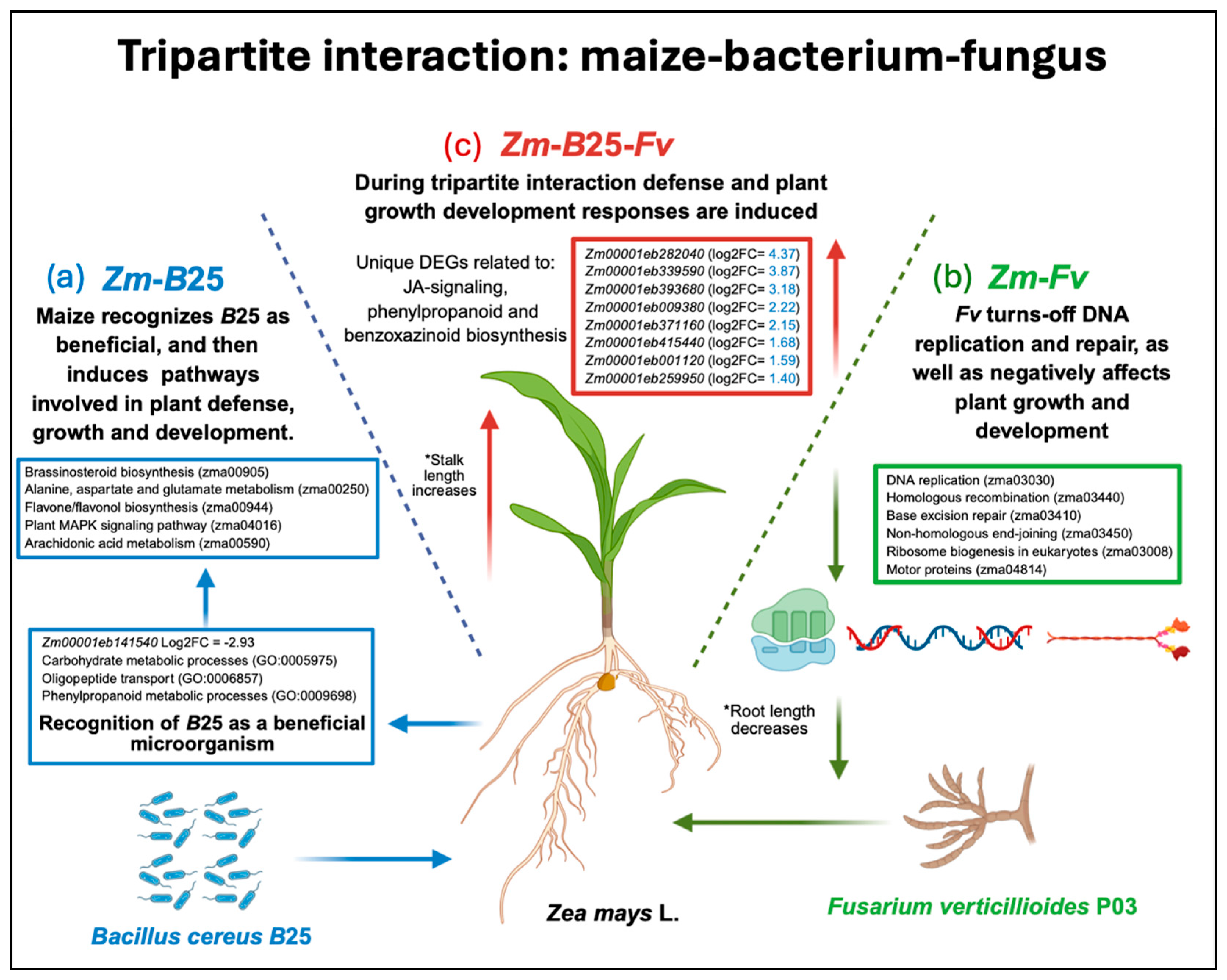
3. Discussion
3.1. Plant–Bacteria Interaction (Zm-B25): B25 Is Recognized by Maize as a Beneficial Microorganism
3.2. Plant–Bacteria Interaction (Zm-B25): B25 Induces the Activation of Pathways Related to Plant Growth, Development, and the Defense Response
3.3. Plant–Pathogenic Fungus Interaction (Zm-Fv): Fv Turns off DNA Replication and Repair, and Negatively Affects Plant Growth and Development
3.4. The Tripartite Interaction Zm-B25-Fv: Induction of Defense Response Genes and Plant Growth Development
3.5. Synthesis and Conceptual Framework
4. Materials and Methods
4.1. Microorganisms and Inoculum Preparation
4.2. Tripartite Assay: Maize–Bacteria–Fungus Interaction
4.3. Maize Root Colonization and Plant Protection Mediated by B25 Against Fv
4.4. RNA Extraction and Sequencing
4.5. Bioinformatic Analysis
4.6. Gene Function Annotation and Overrepresentation Analysis
4.7. Gene Set Enrichment Analyses: Networks and Clustering
4.8. RNA-Seq Validation by qRT-PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Erenstein, O.; Jaleta, M.; Sonder, K.; Mottaleb, K.; Prasanna, B.M. Global Maize Production, Consumption and Trade: Trends and R&D Implications. Food Secur. 2022, 14, 1295–1319. [Google Scholar] [CrossRef]
- Butrón, A.; Reid, L.M.; Santiago, R.; Cao, A.; Malvar, R.A. Inheritance of Maize Resistance to Gibberella and Fusarium Ear Rots and Kernel Contamination with Deoxynivalenol and Fumonisins. Plant Pathol. 2015, 64, 1053–1060. [Google Scholar] [CrossRef]
- Subedi, S. A Review on Important Maize Diseases and Their Management in Nepal. J. Maize Res. Dev. 2015, 1, 28–52. [Google Scholar] [CrossRef]
- Leyva-Madrigal, K.Y.; Larralde-Corona, C.P.; Apodaca-Sánchez, M.A.; Quiroz-Figueroa, F.R.; Mexia-Bolaños, P.A.; Portillo-Valenzuela, S.; Ordaz-Ochoa, J.; Maldonado-Mendoza, I.E. Fusarium Species from the Fusarium fujikuroi Species Complex Involved in Mixed Infections of Maize in Northern Sinaloa, Mexico. J. Phytopathol. 2015, 163, 486–497. [Google Scholar] [CrossRef]
- Bacon, C.W.; Glenn, A.E.; Yates, I.E. Fusarium verticillioides: Managing the Endophytic Association with Maize for Reduced Fumonisins Accumulation. Toxin Rev. 2008, 27, 411–446. [Google Scholar] [CrossRef]
- Deepa, N.; Nagaraja, H.; Sreenivasa, M.Y. Prevalence of Fumonisin Producing Fusarium verticillioides Associated with Cereals Grown in Karnataka (India). Food Sci. Human. Wellness 2016, 5, 156–162. [Google Scholar] [CrossRef]
- Koul, B.; Chopra, M.; Lamba, S. Microorganisms as Biocontrol Agents for Sustainable Agriculture. In Relationship Between Microbes and the Environment for Sustainable Ecosystem Services; Elsevier: Amsterdam, The Netherlands, 2022; Volume 1, pp. 45–68. [Google Scholar] [CrossRef]
- De los Santos Villalobos, S.; Parra Cota, F.I.; Herrera Sepúlveda, A.; Valenzuela Aragón, B.; Estrada Mora, J.C. Colmena: Colección de Microorganismos Edáficos y Endófitos Nativos, para Contribuir a la Seguridad Alimentaria Nacional. Rev. Mex. Cienc. Agric. 2018, 9, 191–202. [Google Scholar] [CrossRef]
- Lizárraga-Sánchez, G.J.; Leyva-Madrigal, K.Y.; Sánchez-Peña, P.; Quiroz-Figueroa, F.R.; Maldonado-Mendoza, I.E. Bacillus cereus sensu lato Strain B25 Controls Maize Stalk and Ear Rot in Sinaloa, Mexico. Field Crops Res. 2015, 176, 11–21. [Google Scholar] [CrossRef]
- Bacon, C.W.; Hinton, D.M. In Planta Reduction of Maize Seedling Stalk Lesions by the Bacterial Endophyte Bacillus mojavensis. Can. J. Microbiol. 2011, 57, 485–492. [Google Scholar] [CrossRef]
- Pal, G.; Kumar, K.; Verma, A.; Verma, S.K. Seed Inhabiting Bacterial Endophytes of Maize Promote Seedling Establishment and Provide Protection against Fungal Disease. Microbiol. Res. 2022, 255, 126926. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, R.; Wu, X.; Xu, T.; Ahmad, S.; Zhang, X.; Zhao, J.; Liu, Y. An Endophytic Strain of the Genus Bacillus Isolated from the Seeds of Maize (Zea mays L.) Has Antagonistic Activity against Maize Pathogenic Strains. Microb. Pathog. 2020, 142, 104074. [Google Scholar] [CrossRef]
- Figueroa-López, A.M.; Cordero-Ramírez, J.D.; Martínez-Álvarez, J.C.; López-Meyer, M.; Lizárraga-Sánchez, G.J.; Félix-Gastélum, R.; Castro-Martínez, C.; Maldonado-Mendoza, I.E. Rhizospheric Bacteria of Maize with Potential for Biocontrol of Fusarium verticillioides. Springerplus 2016, 5, 330. [Google Scholar] [CrossRef] [PubMed]
- Bolivar-Anillo, H.J.; González-Rodríguez, V.E.; Cantoral, J.M.; García-Sánchez, D.; Collado, I.G.; Garrido, C. Endophytic Bacteria Bacillus subtilis, Isolated from Zea mays, as Potential Biocontrol Agent against Botrytis cinerea. Biology 2021, 10, 492. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O. Elucidating Mechanisms of Endophytes Used in Plant Protection and Other Bioactivities with Multifunctional Prospects. Front. Bioeng. Biotechnol. 2020, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Figueroa-López, A.M. Caracterización de los Mecanismos de Antagonismo que Emplea Bacillus cereus Seleccionado para el Control de Fusarium verticillioides. Ph.D. Thesis, Instituto Politécnico Nacional, Guasave, Sinaloa, 2016. [Google Scholar]
- Martínez-Álvarez, J.C.; Castro-Martínez, C.; Sánchez-Peña, P.; Gutiérrez-Dorado, R.; Maldonado-Mendoza, I.E. Development of a Powder Formulation Based on Bacillus cereus sensu lato Strain B25 Spores for Biological Control of Fusarium verticillioides in Maize Plants. World J. Microbiol. Biotechnol. 2016, 32, 75. [Google Scholar] [CrossRef]
- Douriet-Gámez, N.R.; Maldonado-Mendoza, I.E.; Ibarra-Laclette, E.; Blom, J.; Calderón-Vázquez, C.L. Genomic Analysis of Bacillus Sp. Strain B25, a Biocontrol Agent of Maize Pathogen Fusarium verticillioides. Curr. Microbiol. 2018, 75, 247–255. [Google Scholar] [CrossRef]
- Morales-Ruiz, E.; Priego-Rivera, R.; Figueroa-López, A.M.; Cazares-Álvarez, J.E.; Maldonado-Mendoza, I.E. Biochemical Characterization of Two Chitinases from Bacillus cereus sensu lato B25 with Antifungal Activity against Fusarium verticillioides P03. FEMS Microbiol. Lett. 2021, 368, fnaa218. [Google Scholar] [CrossRef]
- Báez-Astorga, P.A.; Cázares-Álvarez, J.E.; Cruz-Mendívil, A.; Quiroz-Figueroa, F.R.; Sánchez-Valle, V.I.; Maldonado-Mendoza, I.E. Molecular and Biochemical Characterisation of Antagonistic Mechanisms of the Biocontrol Agent Bacillus cereus B25 Inhibiting the Growth of the Phytopathogen Fusarium verticillioides P03 during Their Direct Interaction in Vitro. Biocontrol Sci. Technol. 2022, 32, 1074–1094. [Google Scholar] [CrossRef]
- Figueroa-López, A.M.; Leyva-Madrigal, K.Y.; Cervantes-Gámez, R.G.; Beltran-Arredondo, L.I.; Douriet-Gámez, N.R.; Castro-Martínez, C.; Maldonado-Mendoza, I.E. Induction of Bacillus cereus Chitinases as a Response to Lysates of Fusarium verticillioides. Rom. Biotechnol. Lett. 2017, 22, 12722–12731. [Google Scholar]
- Park, Y.S.; Park, J.S.; Lee, S.; Jung, S.H.; Kim, S.K.; Ryu, C.M. Simultaneous Profiling of Arabidopsis thaliana and Vibrio vulnificus MO6-24/O Transcriptomes by Dual RNA-Seq Analysis. Comput. Struct. Biotechnol. J. 2021, 19, 2084–2096. [Google Scholar] [CrossRef]
- Gheler, C.F.; Domingues Zucchi, T.; Soares de Melo, I. Biological Control of Phytopathogenic Fungi by Endophytic Actinomycetes Isolated from Maize (Zea mays L.). Braz. Arch. Biol. Technol. 2013, 56, 948–955. [Google Scholar] [CrossRef]
- Cazares-Álvarez, J.E.; Báez-Astorga, P.A.; Arroyo-Becerra, A.; Maldonado-Mendoza, I.E. Genome-Wide Identification of a Maize Chitinase Gene Family and the Induction of Its Expression by Fusarium verticillioides (Sacc.) Nirenberg (1976) Infection. Genes 2024, 15, 1087. [Google Scholar] [CrossRef]
- Perrot, T.; Pauly, M.; Ramírez, V. Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses. Plants 2022, 11, 1119. [Google Scholar] [CrossRef]
- Villarino, S.H.; Pinto, P.; Jackson, R.B.; Piñeiro, G. Plant Rhizodeposition: A Key Factor for Soil Organic Matter Formation in Stable Fractions. Sci. Adv. 2021, 7, eabd3176. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Gowtham, H.G.; Singh, S.B.; Shilpa, N.; Aiyaz, M.; Nataraj, K.; Udayashankar, A.C.; Amruthesh, K.N.; Murali, M.; Poczai, P.; Gafur, A.; et al. Insight into Recent Progress and Perspectives in Improvement of Antioxidant Machinery upon PGPR Augmentation in Plants under Drought Stress: A Review. Antioxidants 2022, 11, 1763. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, X.; Qiu, H. Flavones and Flavonols: Phytochemistry and Biochemistry. In Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes; Ramawat, K.G., Mérillon, J.-M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1821–1847. ISBN 978-3-642-22144-6. [Google Scholar]
- Singh, P.; Arif, Y.; Bajguz, A.; Hayat, S. The Role of Quercetin in Plants. Plant Physiol. Biochem. 2021, 166, 10–19. [Google Scholar] [CrossRef]
- Wang, L.; Chen, M.; Lam, P.Y.; Dini-Andreote, F.; Dai, L.; Wei, Z. Multifaceted Roles of Flavonoids Mediating Plant-Microbe Interactions. Microbiome 2022, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, D.; Tang, Z.; Zhang, Y.; Zhang, K.; Dong, J.; Wang, F. Exogenous Brassinosteroids Promotes Root Growth, Enhances Stress Tolerance, and Increases Yield in Maize. Plant Signal Behav. 2022, 17, 2095139. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T. Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Franco, A.R.; Gee, M.A.; Guilfoyle, T.J. Induction and Superinduction of Auxin-Responsive MRNAs with Auxin and Protein Synthesis Inhibitors. J. Biol. Chem. 1990, 265, 15845–15849. [Google Scholar] [CrossRef]
- Sowmya, R.S.; Warke, V.G.; Mahajan, G.B.; Annapure, U.S. Effect of Amino Acids on Growth, Elemental Content, Functional Groups, and Essential Oils Composition on Hydroponically Cultivated Coriander under Different Conditions. Ind. Crops Prod. 2023, 197, 116577. [Google Scholar] [CrossRef]
- Colla, G.; Nardi, S.; Cardarelli, M.; Ertani, A.; Lucini, L.; Canaguier, R.; Rouphael, Y. Protein Hydrolysates as Biostimulants in Horticulture. Sci. Hortic. 2015, 196, 28–38. [Google Scholar] [CrossRef]
- Alfosea-Simón, M.; Simón-Grao, S.; Zavala-Gonzalez, E.A.; Cámara-Zapata, J.M.; Simón, I.; Martínez-Nicolás, J.J.; Lidón, V.; García-Sánchez, F. Physiological, Nutritional and Metabolomic Responses of Tomato Plants After the Foliar Application of Amino Acids Aspartic Acid, Glutamic Acid and Alanine. Front. Plant Sci. 2021, 11, 581234. [Google Scholar] [CrossRef]
- Islam, M.M.; Hassan, M.U.; Ishfaq, M.; Ripa, F.A.; Nadeem, F.; Ahmad, Z.; Xu, J.; Ning, P.; Li, X. Foliar Glutamine Application Improves Grain Yield and Nutritional Quality of Field-Grown Maize (Zea mays L.) Hybrid ZD958. J. Plant Growth Regul. 2024, 43, 624–637. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S. Mitogen-Activated Protein Kinase Cascades in Signaling Plant Growth and Development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Wu, J.; Jiang, M.; Wang, Y. Plant Mitogen-activated Protein Kinase Cascades in Environmental Stresses. Int. J. Mol. Sci. 2021, 22, 1543. [Google Scholar] [CrossRef]
- Dedyukhina, E.G.; Kamzolova, S.V.; Vainshtein, M.B. Arachidonic Acid as an Elicitor of the Plant Defense Response to Phytopathogens. Chem. Biol. Technol. Agric. 2014, 1, 2–7. [Google Scholar] [CrossRef]
- Savchenko, T.; Walley, J.W.; Chehab, E.W.; Xiao, Y.; Kaspi, R.; Pye, M.F.; Mohamed, M.E.; Lazarus, C.M.; Bostock, R.M.; Dehesh, K. Arachidonic Acid: An Evolutionarily Conserved Signaling Molecule Modulates Plant Stress Signaling Networks. Plant Cell 2010, 22, 3193–3205. [Google Scholar] [CrossRef]
- Dye, S.M.; Bostock, R.M. Eicosapolyenoic Fatty Acids Induce Defense Responses and Resistance to Phytophthora capsici in Tomato and Pepper. Physiol. Mol. Plant Pathol. 2021, 114, 101642. [Google Scholar] [CrossRef]
- Lewis, D.C.; van der Zwan, T.; Richards, A.; Little, H.; Coaker, G.L.; Bostock, R.M. The Oomycete Microbe-Associated Molecular Pattern, Arachidonic Acid, and an Ascophyllum nodosum-Derived Plant Biostimulant Induce Defense Metabolome Remodeling in Tomato. Phytopathology 2023, 113, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Hyong, W.C.; Byung, G.L.; Nak, H.K.; Park, Y.; Chae, W.L.; Hyun, K.S.; Byung, K.H. A Role for a Menthone Reductase in Resistance against Microbial Pathogens in Plants. Plant Physiol. 2008, 148, 383–401. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, L.J.; Wang, Y.; Liu, S.; Geng, Z.; Song, A.; Jiang, J.; Chen, S.; Chen, F. Functional Identification of a Flavone Synthase and a Flavonol Synthase Genes Affecting Flower Color Formation in Chrysanthemum morifolium. Plant Physiol. Biochem. 2021, 166, 1109–1120. [Google Scholar] [CrossRef]
- Alseekh, S.; Perez de Souza, L.; Benina, M.; Fernie, A.R. The Style and Substance of Plant Flavonoid Decoration; towards Defining both Structure and Function. Phytochemistry 2020, 174, 112347. [Google Scholar] [CrossRef]
- Jiménez-Gómez, A.; García-Estévez, I.; García-Fraile, P.; Escribano-Bailón, M.T.; Rivas, R. Increase in Phenolic Compounds of Coriandrum sativum L. after the Application of a Bacillus Halotolerans Biofertilizer. J. Sci. Food Agric. 2020, 100, 2742–2749. [Google Scholar] [CrossRef]
- Dauwe, R.; Morreel, K.; Goeminne, G.; Gielen, B.; Rohde, A.; Van Beeumen, J.; Ralph, J.; Boudet, A.-M.; Kopka, J.; Rochange, S.F.; et al. Molecular Phenotyping of Lignin-Modified Tobacco Reveals Associated Changes in Cell-Wall Metabolism, Primary Metabolism, Stress Metabolism and Photorespiration. Plant J. 2007, 52, 263–285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, C.; Zhang, S.; Yu, H.; Pan, H.; Zhang, H. Klebsiella Jilinsis 2N3 Promotes Maize Growth and Induces Resistance to Northern Corn Leaf Blight. Biol. Control. 2021, 156, 104554. [Google Scholar] [CrossRef]
- Manova, V.; Gruszka, D. DNA Damage and Repair in Plants—From Models to Crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef]
- Nisa, M.U.; Huang, Y.; Benhamed, M.; Raynaud, C. The Plant DNA Damage Response: Signaling Pathways Leading to Growth Inhibition and Putative Role in Response to Stress Conditions. Front. Plant Sci. 2019, 10, 653. [Google Scholar] [CrossRef]
- Grin, I.R.; Petrova, D.V.; Endutkin, A.V.; Ma, C.; Yu, B.; Li, H.; Zharkov, D.O. Base Excision DNA Repair in Plants: Arabidopsis and Beyond. Int. J. Mol. Sci. 2023, 24, 14746. [Google Scholar] [CrossRef] [PubMed]
- Szurman-Zubrzycka, M.; Jędrzejek, P.; Szarejko, I. How Do Plants Cope with DNA Damage? A Concise Review on the DDR Pathway in Plants. Int. J. Mol. Sci. 2023, 24, 2404. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, X.; Li, Y.; Xing, A.; Wu, C.; Li, D.; Wang, C.; de Bures, A.; Zhang, Y.; Guo, S.; et al. Arabidopsis SMO2 Modulates Ribosome Biogenesis by Maintaining the RID2 Abundance during Organ Growth. Plant J. 2023, 114, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Nebenführ, A.; Dixit, R. Kinesins and Myosins: Molecular Motors That Coordinate Cellular Functions in Plants. Annu. Rev. Plant Biol. 2018, 69, 329–361. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Chong, K. The Novel Functions of Kinesin Motor Proteins in Plants. Protoplasma 2012, 249, 95–100. [Google Scholar] [CrossRef]
- Ryan, J.M.; Nebenführ, A. Update on Myosin Motors: Molecular Mechanisms and Physiological Functions. Plant Physiol. 2018, 176, 119–127. [Google Scholar] [CrossRef]
- Mao, L.; Ge, L.; Ye, X.; Xu, L.; Si, W.; Ding, T. ZmGLP1, a Germin-like Protein from Maize, Plays an Important Role in the Regulation of Pathogen Resistance. Int. J. Mol. Sci. 2022, 23, 14316. [Google Scholar] [CrossRef]
- Khare, D.; Choi, H.; Huh, S.U.; Bassin, B.; Kim, J.; Martinoia, E.; Sohn, K.H.; Paek, K.H.; Lee, Y.; Chrispeels, M.J. Arabidopsis ABCG34 Contributes to Defense against Necrotrophic Pathogens by Mediating the Secretion of Camalexin. Proc. Natl. Acad. Sci. USA 2017, 114, E5712–E5720. [Google Scholar] [CrossRef]
- Li, M.R.; Qi, X.; Li, D.; Wu, Z.; Liu, M.; Yang, W.; Zang, Z.; Jiang, L. Comparative Transcriptome Analysis Highlights Resistance Regulatory Networks of Maize in Response to Exserohilum turcicum Infection at the Early Stage. Physiol. Plant 2024, 176, e14615. [Google Scholar] [CrossRef]
- Shumayla; Madhu; Singh, K.; Upadhyay, S.K. LysM Domain-Containing Proteins Modulate Stress Response and Signalling in Triticum aestivum L. Environ. Exp. Bot. 2021, 189, 104558. [Google Scholar] [CrossRef]
- Giovannoni, M.; Lironi, D.; Marti, L.; Paparella, C.; Vecchi, V.; Gust, A.A.; De Lorenzo, G.; Nürnberger, T.; Ferrari, S. The Arabidopsis thaliana LysM-Containing Receptor-Like Kinase 2 Is Required for Elicitor-Induced Resistance to Pathogens. Plant Cell Environ. 2021, 44, 3545–3562. [Google Scholar] [CrossRef]
- Kumar, R.; Meghwanshi, G.K.; Marcianò, D.; Ullah, S.F.; Bulone, V.; Toffolatti, S.L.; Srivastava, V. Sequence, Structure and Functionality of Pectin Methylesterases and Their Use in Sustainable Carbohydrate Bioproducts: A Review. Int. J. Biol. Macromol. 2023, 244, 125385. [Google Scholar] [CrossRef]
- Zhou, S.; Richter, A.; Jander, G. Beyond Defense: Multiple Functions of Benzoxazinoids in Maize Metabolism. Plant Cell Physiol. 2018, 59, 1528–1533. [Google Scholar] [CrossRef]
- Lee, J.R.; Boltz, K.A.; Lee, S.Y. Molecular Chaperone Function of Arabidopsis thaliana Phloem Protein 2-A1, Encodes a Protein Similar to Phloem Lectin. Biochem. Biophys. Res. Commun. 2014, 443, 18–21. [Google Scholar] [CrossRef]
- Zheng, L.; Zheng, H.; Zheng, X.; Duan, Y.; Yu, X. PP2 Gene Family in Phyllostachys edulis: Identification, Characterization, and Expression Profiles. BMC Genom. 2024, 25, 1081. [Google Scholar] [CrossRef]
- Bao, Y.; Song, W.M.; Pan, J.; Jiang, C.M.; Srivastava, R.; Li, B.; Zhu, L.Y.; Su, H.Y.; Gao, X.S.; Liu, H.; et al. Overexpression of the NDR1/HIN1-Like Gene NHL6 Modifies Seed Germination in Response to Abscisic Acid and Abiotic Stresses in Arabidopsis. PLoS ONE 2016, 11, e0148572. [Google Scholar] [CrossRef]
- Maldonado, A.; Youssef, R.; McDonald, M.; Brewer, E.; Beard, H.; Matthews, B. Overexpression of Four Arabidopsis thaliana NHL Genes in Soybean (Glycine max) Roots and Their Effect on Resistance to the Soybean Cyst Nematode (Heterodera glycines). Physiol. Mol. Plant Pathol. 2014, 86, 1–10. [Google Scholar] [CrossRef]
- Block, A.K.; Tang, H.V.; Hopkins, D.; Mendoza, J.; Solemslie, R.K.; du Toit, L.J.; Christensen, S.A. A Maize Leucine-Rich Repeat Receptor-like Protein Kinase Mediates Responses to Fungal Attack. Planta 2021, 254, 73. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Zhao, Y.; Wang, W.; Liu, X.; Shi, W.; Zhang, D.; Chen, J.; Ma, S.; Sun, Q.; Wang, T.; et al. Transcriptome Analysis in Contrasting Maize Inbred Lines and Functional Analysis of Five Maize NAC Genes under Drought Stress Treatment. Front. Plant Sci. 2023, 13, 1097719. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, M.; Wang, D.; Gong, Y.; Sha, Q.; Lv, P.; Yang, J.; Chu, P.; Guo, S. Research Progress on the Roles of Actin-Depolymerizing Factor in Plant Stress Responses. Front. Plant Sci. 2023, 14, 1278311. [Google Scholar] [CrossRef]
- Shigeyama, T.; Watanabe, A.; Tokuchi, K.; Toh, S.; Sakurai, N.; Shibuya, N.; Kawakami, N. α-Xylosidase Plays Essential Roles in Xyloglucan Remodelling, Maintenance of Cell Wall Integrity, and Seed Germination in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 5615–5629. [Google Scholar] [CrossRef] [PubMed]
- White-Gilbertson, S.; Rubinchik, S.; Voelkel-Johnson, C. Transformation, Translation and TRAIL: An Unexpected Intersection. Cytokine Growth Factor. Rev. 2008, 19, 167–172. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Y.; Yu, X.; Lin, Y.; Zhu, Y.; Chen, J.; Xie, H.; Zhang, Q.; Wang, L.; Wei, Y.; et al. SH3P2, an SH3 Domain-Containing Protein That Interacts with Both Pib and AvrPib, Suppresses Effector-Triggered, Pib-Mediated Immunity in Rice. Mol. Plant 2022, 15, 1931–1946. [Google Scholar] [CrossRef]
- Warham, E.J.; Butler, L.D.; Sutton, B.C. Seed Testing of Maize and Wheat: A Laboratory Guide; CIMMYT: Texcoco, Mexico, 1996; 84p. [Google Scholar]
- Jasim, B.; Joseph, A.A.; John, C.J.; Mathew, J.; Radhakrishnan, E.K. Isolation and Characterization of Plant Growth Promoting Endophytic Bacteria from the Rhizome of Zingiber officinale. 3 Biotech 2014, 4, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Aranda, P.S.; Lajoie, D.M.; Jorcyk, C.L. Bleach Gel: A Simple Agarose Gel for Analyzing RNA Quality. Electrophoresis 2012, 33, 366–369. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, M.R.; Cannon, E.K.; Portwood, J.L.; Harper, L.C.; Gardiner, J.M.; Schaeffer, M.L.; Andorf, C.M. A Pan-Genomic Approach to Genome Databases Using Maize as a Model System. BMC Plant Biol. 2021, 21, 385. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2023, 51, D933–D941. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO Analysis Toolkit for the Agricultural Community, 2017 Update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lin, F.; Jiang, L.; Liu, Y.; Lv, Y.; Dai, H.; Zhao, H. Genome-Wide Identification of Housekeeping Genes in Maize. Plant Mol. Biol. 2014, 86, 543–554. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Portwood, J.L., II; Woodhouse, M.R.; Cannon, E.K.; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Walsh, J.R.; Sen, T.Z.; Cho, K.T.; Schott, D.A.; et al. MaizeGDB 2018: The Maize Multi-Genome Genetics and Genomics Database. Nucleic Acids Res. 2019, 47, D1146–D1154. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.G.O. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L.; et al. The Pfam Protein Families Database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Mitchell, A.; Chang, H.-Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro Protein Families Database: The Classification Resource after 15 Years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]

| Condition | Length (cm) | Fresh Weight (g) | ||
|---|---|---|---|---|
| Leaf | Root | Leaf | Root | |
| Zm | 11.92 b | 13.07 a | 0.38 b | 0.78 a |
| Zm-B25 | 11.27 b | 12.52 a | 0.34 b | 0.78 a |
| Zm-Fv | 11.52 b | 9.86 b | 0.34 b | 0.64 b |
| Zm-B25-Fv | 14.28 a | 13.80 a | 0.47 a | 0.77 a |
| Sample | Raw Reads | Filtered Reads (Q > 20) | Unique Mapped Reads | ||
|---|---|---|---|---|---|
| Zm-2 | 13,742,216 | 13,534,709 | (98.49%) | 12,542,341 | (91.27%) |
| Zm-3 | 21,273,970 | 20,839,981 | (97.96%) | 17,042,555 | (80.11%) |
| Zm-B25-1 | 22,841,141 | 22,498,524 | (98.50%) | 19,719,075 | (86.33%) |
| Zm-B25-2 | 27,395,643 | 27,003,885 | (98.57%) | 24,442,305 | (89.22%) |
| Zm-Fv-1 | 19,155,891 | 18,880,046 | (98.56%) | 17,497,335 | (91.34%) |
| Zm-Fv-3 | 18,080,786 | 17,815,902 | (98.54%) | 14,513,113 | (80.27%) |
| Zm-B25-Fv-1 | 18,442,655 | 18.199,212 | (98.68%) | 16,495,370 | (89.44%) |
| Zm-B25-Fv-3 | 21,465,145 | 21,166,779 | (98.61%) | 19,373,477 | (90.26%) |
| Average | 20,299,681 | 19,992,380 | (98.49%) | 17,703,196 | (87.28%) |
| Condition | Total EGs | Percentage of the Genome a |
|---|---|---|
| Zm | 23,625 | 53.33% |
| Zm-B25 | 25,412 | 57.36% |
| Zm-Fv | 24,668 | 55.68% |
| Zm-B25-Fv | 24,808 | 56.00% |
| Average | 24,628 | 55.59% |
| Condition | Total DEGs | Up-Regulated | Down-Regulated |
|---|---|---|---|
| Zm-B25 | 4331 | 3884 (89.68%) | 447 (10.32%) |
| Zm-Fv | 5473 | 4076 (74.47%) | 1397 (25.53%) |
| Zm-B25-Fv | 2334 | 2077 (88.98%) | 257 (11.02%) |
| Condition | ID a | Gene | Fold Change vs. Control | |
|---|---|---|---|---|
| FC RNA-seq | FC qRT-PCR | |||
| Zm-B25-Fv | Zm00001eb375460 | Glutaredoxin | 11.49 | 4.11 |
| Zm00001eb229260 | Protein kinase domain | 2.18 | 2.24 | |
| Zm00001eb251380 | Cyclic nucleotide-binding domain | −2.23 | −7.56 | |
| Zm00001eb029490 | Gdt1 family | −2.41 | −4.37 | |
| Zm00001eb397080 | CRAL/TRIO, N-terminal domain | −2.61 | −2.74 | |
| Zm-B25 | Zm00001eb124940 | Multi antimicrobial extrusion protein | 2.87 | 2.92 |
| Zm-Fv | Zm00001eb041100 | Hydroxycinnamoyltransferase 9 | 13.1 | 7.3 |
| Zm00001eb419890 | Deoxymugineic acid synthase 6 | 5.09 | 5.57 | |
| Zm00001eb285030 | Putative cytochrome P450 superfamily protein | 7.07 | 3.72 | |
| Zm00001eb241870 | Small auxin up RNA54 | 9.69 | 24.98 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Báez-Astorga, P.A.; Cruz-Mendívil, A.; Figueroa-Castro, J.L.; López-Soto, I.G.; Cazares-Álvarez, J.E.; Gregorio-Jorge, J.; Calderón-Vázquez, C.L.; Maldonado-Mendoza, I.E. Transcriptional Analysis of a Tripartite Interaction Between Maize (Zea mays, L.) Roots Inoculated with the Pathogenic Fungus Fusarium verticillioides and Its Bacterial Control Agent Bacillus cereus sensu lato Strain B25. Plants 2025, 14, 3661. https://doi.org/10.3390/plants14233661
Báez-Astorga PA, Cruz-Mendívil A, Figueroa-Castro JL, López-Soto IG, Cazares-Álvarez JE, Gregorio-Jorge J, Calderón-Vázquez CL, Maldonado-Mendoza IE. Transcriptional Analysis of a Tripartite Interaction Between Maize (Zea mays, L.) Roots Inoculated with the Pathogenic Fungus Fusarium verticillioides and Its Bacterial Control Agent Bacillus cereus sensu lato Strain B25. Plants. 2025; 14(23):3661. https://doi.org/10.3390/plants14233661
Chicago/Turabian StyleBáez-Astorga, Paúl Alán, Abraham Cruz-Mendívil, Juan Luis Figueroa-Castro, Itzel Guadalupe López-Soto, Jesús Eduardo Cazares-Álvarez, Josefat Gregorio-Jorge, Carlos Ligne Calderón-Vázquez, and Ignacio Eduardo Maldonado-Mendoza. 2025. "Transcriptional Analysis of a Tripartite Interaction Between Maize (Zea mays, L.) Roots Inoculated with the Pathogenic Fungus Fusarium verticillioides and Its Bacterial Control Agent Bacillus cereus sensu lato Strain B25" Plants 14, no. 23: 3661. https://doi.org/10.3390/plants14233661
APA StyleBáez-Astorga, P. A., Cruz-Mendívil, A., Figueroa-Castro, J. L., López-Soto, I. G., Cazares-Álvarez, J. E., Gregorio-Jorge, J., Calderón-Vázquez, C. L., & Maldonado-Mendoza, I. E. (2025). Transcriptional Analysis of a Tripartite Interaction Between Maize (Zea mays, L.) Roots Inoculated with the Pathogenic Fungus Fusarium verticillioides and Its Bacterial Control Agent Bacillus cereus sensu lato Strain B25. Plants, 14(23), 3661. https://doi.org/10.3390/plants14233661






