The Phenylpropanoid Gatekeeper CtPAL1 Coordinates ABA-Induced Flavonoid Biosynthesis and Oxidative Stress Tolerance in Safflower (Carthamus tinctorius L.)
Abstract
1. Introduction
2. Results
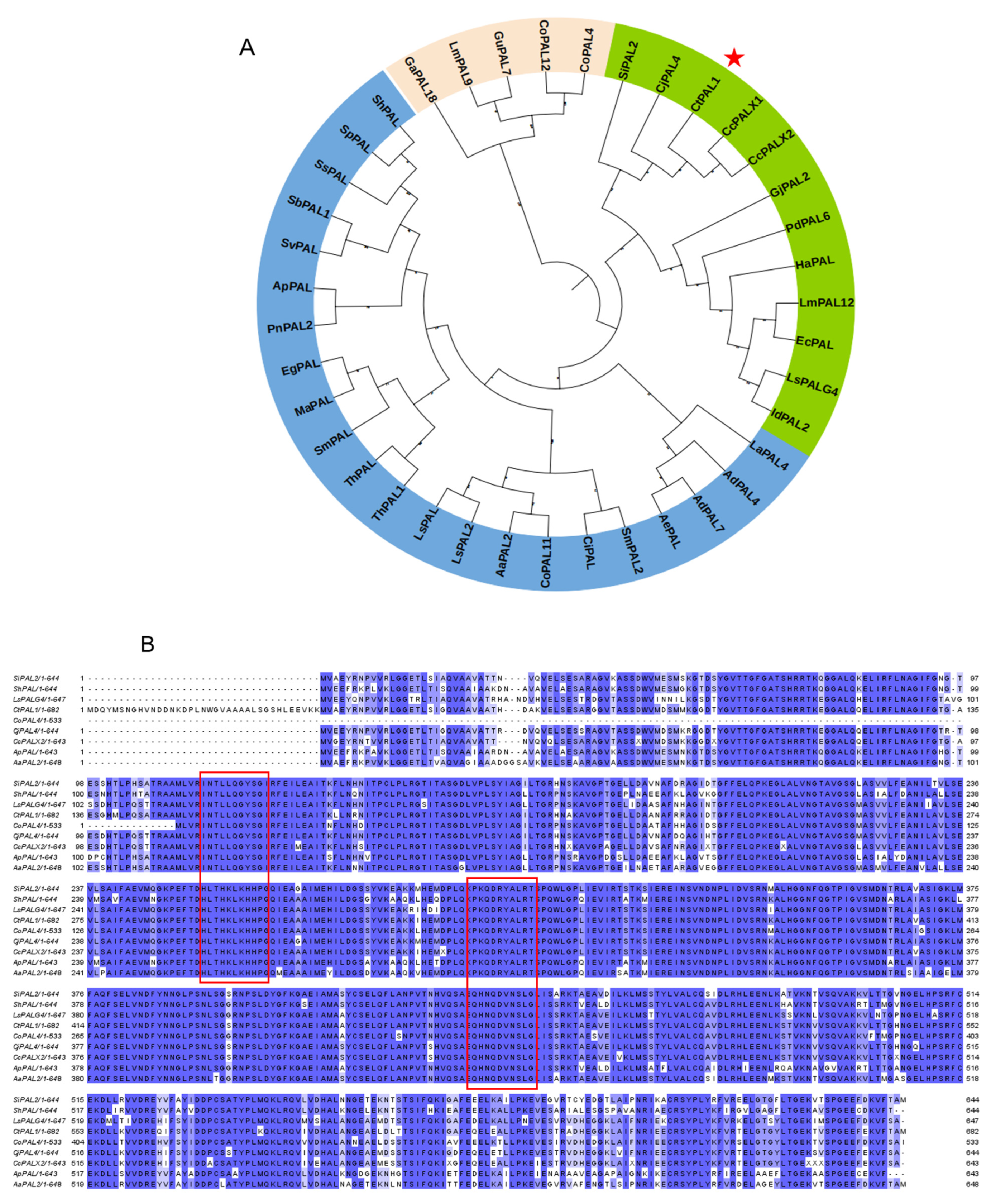
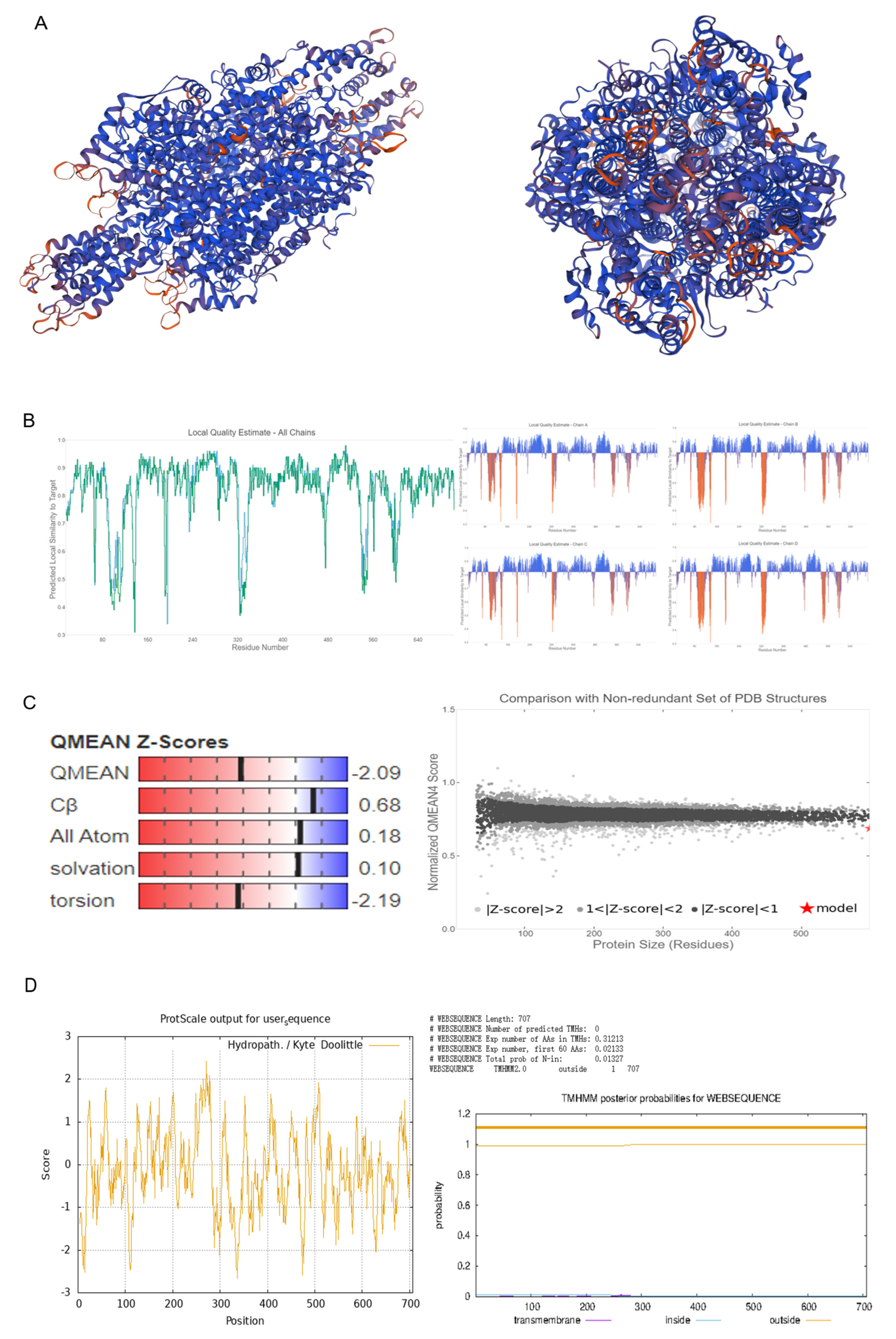
2.1. Isolation, Evolutionary Insights, and Structural Prediction of CtPAL1
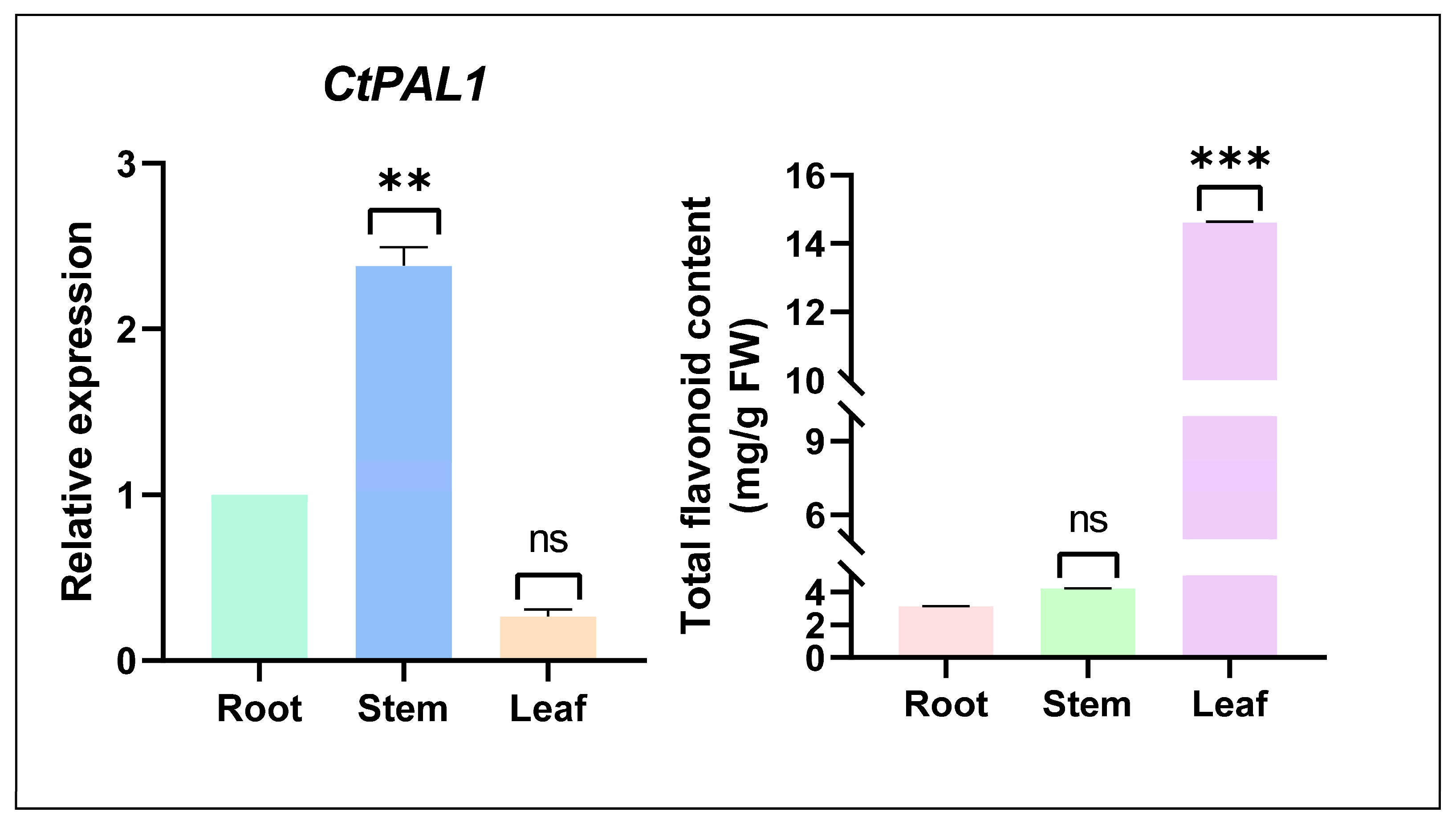
2.2. Basal Expression Patterns of CtPAL1 Reveal Tissue-Specific but Nonlinear Correlation with Flavonoid Accumulation Under Normal Condition
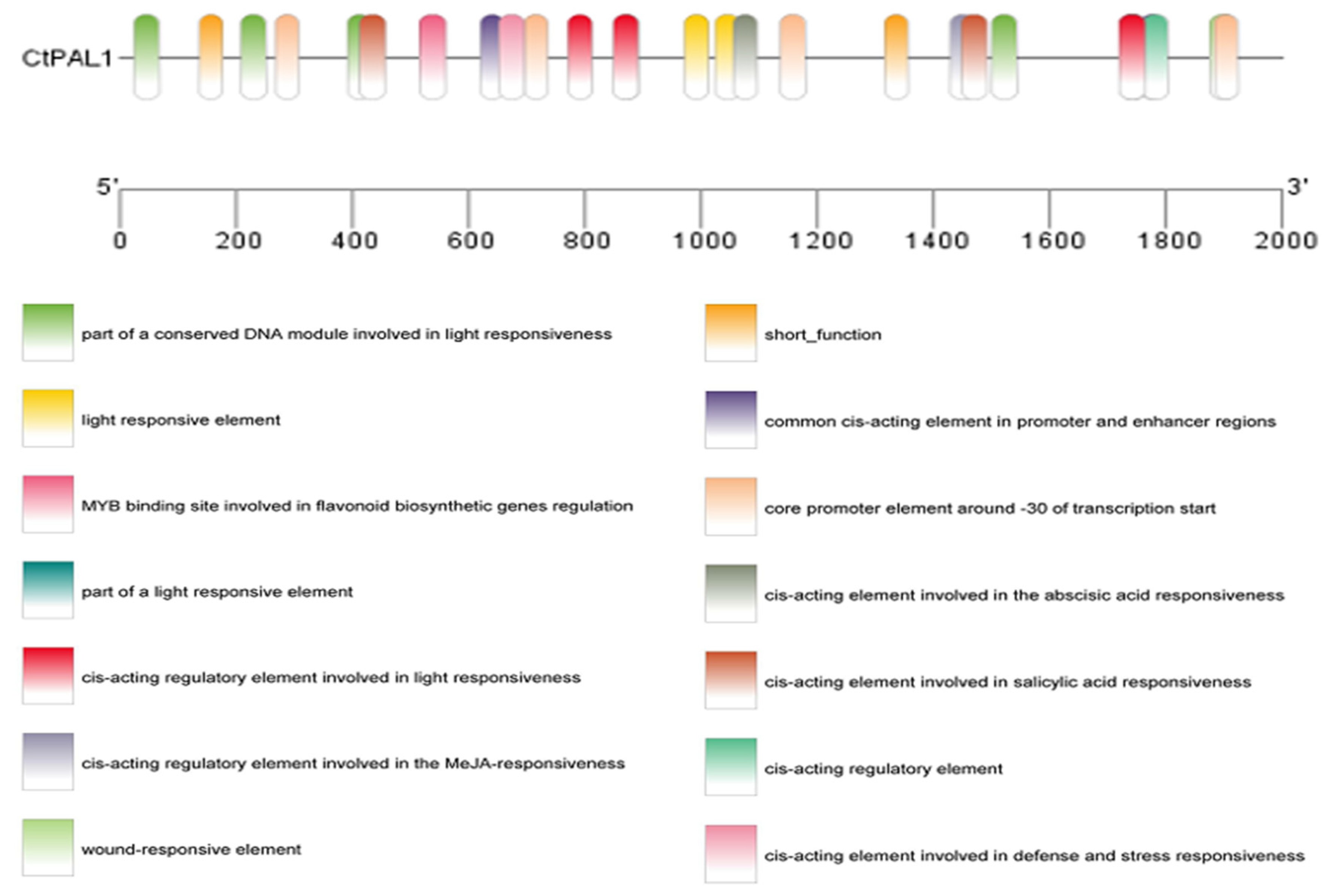
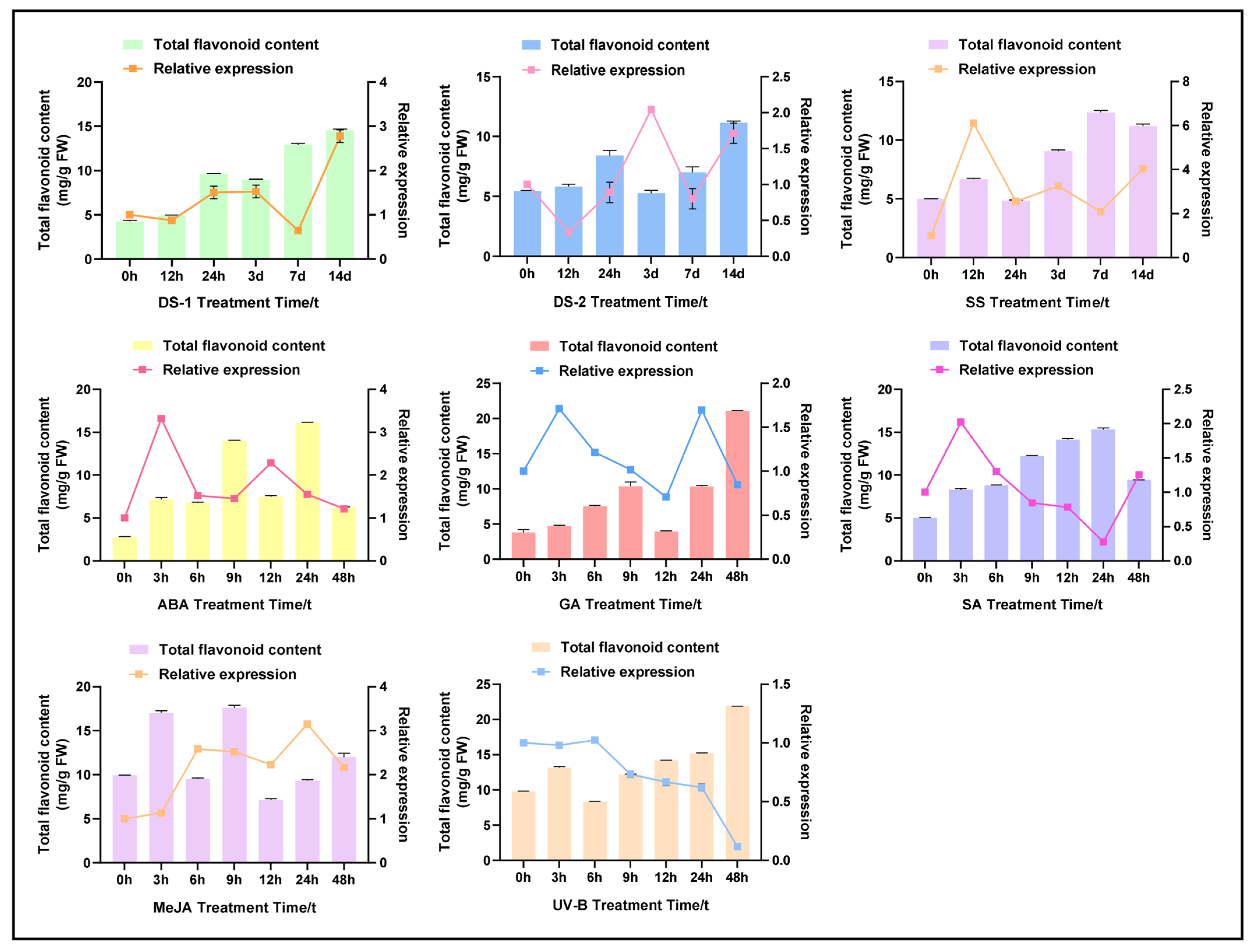
2.3. Abiotic Stress and Hormonal-Induced CtPAL1 Expression Suggested Modulation in Flavonoid Accumulation
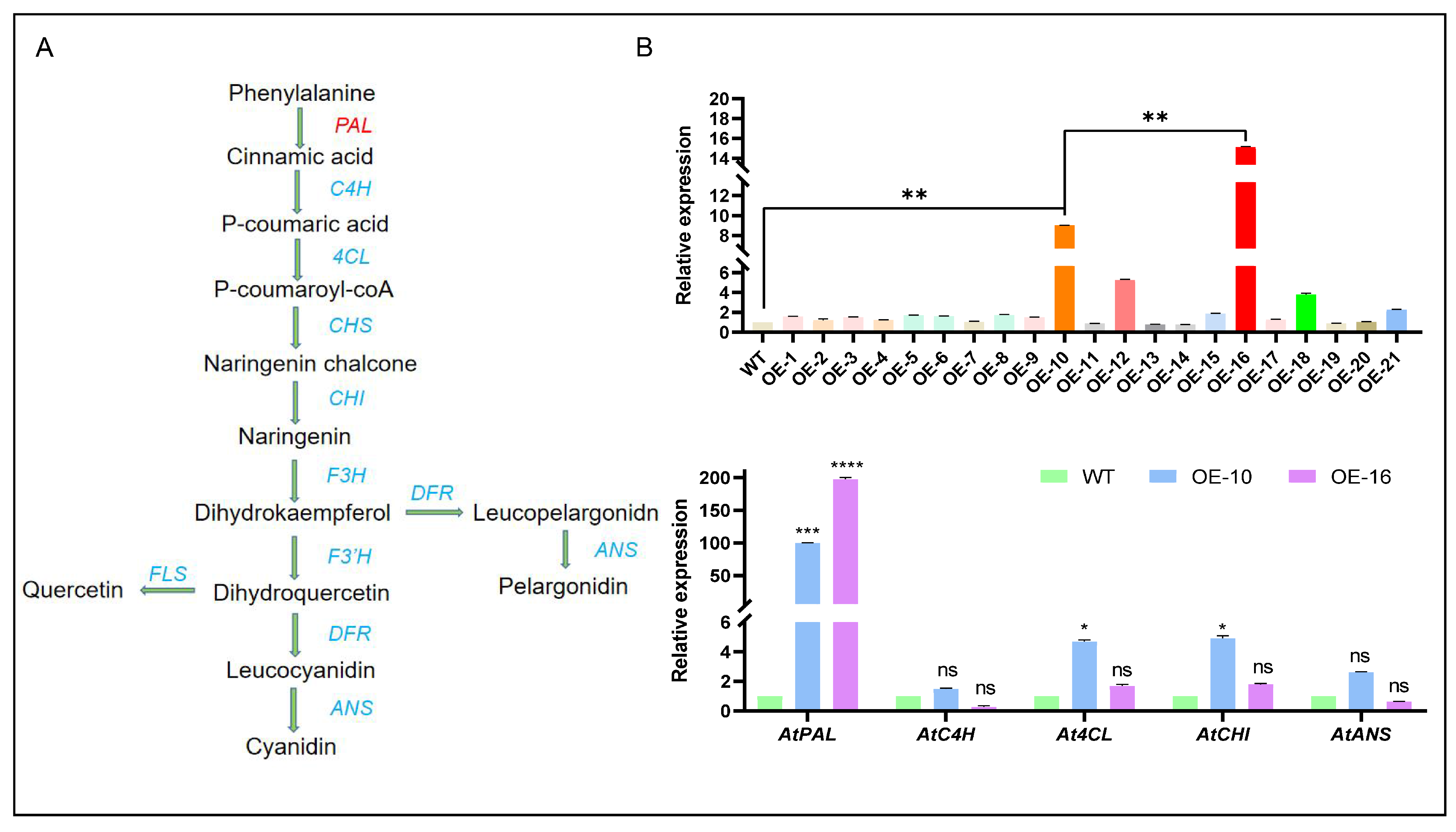
2.4. Analysis of CtPAL1 Expression and Its Impact on Flavonoid Pathway-Related Genes in Transgenic Arabidopsis
2.5. CtPAL1 Overexpression Significantly Modulates Flavonoid Accumulation in Transgenic Arabidopsis
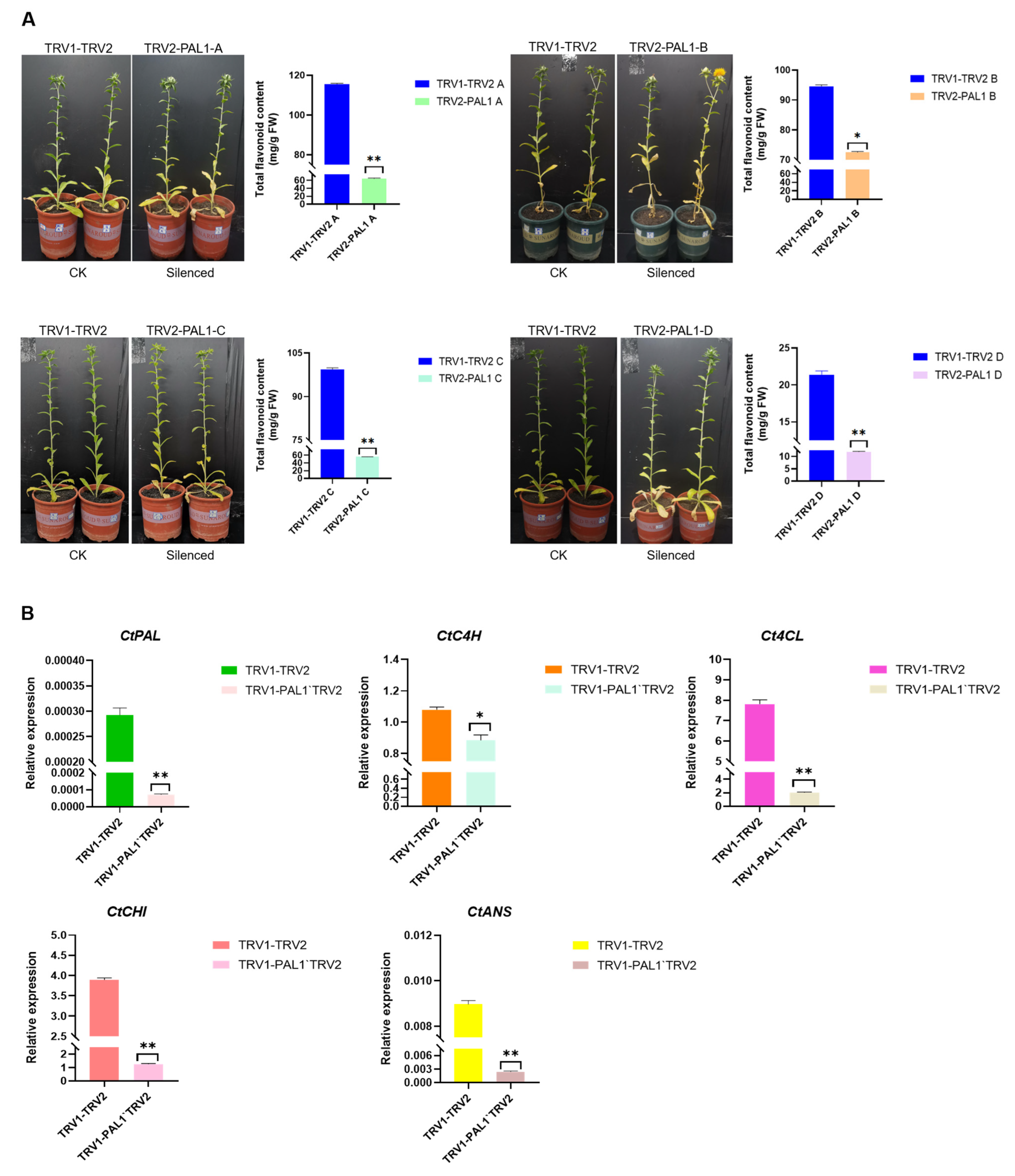
2.6. CtPAL1 Silencing Leads to Impaired Flavonoid Accumulation and Altered Expression of Flavonoid-Pathway-Related Genes in Transgenic Safflower
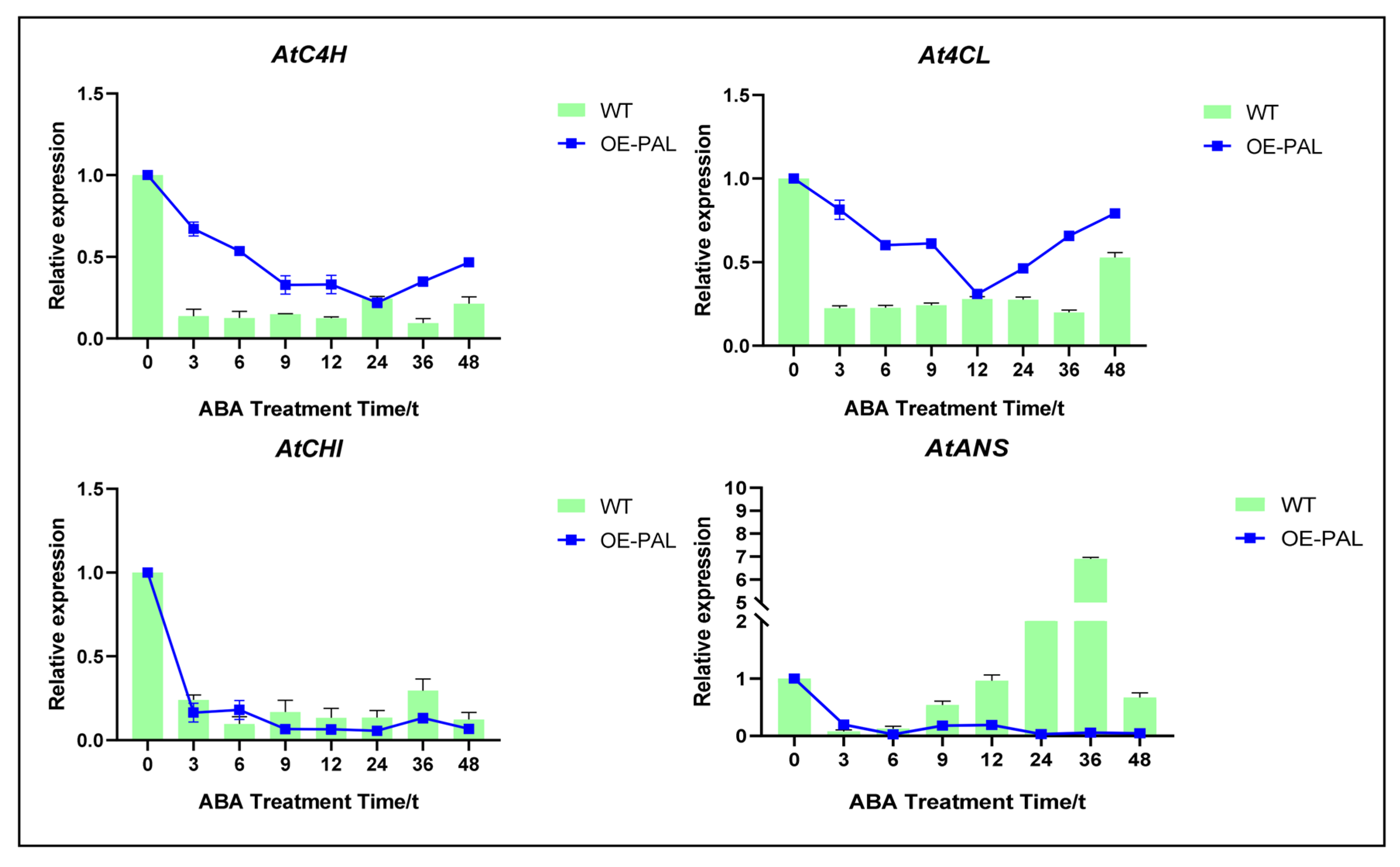
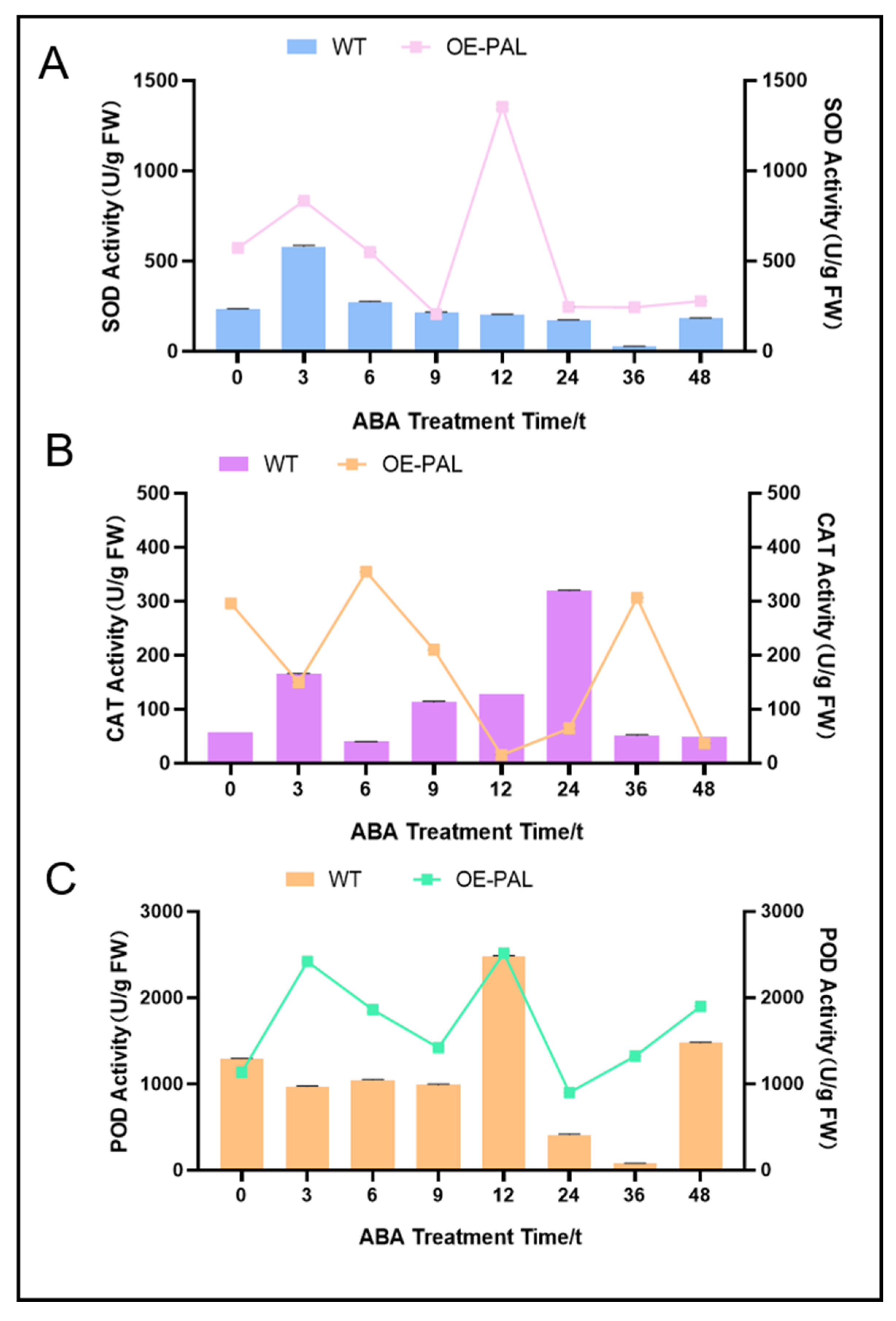
2.7. CtPAL1 Overexpression Enhances ABA-Induced Antioxidant Defense by Modulating Flavonoid Biosynthesis in Arabidopsis
2.8. CtPAL1-Overexpressed Transgenic Plants Under ABA Showed Enhanced Antioxidant Defense by Reducing Oxidative Damage
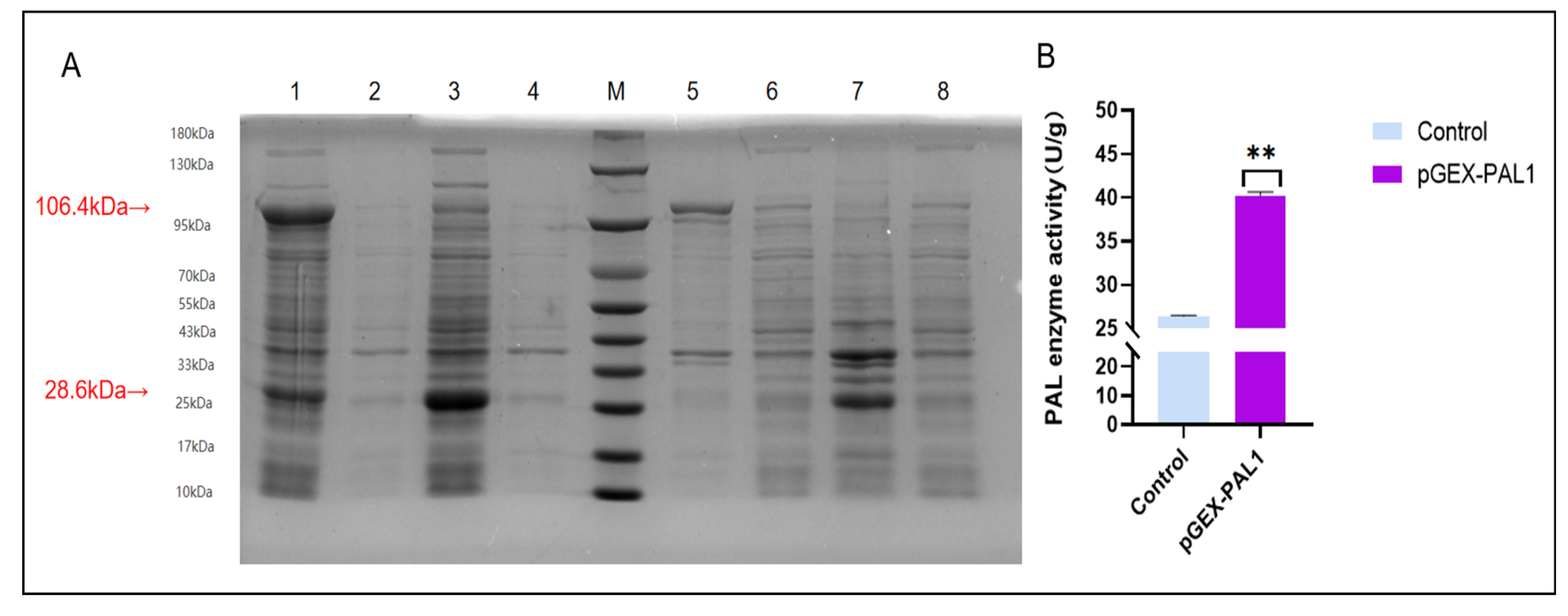
2.9. Protein Expression and In Vitro Enzymatic Activity of CtPAL1
3. Discussion
3.1. CtPAL1 Functions as Catalytic Gatekeeper in the Flavonoid Pathway
3.2. ABA as Key External Signal for Modulation of CtPAL1-Mediated Flavonoid Biosynthesis and Antioxidant Defense
3.3. Crosstalk Between CtPAL1 and Downstream Flavonoid Pathway Genes
4. Materials and Methods
4.1. Plant Materials, Treatments, and Growing Conditions
4.2. Protein Structure Analysis
4.3. Multiple Sequence Alignment and Phylogenetic Analysis
4.4. Construction of Prokaryotic Expression Vector of CtPAL1 Gene and Determination of Enzymatic Activity
4.5. Generation of T3 Transgenic Arabidopsis and Screening of High-Expression Lines
4.6. RNA Isolation and Real-Time Fluorescence Quantitative PCR Analysis
4.7. Determination of Total Flavonoid Content and Physiological and Biochemical Indices
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ahmad, N.; Li, T.; Liu, Y.; Hoang, N.Q.V.; Ma, X.; Zhang, X.; Liu, J.; Yao, N.; Liu, X.; Li, H. Molecular and biochemical rhythms in dihydroflavonol 4-reductase-mediated regulation of leucoanthocyanidin biosynthesis in Carthamus tinctorius L. Ind. Crops Prod. 2020, 156, 112838. [Google Scholar] [CrossRef]
- Asgarpanah, J.; Kazemivash, N. Phytochemistry, pharmacology and medicinal properties of Carthamus tinctorius L. Chin. J. Integr. Med. 2013, 19, 153–159. [Google Scholar] [CrossRef]
- Liu, J.; Yue, S.; Yang, Z.; Feng, W.; Meng, X.; Wang, A.; Peng, C.; Wang, C.; Yan, D. Oral hydroxysafflor yellow A reduces obesity in mice by modulating the gut microbiota and serum metabolism. Pharmacol. Res. 2018, 134, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-M.; Xiao, J.-H.; Wang, Y.; Tang, Y.-P.; Yue, S.-J. Research progress in chemical constituents and pigment extraction process of Carthami Flos. Zhongguo Zhong Yao Za Zhi = Zhongguo Zhongyao Zazhi = China J. Chin. Mater. Medica 2024, 49, 1725–1740. [Google Scholar]
- Adamska, I.; Biernacka, P. Bioactive Substances in Safflower Flowers and Their Applicability in Medicine and Health—Promoting Foods. Int. J. Food Sci. 2021, 2021, 6657639. [Google Scholar] [CrossRef] [PubMed]
- Gomashe, S.S.; Ingle, K.P.; Sarap, Y.A.; Chand, D.; Rajkumar, S. Safflower (Carthamus tinctorius L.): An underutilized crop with potential medicinal values. Ann. Phytomed. 2021, 10, 242–248. [Google Scholar] [CrossRef]
- Ma, X.; Hou, Y.; Umar, A.W.; Wang, Y.; Yu, L.; Ahmad, N.; Yao, N.; Zhang, M.; Liu, X. Safflower CtFLS1-induced drought tolerance by stimulating the accumulation of flavonols and anthocyanins in Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 5546. [Google Scholar] [CrossRef]
- Liu, X.; Ahmad, N.; Yang, L.; Fu, T.; Kong, J.; Yao, N.; Dong, Y.; Wang, N.; Li, X.; Wang, F. Molecular cloning and functional characterization of chalcone isomerase from Carthamus tinctorius. AMB Express 2019, 9, 132. [Google Scholar] [CrossRef]
- Ahmad, N.; Jianyu, L.; Xu, T.; Noman, M.; Jameel, A.; Na, Y.; Yuanyuan, D.; Nan, W.; Xiaowei, L.; Fawei, W. Overexpression of a novel cytochrome P450 promotes flavonoid biosynthesis and osmotic stress tolerance in transgenic Arabidopsis. Genes 2019, 10, 756. [Google Scholar] [CrossRef]
- Zhang, Q.; Ahmad, N.; Li, Z.; He, J.; Wang, N.; Naeem, M.; Jin, L.; Yao, N.; Liu, X. CtCYP71A1 promotes drought stress tolerance and lignin accumulation in safflower and Arabidopsis. Environ. Exp. Bot. 2023, 213, 105430. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Ahmad, N.; Sheng, X.; Iqbal, B.; Naeem, M.; Wang, N.; Li, F.; Yao, N.; Liu, X. Unraveling the functional characterization of a jasmonate-induced flavonoid biosynthetic CYP45082G24 gene in Carthamus tinctorius. Funct. Integr. Genom. 2023, 23, 172. [Google Scholar] [CrossRef]
- Ma, X.; Umar, A.W.; Zhou, Y.; Yu, L.; Liu, X.; Ahmad, N.; Zhang, J.; Liu, X. Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase CtHCT061 enhances lignin biosynthesis and drought tolerance in safflower (Carthamus tinctorius L.). Ind. Crops Prod. 2025, 236, 121929. [Google Scholar] [CrossRef]
- Sun, F.; Ahmad, N.; Jin, L.; Xinyue, Z.; Xintong, M.; Hoang, N.Q.; Mallano, A.I.; Wang, N.; Zhuoda, Y.; Liu, X. Genome-wide investigation of Hydroxycinnamoyl CoA: Shikimate Hydroxycinnamoyl Transferase (HCT) gene family in Carthamus tinctorius L. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12489. [Google Scholar]
- Hou, Y.; Wang, Y.; Liu, X.; Ahmad, N.; Wang, N.; Jin, L.; Yao, N.; Liu, X. A cinnamate 4-HYDROXYLASE1 from Safflower promotes flavonoids Accumulation and stimulates antioxidant Defense System in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 5393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ahmad, N.; Zhang, Q.; Wakeel Umar, A.; Wang, N.; Zhao, X.; Zhou, K.; Yao, N.; Liu, X. Safflower flavonoid 3′ 5′ hydroxylase promotes methyl jasmonate-induced anthocyanin accumulation in transgenic plants. Molecules 2023, 28, 3205. [Google Scholar] [CrossRef]
- Hong, Y.; Ahmad, N.; Tian, Y.; Liu, J.; Wang, L.; Wang, G.; Liu, X.; Dong, Y.; Wang, F.; Liu, W. Genome-wide identification, expression analysis, and subcellular localization of Carthamus tinctorius bHLH transcription factors. Int. J. Mol. Sci. 2019, 20, 3044. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, L.; ShangGuan, G.; Jia, C.; Deng, S.; Noman, M.; Liu, Y.; Guo, Y.; Han, L.; Zhang, X. Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctorius L. Sci. Rep. 2020, 10, 15521. [Google Scholar]
- Hong, Y.; Ahmad, N.; Zhang, J.; Lv, Y.; Zhang, X.; Ma, X.; Xiuming, L.; Na, Y. Genome-wide analysis and transcriptional reprogrammings of MYB superfamily revealed positive insights into abiotic stress responses and anthocyanin accumulation in Carthamus tinctorius L. Mol. Genet. Genom. 2022, 297, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Yufei, W.; Ahmad, N.; Jiaxin, C.; Lili, Y.; Yuying, H.; Nan, W.; Min, Z.; Libo, J.; Na, Y.; Xiuming, L. CtDREB52 transcription factor regulates UV-B-induced flavonoid biosynthesis by transactivating CtMYB and CtF3′ H in Safflower (Carthamus tinctorius L.). Plant Stress 2024, 11, 100384. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Jia, J.; Wang, C.; Fu, Y. Flavonoid—Lignin Crosstalk: Engineering Metabolic Flux for Optimised Plant Growth and Stress Resilience. Plant Cell Environ. 2025, 48, 8141–8160. [Google Scholar] [CrossRef]
- Amjad, M.; Wang, Y.; Han, S.; Haider, M.Z.; Sami, A.; Batool, A.; Shafiq, M.; Ali, Q.; Dong, J.; Sabir, I.A. Genome wide identification of phenylalanine ammonia-lyase (PAL) gene family in Cucumis sativus (cucumber) against abiotic stress. BMC Genom. Data 2024, 25, 76. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.-Q. Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RSC Adv. 2015, 5, 62587–62603. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.-J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Lavhale, S.G.; Kalunke, R.M.; Giri, A.P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 2018, 248, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Guo, L.; Wang, Y.; Li, Y.; Jiang, X.; Liu, Y.; Xie, D.-Y.; Gao, L.; Xia, T. Molecular and biochemical characterization of two 4-coumarate: CoA ligase genes in tea plant (Camellia sinensis). Plant Mol. Biol. 2022, 109, 579–593. [Google Scholar] [CrossRef]
- Ehlting, J.; Hamberger, B.; Million-Rousseau, R.; Werck-Reichhart, D. Cytochromes P450 in phenolic metabolism. Phytochem. Rev. 2006, 5, 239–270. [Google Scholar] [CrossRef]
- Longo, N.; Harding, C.O.; Burton, B.K.; Grange, D.K.; Vockley, J.; Wasserstein, M.; Rice, G.M.; Dorenbaum, A.; Neuenburg, J.K.; Musson, D.G. Single-dose, subcutaneous recombinant phenylalanine ammonia lyase conjugated with polyethylene glycol in adult patients with phenylketonuria: An open-label, multicentre, phase 1 dose-escalation trial. Lancet 2014, 384, 37–44. [Google Scholar] [CrossRef]
- Babich, O.O.; Pokrovsky, V.S.; Anisimova, N.Y.; Sokolov, N.N.; Prosekov, A.Y. Recombinant l-phenylalanine ammonia lyase from Rhodosporidium toruloides as a potential anticancer agent. Biotechnol. Appl. Biochem. 2013, 60, 316–322. [Google Scholar] [CrossRef]
- Rasool, F.; Uzair, M.; Naeem, M.K.; Rehman, N.; Afroz, A.; Shah, H.; Khan, M.R. Phenylalanine ammonia-lyase (PAL) genes family in wheat (Triticum aestivum L.): Genome-wide characterization and expression profiling. Agronomy 2021, 11, 2511. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Khan, N. Exploring plant resilience through secondary metabolite profiling: Advances in stress response and crop improvement. Plant Cell Environ. 2025, 48, 4823–4837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, W.; Wang, H.; Shu, C.; Chen, L.; Cao, J.; Jiang, W. The combination treatment of chlorogenic acid and sodium alginate coating could accelerate the wound healing of pear fruit by promoting the metabolic pathway of phenylpropane. Food Chem. 2023, 414, 135689. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef]
- Howles, P.A.; Sewalt, V.J.; Paiva, N.L.; Elkind, Y.; Bate, N.J.; Lamb, C.; Dixon, R.A. Overexpression of L-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol. 1996, 112, 1617–1624. [Google Scholar] [CrossRef]
- Kumar, A.; Ellis, B.E. The phenylalanine ammonia-lyase gene family in raspberry. Structure, expression, and evolution. Plant Physiol. 2001, 127, 230–239. [Google Scholar] [CrossRef]
- Saleem, M.H.; Mfarrej, M.F.B.; Khan, K.A.; Ercisli, S.; Elsharkawy, M.M.; Fahad, S. Advances in physiochemical and molecular mechanisms of abiotic stress tolerance in plants. J. Crop Health 2024, 76, 753–767. [Google Scholar] [CrossRef]
- Varadharajan, V.; Rajendran, R.; Muthuramalingam, P.; Runthala, A.; Madhesh, V.; Swaminathan, G.; Murugan, P.; Srinivasan, H.; Park, Y.; Shin, H. Multi-Omics Approaches Against Abiotic and Biotic Stress—A Review. Plants 2025, 14, 865. [Google Scholar] [CrossRef]
- Thingujam, D.; Gouli, S.; Cooray, S.P.; Chandran, K.B.; Givens, S.B.; Gandhimeyyan, R.V.; Tan, Z.; Wang, Y.; Patam, K.; Greer, S.A. Climate-Resilient Crops: Integrating AI, Multi-Omics, and Advanced Phenotyping to Address Global Agricultural and Societal Challenges. Plants 2025, 14, 2699. [Google Scholar] [CrossRef]
- Singh, A.; Roychoudhury, A. Abscisic acid in plants under abiotic stress: Crosstalk with major phytohormones. Plant Cell Rep. 2023, 42, 961–974. [Google Scholar] [CrossRef]
- Liu, H.; Song, S.; Zhang, H.; Li, Y.; Niu, L.; Zhang, J.; Wang, W. Signaling transduction of ABA, ROS, and Ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci. 2022, 23, 14824. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.-H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Q.; Wang, Y.; Xu, Y.; Li, J.; Zhao, S.; Wang, D.; Ma, Z.; Yan, F.; Liu, Y. Combined transcriptomic and metabolomic analysis reveals the role of phenylpropanoid biosynthesis pathway in the salt tolerance process of Sophora alopecuroides. Int. J. Mol. Sci. 2021, 22, 2399. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lai, W.; Long, L.; Gao, W.; Xu, F.; Li, P.; Zhou, S.; Ding, Y.; Hu, H. Comparative proteomic analysis identified proteins and the phenylpropanoid biosynthesis pathway involved in the response to ABA treatment in cotton fiber development. Sci. Rep. 2023, 13, 1488. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Gong, F.; Dong, J.; Lin, X.; Cao, K.; Xu, H.; Zhou, X. Abscisic Acid Affects Phenolic Acid Content to Increase Tolerance to UV-B Stress in Rhododendron chrysanthum Pall. Int. J. Mol. Sci. 2024, 25, 1234. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Park, N.I.; Li, X.; Xu, H.; Kim, H.H.; Park, S.U. Molecular cloning and characterization of phenylalanine ammonia-lyase and cinnamate 4-hydroxylase in the phenylpropanoid biosynthesis pathway in garlic (Allium sativum). J. Agric. Food Chem. 2010, 58, 10911–10917. [Google Scholar] [CrossRef]
- Rao, M.J.; Zheng, B. The role of polyphenols in abiotic stress tolerance and their antioxidant properties to scavenge reactive oxygen species and free radicals. Antioxidants 2025, 14, 74. [Google Scholar] [CrossRef]
- Sakihama, Y.; Cohen, M.F.; Grace, S.C.; Yamasaki, H. Plant phenolic antioxidant and prooxidant activities: Phenolics-induced oxidative damage mediated by metals in plants. Toxicology 2002, 177, 67–80. [Google Scholar] [CrossRef]
- Shomali, A.; Das, S.; Arif, N.; Sarraf, M.; Zahra, N.; Yadav, V.; Aliniaeifard, S.; Chauhan, D.K.; Hasanuzzaman, M. Diverse physiological roles of flavonoids in plant environmental stress responses and tolerance. Plants 2022, 11, 3158. [Google Scholar] [CrossRef]
- Sakihama, Y.; Yamasaki, H. Phytochemical Antioxidants: Past, Present and Future; IntechOpen: London, UK, 2021. [Google Scholar]
- Yamasaki, H. A function of colour. Trends Plant Sci. 1997, 2, 7–8. [Google Scholar] [CrossRef]
- Zagoskina, N.V.; Zubova, M.Y.; Nechaeva, T.L.; Kazantseva, V.V.; Goncharuk, E.A.; Katanskaya, V.M.; Baranova, E.N.; Aksenova, M.A. Polyphenols in plants: Structure, biosynthesis, abiotic stress regulation, and practical applications. Int. J. Mol. Sci. 2023, 24, 13874. [Google Scholar] [CrossRef]
- Rodríguez, B.; Pacheco, L.; Bernal, I.; Piña, M. Mechanisms of action of flavonoids: Antioxidant, antibacterial and antifungal properties. Cienc. Ambiente Clima 2023, 6, 33–66. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sakihama, Y.; Ikehara, N. Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol. 1997, 115, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.-F.; Liu, Y.; Pei, X.-Q.; Wu, Z.-L. Characterization of phenylalanine ammonia lyases from lettuce (Lactuca sativa L.) as robust biocatalysts for the production of D-and L-amino acids. J. Agric. Food Chem. 2023, 71, 2935–2942. [Google Scholar] [CrossRef]
- Hou, X.; Shao, F.; Ma, Y.; Lu, S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: Genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 2013, 40, 4301–4310. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Huang, S.-J.; Lin, S.-Y.; Wang, T.-T.; Hsu, F.-C. Combining acetic acid and ethanol as an anti-browning treatment for lettuce butt discoloration through repression of the activity and expression of phenylalanine ammonia lyase. Postharvest Biol. Technol. 2020, 164, 111151. [Google Scholar] [CrossRef]
- Katayama-Ikegami, A.; Sakamoto, T.; Shibuya, K.; Katayama, T.; Gao-Takai, M. Effects of abscisic acid treatment on berry coloration and expression of flavonoid biosynthesis genes in grape. Am. J. Plant Sci. 2016, 7, 1325. [Google Scholar] [CrossRef]
- Liu, A.; Zhu, Y.; Wang, Y.; Wang, T.; Zhao, S.; Feng, K.; Li, L.; Wu, P. Molecular identification of phenylalanine ammonia lyase-encoding genes EfPALs and EfPAL2-interacting transcription factors in Euryale ferox. Front. Plant Sci. 2023, 14, 1114345. [Google Scholar] [CrossRef]
- Hao, G.; Du, X.; Zhao, F.; Ji, H. Fungal endophytes-induced abscisic acid is required for flavonoid accumulation in suspension cells of Ginkgo biloba. Biotechnol. Lett. 2010, 32, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Gai, Z.; Wang, Y.; Ding, Y.; Qian, W.; Qiu, C.; Xie, H.; Sun, L.; Jiang, Z.; Ma, Q.; Wang, L. Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Sci. Rep. 2020, 10, 12275. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, L.-m.; Feng, N.; Zheng, D.; Shen, X.F.; Zhou, H.; Jiang, W.; Du, Y.; Zhao, H.; Lu, X. Plant growth regulators mitigate oxidative damage to rice seedling roots by NaCl stress. PeerJ 2024, 12, e17068. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Yoshida, K.; Igarashi, E.; Mukai, M.; Hirata, K.; Miyamoto, K. Induction of tolerance to oxidative stress in the green alga, Chlamydomonas reinhardtii, by abscisic acid. Plant Cell Environ. 2003, 26, 451–457. [Google Scholar] [CrossRef]
- Gao, W.; Yin, D.; Han, Z. Effects of ABA on Physiological Characteristics of Tomato under Waterlogging. E3S Web Conf. 2020, 189, 02007. [Google Scholar] [CrossRef]
- Cen, H.; Wang, T.; Liu, H.; Tian, D.; Zhang, Y. Melatonin application improves salt tolerance of alfalfa (Medicago sativa L.) by enhancing antioxidant capacity. Plants 2020, 9, 220. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Aravena-Calvo, J.; Busck-Mellor, S.; Laursen, T. Global organization of phenylpropanoid and anthocyanin pathways revealed by proximity labeling of trans-cinnamic acid 4-hydroxylase in Petunia inflata petal protoplasts. Front. Plant Sci. 2024, 15, 1295750. [Google Scholar] [CrossRef]
- Bhar, A. Integrative homology modeling and structural analyses of Cicer arietinum phenylalanine ammonia lyase (ca PAL) with co-expressed protein trans cinnamate 4 monooxygenase (C4H1) give clues to their interaction during stress response in chickpeas. Vegetos 2025, 38, 707–720. [Google Scholar] [CrossRef]
- Achnine, L.; Blancaflor, E.B.; Rasmussen, S.; Dixon, R.A. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 2004, 16, 3098–3109. [Google Scholar] [CrossRef] [PubMed]
- Sybilska, E.; Haddadi, B.S.; Mur, L.A.; Beckmann, M.; Hryhorowicz, S.; Suszynska-Zajczyk, J.; Knaur, M.; Pławski, A.; Daszkowska-Golec, A. Mapping the molecular signature of ABA-regulated gene expression in germinating barley embryos. BMC Plant Biol. 2025, 25, 619. [Google Scholar] [CrossRef]
- Xu, C.; Fan, X.; Shen, G.; Guo, B. Genome-wide identification of the phenylalanine ammonia-lyase gene from Epimedium Pubescens Maxim. (Berberidaceae): Novel insight into the evolution of the PAL gene family. BMC Plant Biol. 2024, 24, 831. [Google Scholar] [CrossRef]
- Biała, W.; Jasiński, M. The phenylpropanoid case–it is transport that matters. Front. Plant Sci. 2018, 9, 1610. [Google Scholar] [CrossRef]
- Hussain, Q.; Asim, M.; Zhang, R.; Khan, R.; Farooq, S.; Wu, J. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress. Biomolecules 2021, 11, 1159. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, N.; Yang, R.; Zhang, X.; Wang, Y.; Li, Y.; Prusky, D.; Bi, Y.; Han, Y. Essential role of ABA signaling and related transcription factors in phenolic acid and lignin synthesis during muskmelon wound healing. Front. Plant Sci. 2024, 15, 1404477. [Google Scholar] [CrossRef]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef]
- Villalobos-González, L.; Peña-Neira, A.; Ibáñez, F.; Pastenes, C. Long-term effects of abscisic acid (ABA) on the grape berry phenylpropanoid pathway: Gene expression and metabolite content. Plant Physiol. Biochem. 2016, 105, 213–223. [Google Scholar] [CrossRef]
- Koyama, R.; Roberto, S.R.; De Souza, R.T.; Borges, W.F.; Anderson, M.; Waterhouse, A.L.; Cantu, D.; Fidelibus, M.W.; Blanco-Ulate, B. Exogenous abscisic acid promotes anthocyanin biosynthesis and increased expression of flavonoid synthesis genes in Vitis vinifera× Vitis labrusca table grapes in a subtropical region. Front. Plant Sci. 2018, 9, 323. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhang, G.; Dai, M.; Zhao, H.; Ma, W.; Hu, Y.; Yao, N.; Zhang, J.; Ahmad, N.; Liu, X. The Phenylpropanoid Gatekeeper CtPAL1 Coordinates ABA-Induced Flavonoid Biosynthesis and Oxidative Stress Tolerance in Safflower (Carthamus tinctorius L.). Plants 2025, 14, 3606. https://doi.org/10.3390/plants14233606
Liu X, Zhang G, Dai M, Zhao H, Ma W, Hu Y, Yao N, Zhang J, Ahmad N, Liu X. The Phenylpropanoid Gatekeeper CtPAL1 Coordinates ABA-Induced Flavonoid Biosynthesis and Oxidative Stress Tolerance in Safflower (Carthamus tinctorius L.). Plants. 2025; 14(23):3606. https://doi.org/10.3390/plants14233606
Chicago/Turabian StyleLiu, Xiaoyu, Guanyao Zhang, Mingran Dai, Hong Zhao, Wei Ma, Yanli Hu, Na Yao, Jian Zhang, Naveed Ahmad, and Xiuming Liu. 2025. "The Phenylpropanoid Gatekeeper CtPAL1 Coordinates ABA-Induced Flavonoid Biosynthesis and Oxidative Stress Tolerance in Safflower (Carthamus tinctorius L.)" Plants 14, no. 23: 3606. https://doi.org/10.3390/plants14233606
APA StyleLiu, X., Zhang, G., Dai, M., Zhao, H., Ma, W., Hu, Y., Yao, N., Zhang, J., Ahmad, N., & Liu, X. (2025). The Phenylpropanoid Gatekeeper CtPAL1 Coordinates ABA-Induced Flavonoid Biosynthesis and Oxidative Stress Tolerance in Safflower (Carthamus tinctorius L.). Plants, 14(23), 3606. https://doi.org/10.3390/plants14233606







