Genome-Wide Identification and Characterization of the Xyloglucan Endotransglucosylase/Hydrolase (XTH) Gene Family in Camellia oleifera and the Function of CoXTH1 During Drought Stress
Abstract
1. Introduction
2. Results
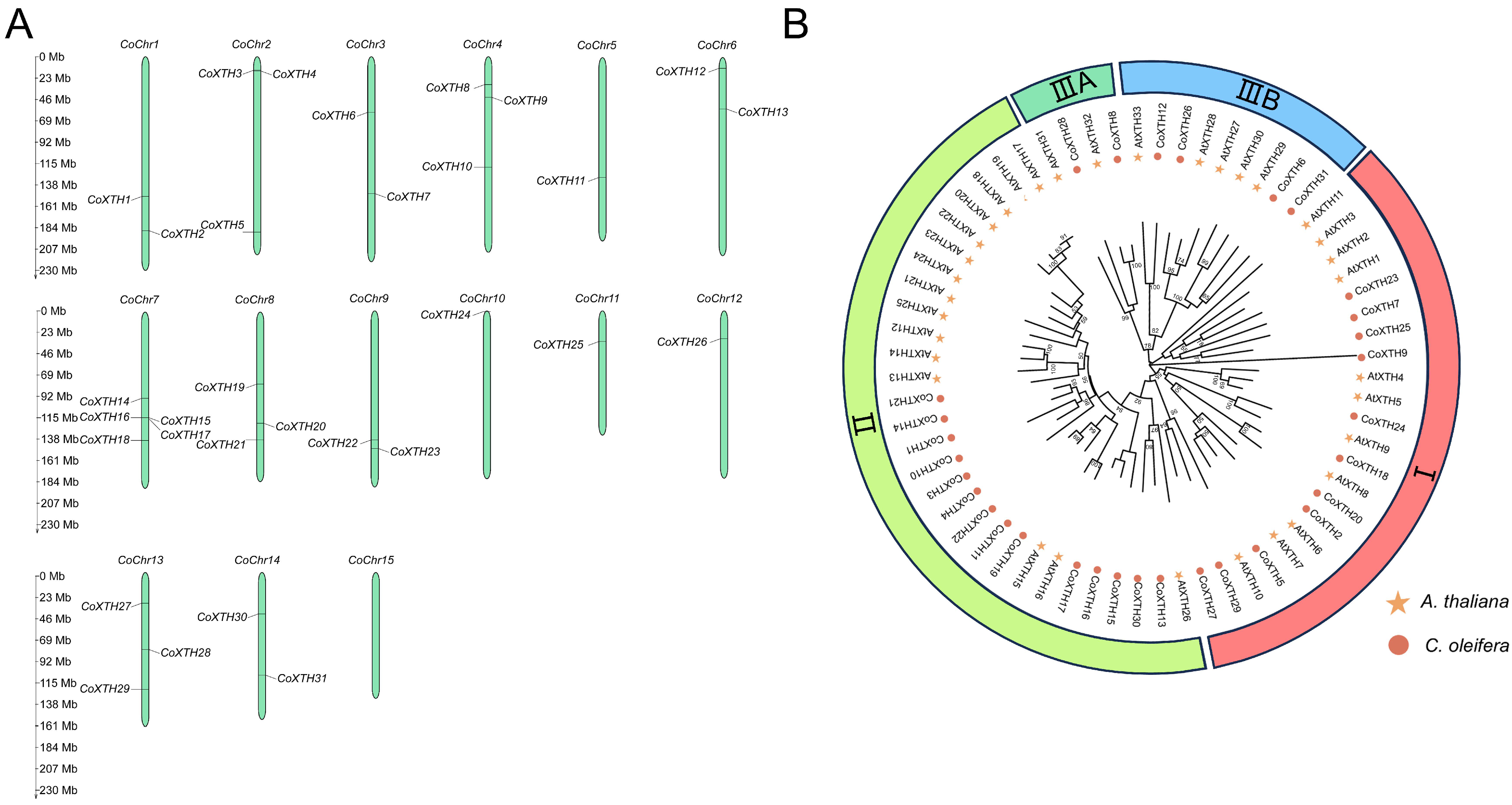
2.1. Identification of CoXTHs
2.2. Phylogenetic Relationships, Gene Structure, and Conserved Motifs of CoXTHs
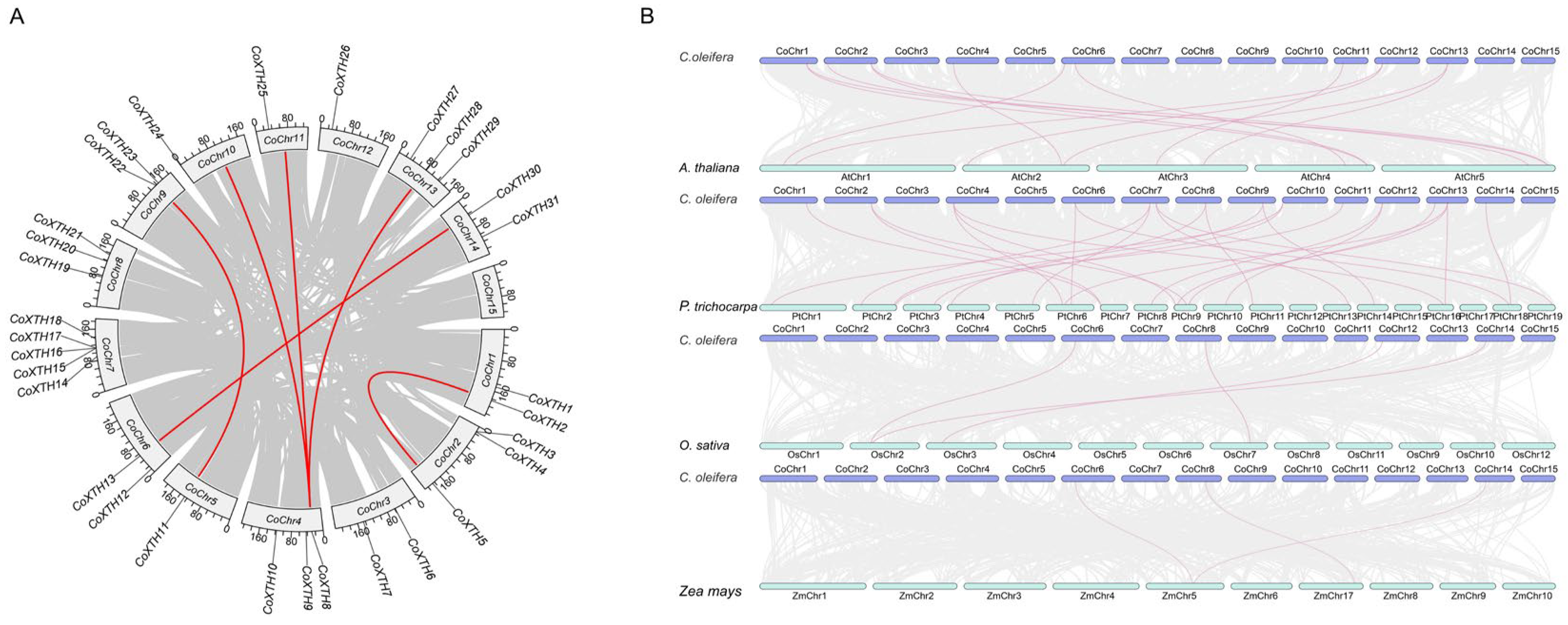
2.3. Synteny of CoXTHs
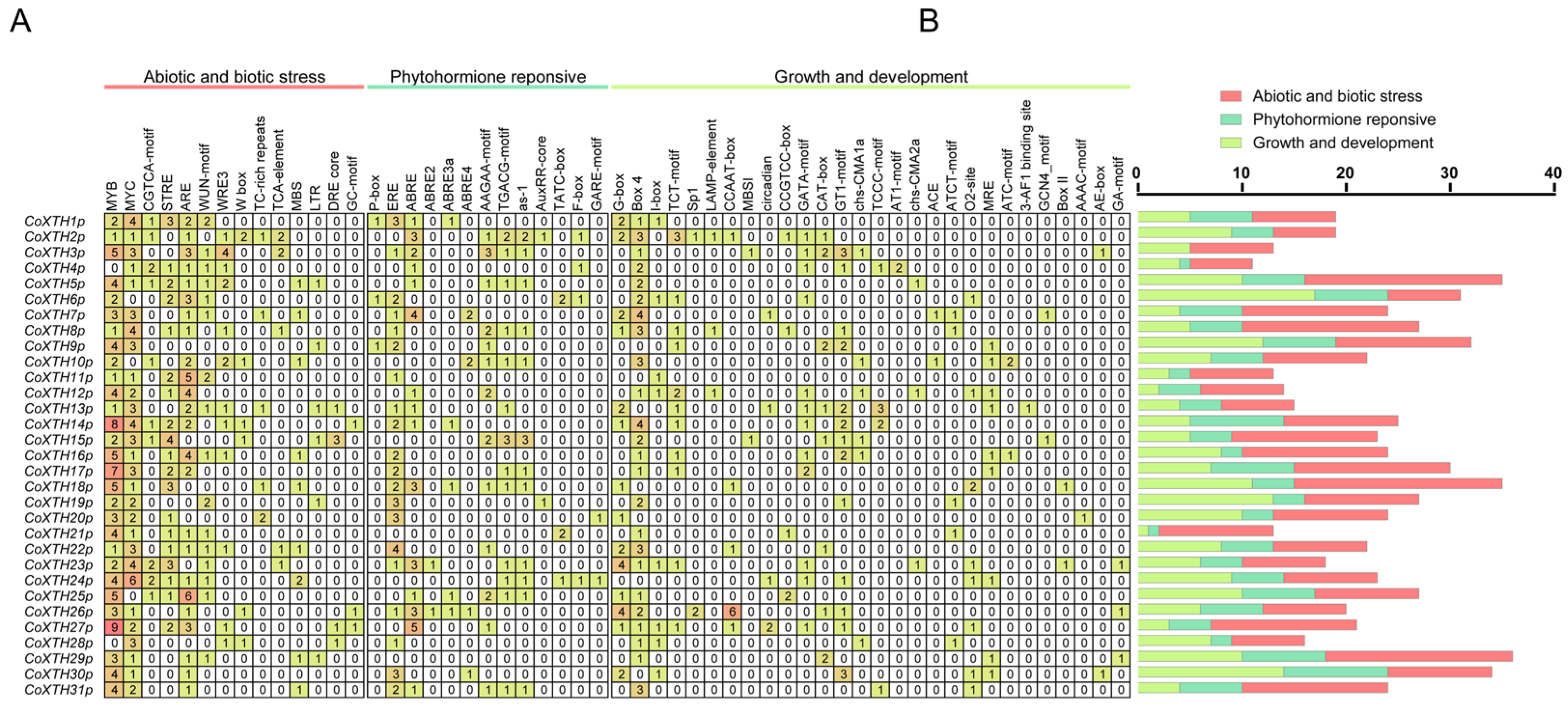
2.4. Cis-Acting Regulatory Elements of CoXTH Promoters
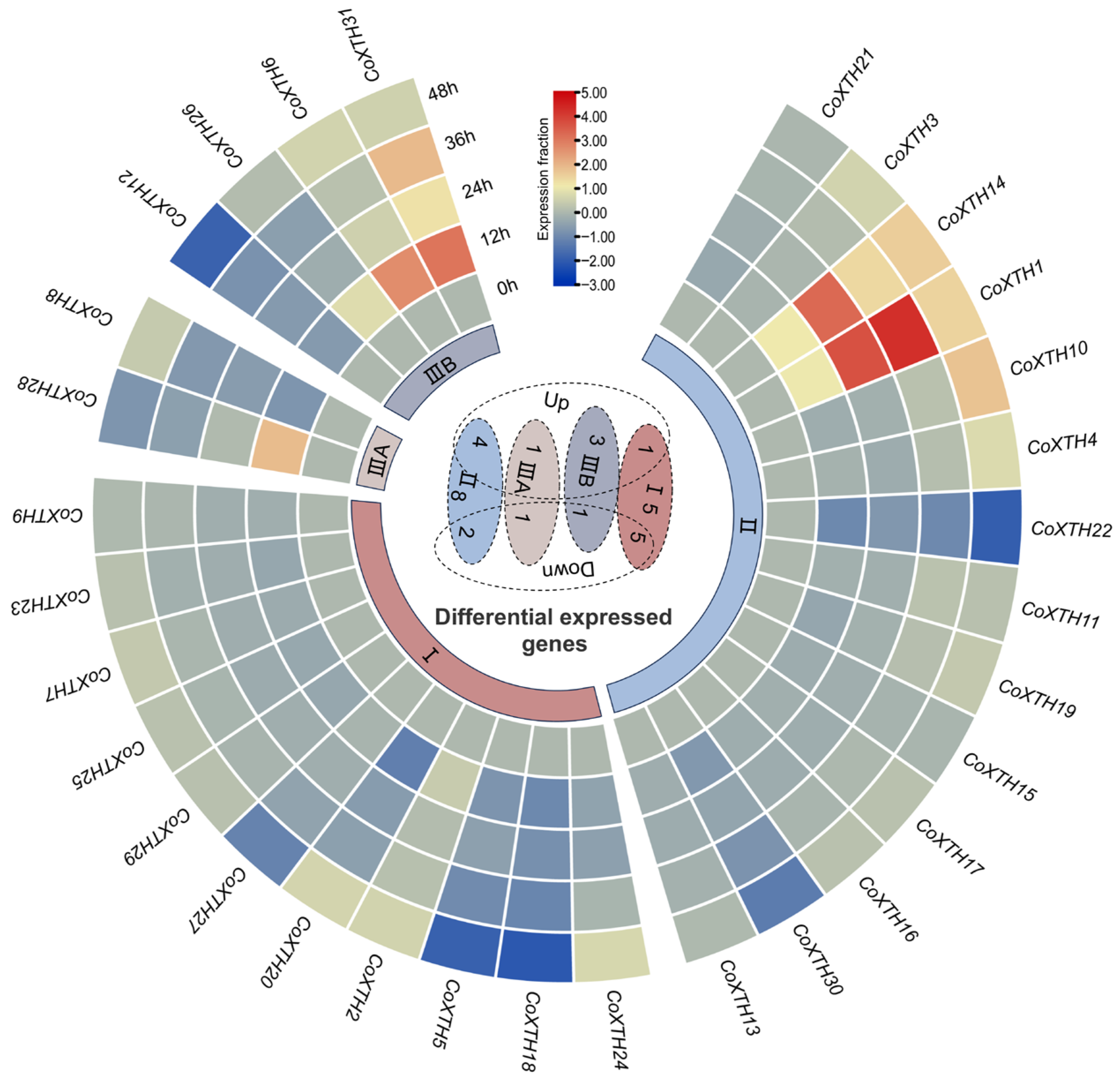
2.5. Expression of CoXTHs Under Drought Stress
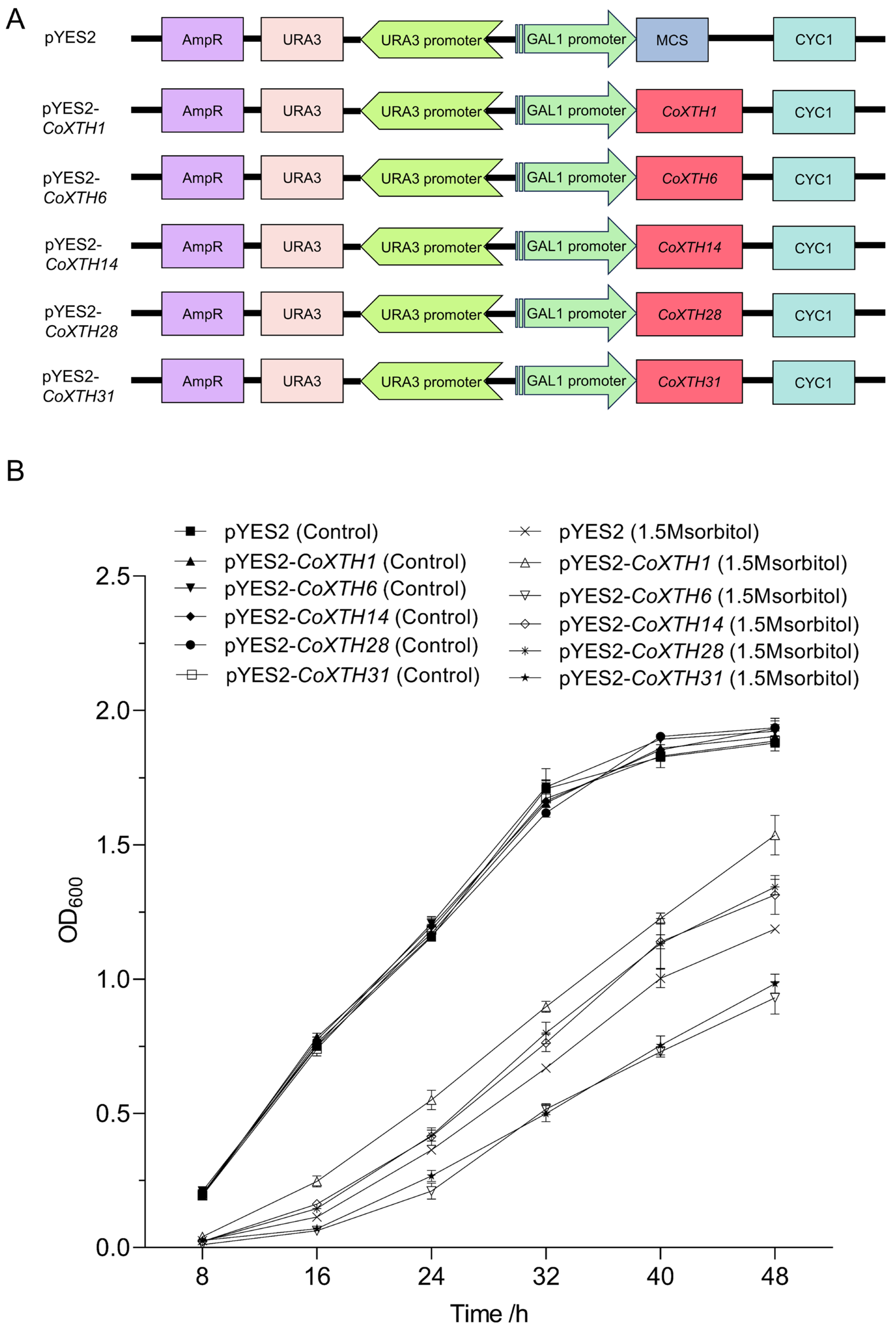
2.6. Drought Resistance of CoXTHs in Yeast
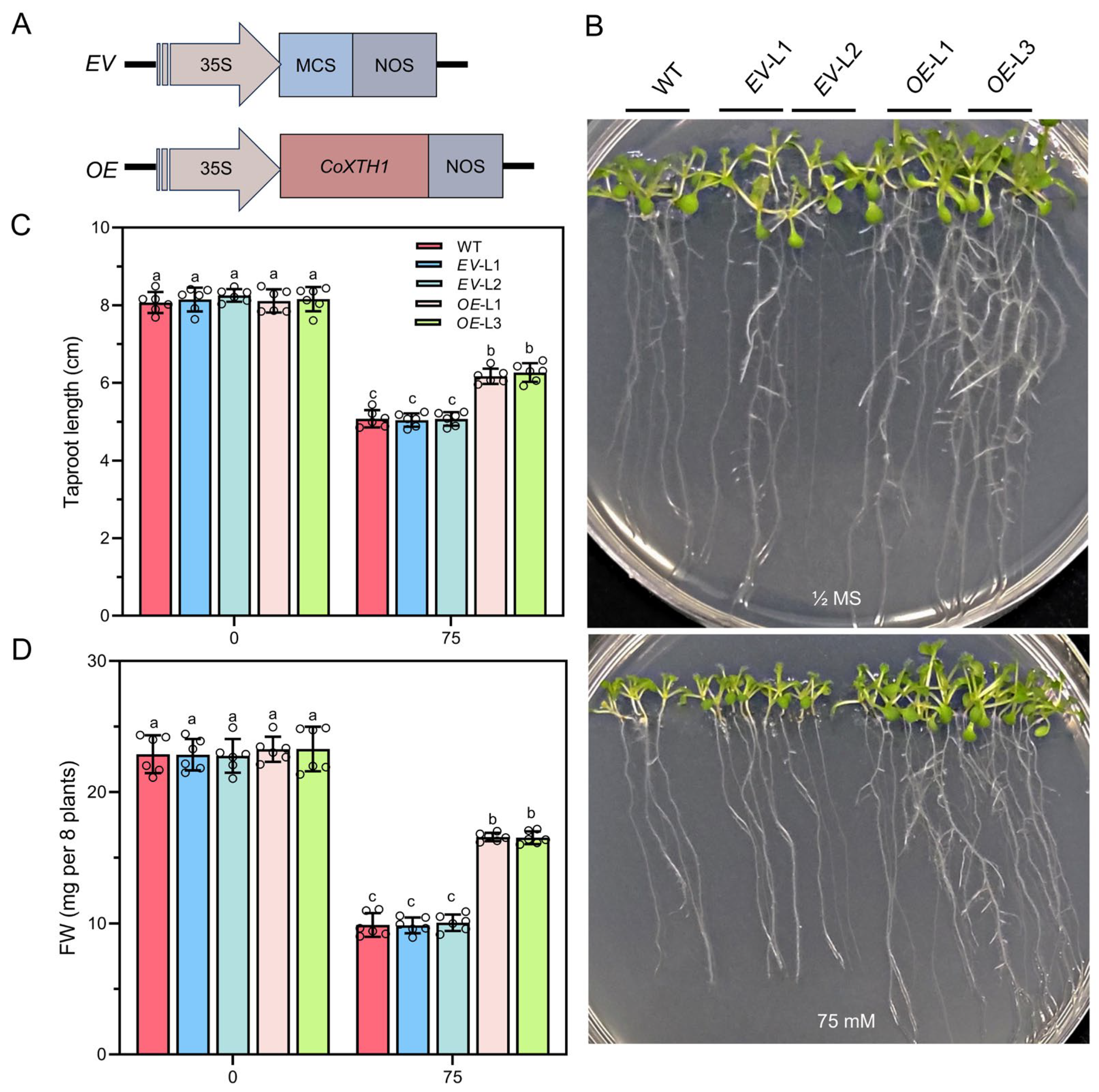
2.7. Overexpressing CoXTH1 in A. thaliana Enhances Its Drought Tolerance
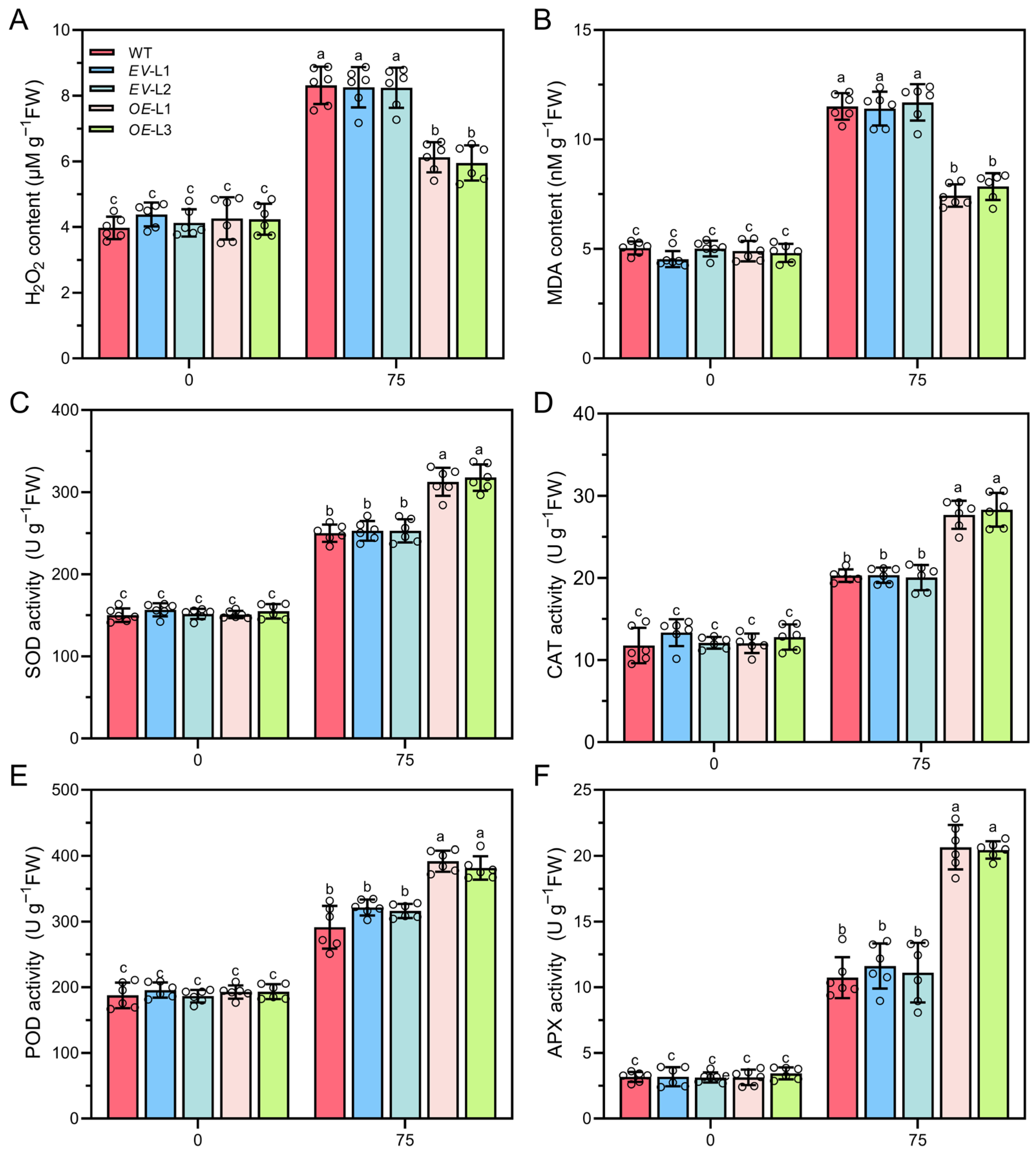
2.8. Overexpressing CoXTH1 Enhances the ROS Scavenging Ability of A. thaliana
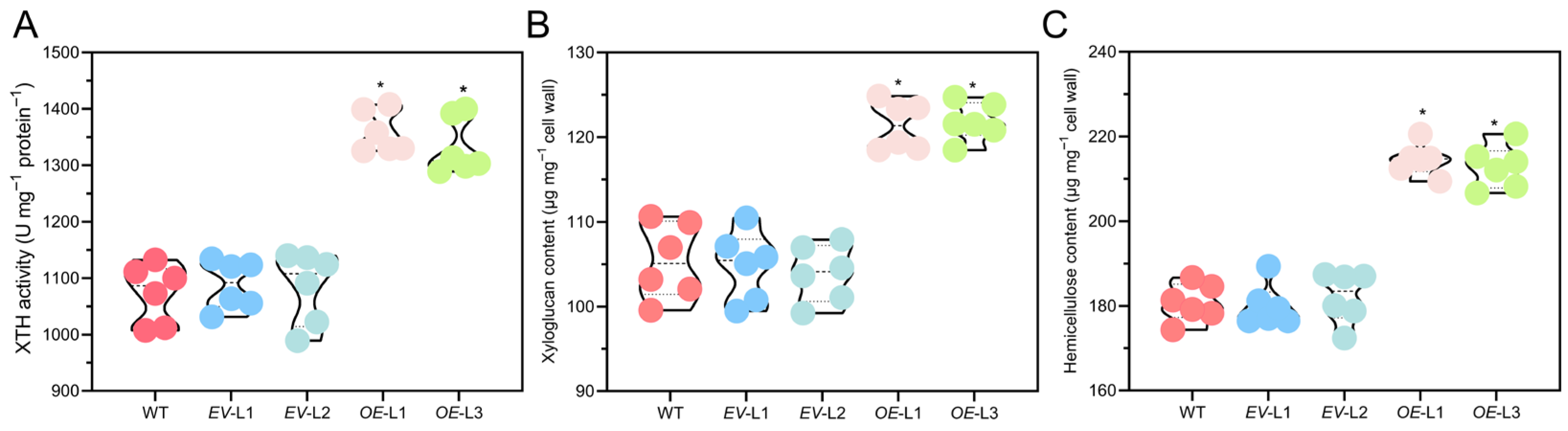
2.9. Overexpressing CoXTH1 Improves XTH Activity to Increase the Hemicellulose Content of A. thaliana
3. Discussion
4. Materials and Methods
4.1. Identification of the CoXTH Gene Family
4.2. Phylogenetic Relationship, Gene Structure, and Conserved Motif Analysis of the CoXTH Gene Family
4.3. Gene Duplication and Collinearity Analysis of the CoXTH Gene Family
4.4. Cis-Element Analysis of the CoXTH Gene Family Promoter
4.5. Expression Analysis of the CoXTH Gene Family Under Drought Stress
4.6. Heterologous Expression and Drought Resistance Assays of CoXTHs in Yeast
4.7. A. thaliana Transformation and Drought Treatment
4.8. Determination of H2O2, MDA, and Antioxidant Enzyme Activity
4.9. Measurement of Cell Wall Fraction and Xyloglucan Content
4.10. XTH Activities Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gebrechorkos, S.H.; Peng, J.; Dyer, E.; Miralles, D.G.; Vicente-Serrano, S.M.; Funk, C.; Beck, H.E.; Asfaw, D.T.; Singer, M.B.; Dadson, S.J. Global high-resolution drought indices for 1981–2022. Earth Syst. Sci. Data 2023, 15, 5449–5466. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological responses to drought, salinity, and heat stress in plants: A review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, W.; Ma, J.; Cheng, Z.; Zhang, X.; Liu, X.; Wu, X.; Zhang, J. An Integrated Framework for Drought Stress in Plants. Int. J. Mol. Sci. 2024, 25, 9347. [Google Scholar] [CrossRef]
- Ali, S.; Mir, R.A.; Haque, M.A.; Danishuddin; Almalki, M.A.; Alfredan, M.; Khalifa, A.; Mahmoudi, H.; Shahid, M.; Tyagi, A.; et al. Exploring physiological and molecular dynamics of drought stress responses in plants: Challenges and future directions. Front. Plant Sci. 2025, 16, 1565635. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Mishra, A.K.; Leung, R.; Cook, B. Global droughts connected by linkages between drought hubs. Nat. Commun. 2023, 14, 144. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Piao, S.; Huntingford, C.; Peñuelas, J.; Yang, H.; Xu, H.; Chen, A.; Friedlingstein, P.; Keenan, T.F.; Stephen Sitch, S.; et al. Global variations in critical drought thresholds that impact vegetation. Natl. Sci. Rev. 2023, 10, nwad049. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Folz, J.; Li, C.; Zhang, Y.; Hou, Z.; Zhang, L.; Su, S. Response of metabolic and lipid synthesis gene expression changes in Camellia oleifera to mulched ecological mat under drought conditions. Sci. Total Environ. 2021, 795, 148856. [Google Scholar] [CrossRef]
- Ma, X.; Sheng, L.; Li, F.; Zhou, T.; Guo, J.; Chang, Y.; Yang, J.; Jin, Y.; Chen, Y.; Lu, X. Seasonal drought promotes citrate accumulation in citrus fruit through the CsABF3-activated CsAN1-CsPH8 pathway. New Phytol. 2024, 242, 1131–1145. [Google Scholar] [CrossRef]
- Chieb, M.; Gachomo, E.W. The role of plant growth promoting rhizobacteria in plant drought stress responses. BMC Plant Biol. 2023, 23, 407. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Jie, H.; Zhao, L.; He, P.; Lv, X.; Xu, Y.; Zhang, Y.; Xing, H.; Jie, Y. BnXTH1 regulates cadmium tolerance by modulating vacuolar compartmentalization and the cadmium binding capacity of cell walls in ramie (Boehmeria nivea). J. Hazard. Mater. 2024, 470, 134172. [Google Scholar] [CrossRef] [PubMed]
- Oelmüller, R.; Tseng, Y.H.; Gandhi, A. Signals and their perception for remodelling, adjustment and repair of the plant cell wall. Int. J. Mol. Sci. 2023, 24, 7417. [Google Scholar] [CrossRef]
- Ma, Y.; Jie, H.; Tang, Y.; Xing, H.; Jie, Y. The role of hemicellulose in cadmium tolerance in ramie (Boehmeria nivea (L.) Gaud.). Plants 2022, 11, 1941. [Google Scholar] [CrossRef] [PubMed]
- Viborg, A.H.; Terrapon, N.; Lombard, V.; Michel, G.; Czjzek, M.; Henrissat, B.; Brumer, H. A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J. Biol. Chem. 2019, 294, 15973–15986. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Bio. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.S.; Jie, H.D.; Zhao, L.; Lv, X.Y.; Liu, X.C.; Tang, Y.Y.; Zhang, Y.; He, P.L.; Xing, H.C.; Jie, Y.C. Identification of the Xyloglucan Endotransglycosylase/Hydrolase (XTH) gene family members expressed in Boehmeria nivea in response to cadmium stress. Int. J. Mol. Sci. 2022, 23, 16104. [Google Scholar] [CrossRef]
- Yokoyama, R.; Rose, J.K.; Nishitani, K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004, 134, 1088–1099. [Google Scholar] [CrossRef]
- Yokoyama, R.; Nishitani, K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar] [CrossRef]
- Geisler-Lee, J.; Geisler, M.; Coutinho, P.M.; Segerman, B.; Nishikubo, N.; Takahashi, J.; Aspeborg, H.; Djerbi, S.; Master, E.; Andersson-Gunnerås, S.; et al. Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 2006, 140, 946–962. [Google Scholar] [CrossRef]
- Esmaeilzadeh-Moridani, M.; Esfahani, M.; Aalami, A.; Moumeni, A.; Khaledian, M.; Chaleshtori, M.H. Expression profiling of yield related genes in rice cultivars under terminal drought stress. Mol. Biol. Rep. 2023, 50, 8867–8875. [Google Scholar] [CrossRef]
- Han, Y.; Han, S.; Ban, Q.; He, Y.; Jin, M.; Rao, J. Overexpression of persimmon DkXTH1 enhanced tolerance to abiotic stress and delayed fruit softening in transgenic plants. Plant Cell Rep. 2017, 36, 583–596. [Google Scholar] [CrossRef]
- Fu, W.; Fan, D.; Zhang, Y. Genome-wide identification and characterization of xyloglucan endotransglucosylase/hydrolase gene family in maize (Zea mays L.) and the function of ZmXTH30 in response to drought stress. Environ. Exp. Bot. 2024, 222, 105744. [Google Scholar] [CrossRef]
- Bi, H.; Liu, Z.; Liu, S.; Qiao, W.; Zhang, K.; Zhao, M.; Wang, D. Genome-wide analysis of wheat xyloglucan endotransglucosylase/hydrolase (XTH) gene family revealed TaXTH17 involved in abiotic stress responses. BMC Plant Biol. 2024, 24, 640. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.M.; Cao, F.; Qiu, C.W.; Liu, C.; Tong, T.; Feng, X.; Cai, S.; Chen, Z.H.; Wu, F. Xyloglucan endotransglucosylase-hydrolase 1 is a negative regulator of drought tolerance in barley via modulating lignin biosynthesis and stomatal closure. Plant Physiol. Biochem. 2024, 216, 109171. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Lu, C.; Tang, Y.; Jiang, X.; Gai, Y. Isolation and characterization of Populus xyloglucan endotransglycosylase/hydrolase (XTH) involved in osmotic stress responses. Int. J. Biol. Macromol. 2020, 155, 1277–1287. [Google Scholar] [CrossRef]
- Wu, D.; Liu, A.; Qu, X.; Liang, J.; Song, M. Genome-wide identification, and phylogenetic and expression profiling analyses, of XTH gene families in Brassica rapa L. and Brassica oleracea L. BMC Genom. 2020, 21, 782. [Google Scholar] [CrossRef]
- Campbell, P.; Braam, J. Co- and/or post-translational modifications are critical for TCH4 XET activity. Plant J. 1998, 15, 553–561. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; International Wheat Genome Sequencing Consortium; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, X.; Yao, W.; Gao, Y.; Zhao, K.; Guo, Q.; Zhou, B.; Jiang, T. Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in poplar. BMC Genom. 2021, 22, 804. [Google Scholar] [CrossRef]
- Zhang, J.Z.; He, P.W.; Xu, X.M.; Lü, Z.F.; Cui, P.; George, M.S.; Lu, G.Q. Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in sweet potato [Ipomoea batatas (L.) Lam]. Int. J. Mol. Sci. 2023, 24, 775. [Google Scholar] [CrossRef]
- He, F.; Shi, Y.J.; Li, J.L.; Lin, T.T.; Zhao, K.J.; Chen, L.H.; Mi, J.X.; Zhang, F.; Zhong, Y.; Lu, M.M.; et al. Genome-wide analysis and expression profiling of Cation/H+ exchanger (CAX) family genes reveal likely functions in cadmium stress responses in poplar. Int. J. Biol. Macromol. 2022, 204, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Wei, Z.; Zhai, H.; Xing, S.; Wang, Y.; He, S.; Gao, S.; Zhao, N.; Zhang, H.; Liu, Q. The IbPYL8–IbbHLH66–IbbHLH118 complex mediates the abscisic acid-dependent drought response in sweet potato. New Phytol. 2022, 236, 2151–2171. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Xu, P.; Fang, S.; Chen, H.; Cai, W. The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 2020, 104, 59–75. [Google Scholar] [CrossRef]
- Wang, P.; Liu, W.C.; Han, C.; Wang, S.; Bai, M.Y.; Song, C.P. Reactive oxygen species: Multidimensional regulators of plant adaptation to abiotic stress and development. J. Integr. Plant Biol. 2024, 66, 330–367. [Google Scholar] [CrossRef]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Zhang, T.; Zheng, Y.; Cosgrove, D.J.; Anderson, C.T. Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol. 2016, 170, 234–249. [Google Scholar] [CrossRef]
- Liu, Y.B.; Lu, S.M.; Zhang, J.F.; Liu, S.; Lu, Y.T. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 2007, 226, 1547–1560. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Huang, Y.; He, H.; Han, T.; Di, P.; Sechet, J.; Fang, L.; Liang, Y.; Scheller, H.V.; Mortimer, J.C.; et al. Xyloglucan endotransglucosylase-hydrolase 30 negatively affects salt tolerance in Arabidopsis. J. Exp. Bot. 2019, 70, 5495–5506. [Google Scholar] [CrossRef]
- Gong, W.; Xiao, S.; Wang, L.; Liao, Z.; Chang, Y.; Mo, W.; Hu, G.; Li, W.; Zhao, G.; Zhu, H.; et al. Chromosome-level genome of Camellia lanceoleosa provides a valuable resource for understanding genome evolution and self-incompatibility. Plant J. 2022, 110, 881–898. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, Springer Protocols Handbooks; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Dong, B.; Wu, B.; Hong, W.; Li, X.; Li, Z.; Xue, L.; Huang, Y. Transcriptome analysis of the tea oil camellia (Camellia oleifera) reveals candidate drought stress genes. PLoS ONE 2017, 12, e0181835. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Li, X.Y.; Chow, C.K. An improved method for the measurement of malondialdehyde in biological samples. Lipids 1994, 29, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Gebicki, J.M.; Collins, J.; Baoutina, A.; Phair, P. The limitations of an iodometric aerobic assay for peroxides. Anal. Biochem. 1996, 240, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.C.; Desborough, S.L. Rapid estimation of potato tuber total protein content with coomassie brilliant blue G-250. Theor. Appl. Genet. 1978, 52, 135–139. [Google Scholar] [CrossRef] [PubMed]
| Name | Genome Location | PI | Mw (kDa) | Peptide Residue (aa) | Gravy | Aliphatic Index | CDS Length (bp) | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| CoXTH1 | Chr1:148173832-148175859 | 6.81 | 32.24 | 287 | −0.322 | 73.41 | 864 | Cell wall or Cytoplasm |
| CoXTH2 | Chr1:184263085-184266113 | 7.04 | 32.90 | 287 | −0.309 | 74.70 | 864 | Cell wall |
| CoXTH3 | Chr2:14125984-14127001 | 6.59 | 32.01 | 283 | −0.226 | 74.81 | 852 | Cell wall or Cytoplasm |
| CoXTH4 | Chr2:15002727-15005660 | 4.73 | 33.43 | 297 | −0.374 | 75.22 | 894 | Cell wall |
| CoXTH5 | Chr2:185621304-185624087 | 6.44 | 33.39 | 293 | −0.363 | 68.91 | 882 | Cell wall |
| CoXTH6 | Chr3:144786121-144788984 | 8.97 | 39.27 | 341 | −0.549 | 67.18 | 1026 | Cell wall |
| CoXTH7 | Chr3:58816285-58819353 | 6.04 | 31.60 | 276 | −0.471 | 79.29 | 831 | Cell wall |
| CoXTH8 | Chr4:29353162-29355745 | 9.31 | 33.75 | 294 | −0.339 | 63.06 | 885 | Cell wall |
| CoXTH9 | Chr4:43136898-43148390 | 9.11 | 36.83 | 319 | −0.461 | 81.25 | 960 | Cell wall |
| CoXTH10 | Chr4:117068255-117071186 | 4.81 | 28.63 | 256 | −0.198 | 80.43 | 771 | Cell wall |
| CoXTH11 | Chr5:127002401-127004274 | 8.45 | 32.49 | 284 | −0.367 | 64.51 | 855 | Cell wall or Cytoplasm |
| CoXTH12 | Chr6:11102489-11105784 | 6.13 | 35.19 | 311 | −0.188 | 77.72 | 936 | Cell wall |
| CoXTH13 | Chr6:54452832-54463757 | 8.21 | 33.53 | 291 | −0.449 | 60.72 | 876 | Cell wall |
| CoXTH14 | Chr7:91156960-91242883 | 5.36 | 32.07 | 288 | −0.341 | 69.10 | 867 | Cell wall or Cytoplasm |
| CoXTH15 | Chr7:112011888-112012997 | 7.58 | 32.53 | 289 | −0.252 | 72.56 | 870 | Cell wall |
| CoXTH16 | Chr7:112063339-112064355 | 9.25 | 30.10 | 261 | −0.526 | 63.87 | 786 | Cell wall |
| CoXTH17 | Chr7:112080286-112081540 | 8.58 | 38.44 | 336 | −0.372 | 68.18 | 1011 | Cell wall or Cytoplasm |
| CoXTH18 | Chr7:136193032-136194628 | 5.99 | 33.79 | 297 | −0.308 | 67.58 | 894 | Cell wall |
| CoXTH19 | Chr8:76121722-76124240 | 8.82 | 33.21 | 293 | −0.348 | 64.91 | 882 | Cell wall or Cytoplasm |
| CoXTH20 | Chr8:117607450-117609597 | 5.45 | 39.09 | 337 | −0.557 | 58.46 | 1014 | Cell wall |
| CoXTH21 | Chr8:135166938-135167942 | 8.80 | 38.18 | 334 | −0.500 | 58.71 | 1005 | Cell wall or Cytoplasm |
| CoXTH22 | Chr9:136941105-136943159 | 9.25 | 32.08 | 281 | −0.326 | 71.78 | 846 | Cell wall or Cytoplasm |
| CoXTH23 | Chr9:145532451-145534317 | 8.72 | 33.11 | 286 | −0.490 | 67.13 | 861 | Cell wall |
| CoXTH24 | Chr10:891170-894544 | 7.65 | 34.12 | 295 | −0.465 | 65.42 | 888 | Cell wall or Cytoplasm |
| CoXTH25 | Chr11:32609393-32611893 | 5.41 | 32.29 | 282 | −0.661 | 59.82 | 849 | Cell wall |
| CoXTH26 | Chr12:29226967-29229771 | 6.17 | 36.39 | 320 | −0.495 | 71.84 | 963 | Cell wall |
| CoXTH27 | Chr13:32352163-32354989 | 8.23 | 35.13 | 302 | −0.374 | 72.95 | 909 | Cell wall |
| CoXTH28 | Chr13:81713234-81715882 | 5.33 | 32.84 | 291 | −0.453 | 61.65 | 876 | Cell wall |
| CoXTH29 | Chr13:123918650-123921005 | 5.88 | 34.71 | 298 | −0.412 | 74.90 | 897 | Cell wall |
| CoXTH30 | Chr14:43790930-43793214 | 6.20 | 33.50 | 289 | −0.398 | 63.11 | 870 | Cell wall |
| CoXTH31 | Chr14:108417298-108423064 | 9.11 | 40.14 | 352 | −0.414 | 68.47 | 1059 | Cell wall |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Zhang, Y.; Zhang, Z.; He, Z.; Xun, C.; Wang, X.; Zhang, Y.; Wang, R.; Chen, Y. Genome-Wide Identification and Characterization of the Xyloglucan Endotransglucosylase/Hydrolase (XTH) Gene Family in Camellia oleifera and the Function of CoXTH1 During Drought Stress. Plants 2025, 14, 3605. https://doi.org/10.3390/plants14233605
Ma Y, Zhang Y, Zhang Z, He Z, Xun C, Wang X, Zhang Y, Wang R, Chen Y. Genome-Wide Identification and Characterization of the Xyloglucan Endotransglucosylase/Hydrolase (XTH) Gene Family in Camellia oleifera and the Function of CoXTH1 During Drought Stress. Plants. 2025; 14(23):3605. https://doi.org/10.3390/plants14233605
Chicago/Turabian StyleMa, Yushen, Ying Zhang, Zhen Zhang, Zhilong He, Chengfeng Xun, Xiangnan Wang, Yufeng Zhang, Rui Wang, and Yongzhong Chen. 2025. "Genome-Wide Identification and Characterization of the Xyloglucan Endotransglucosylase/Hydrolase (XTH) Gene Family in Camellia oleifera and the Function of CoXTH1 During Drought Stress" Plants 14, no. 23: 3605. https://doi.org/10.3390/plants14233605
APA StyleMa, Y., Zhang, Y., Zhang, Z., He, Z., Xun, C., Wang, X., Zhang, Y., Wang, R., & Chen, Y. (2025). Genome-Wide Identification and Characterization of the Xyloglucan Endotransglucosylase/Hydrolase (XTH) Gene Family in Camellia oleifera and the Function of CoXTH1 During Drought Stress. Plants, 14(23), 3605. https://doi.org/10.3390/plants14233605






