A Comprehensive Profiling of the Rice LATERAL ORGAN BOUNDARIES DOMAIN (LBD) Gene Family: Structure, Evolution, and Expressional Dynamics
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Characterization of OsLBD Genes
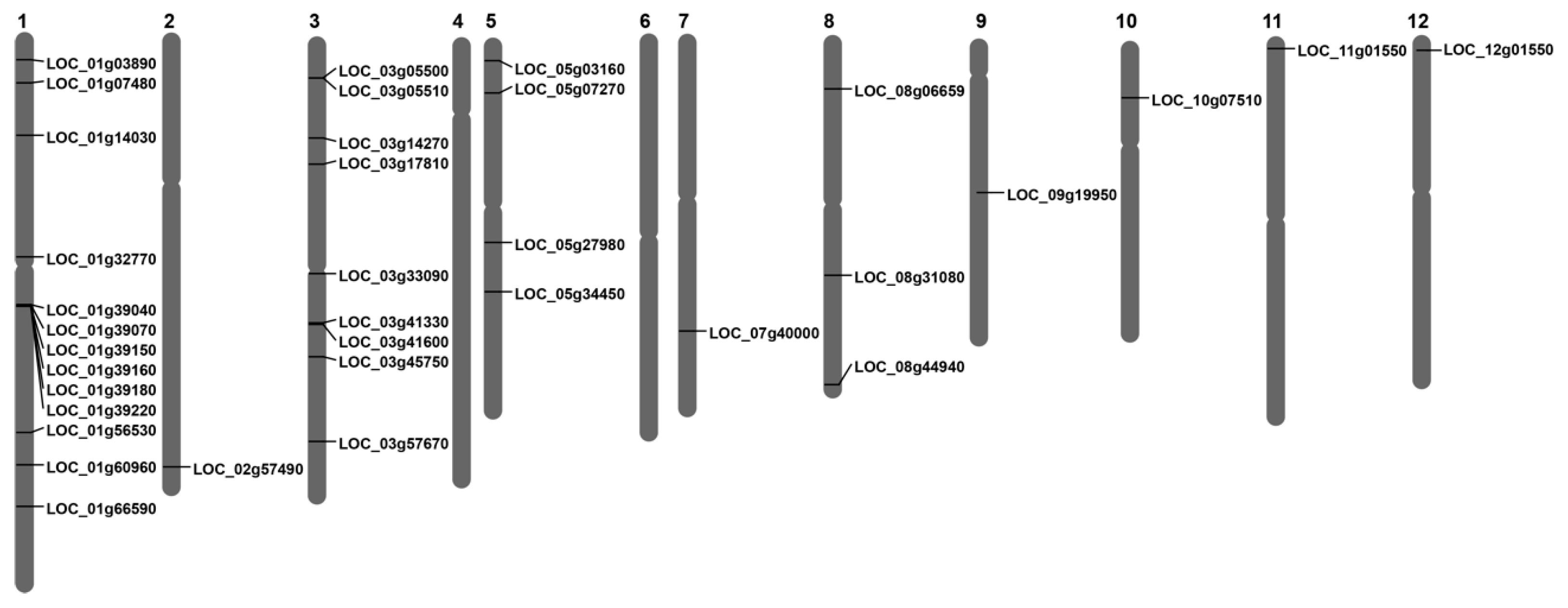
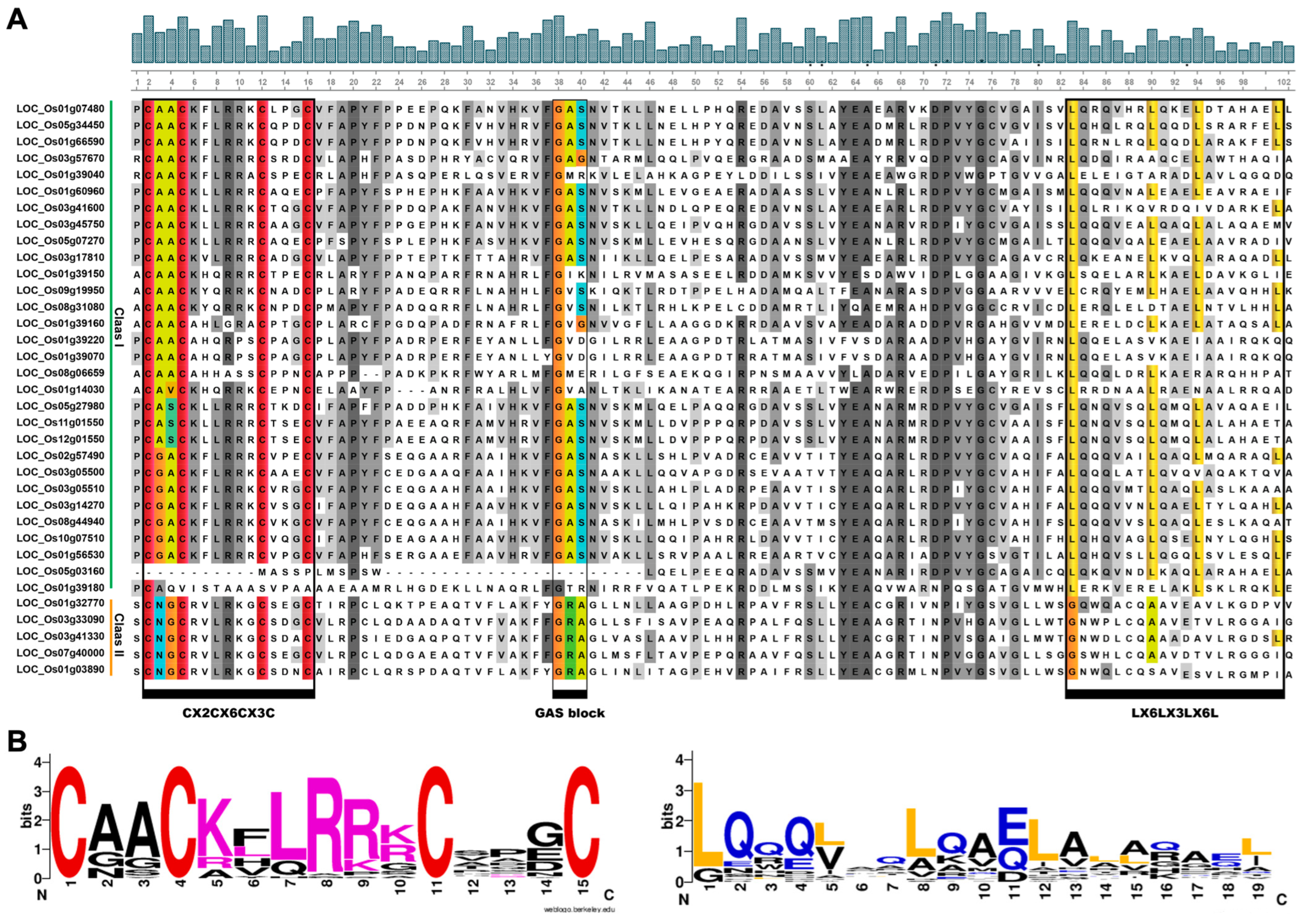
2.2. Chromosomal Distribution and Multiple Sequence Alignment of OsLBD Genes
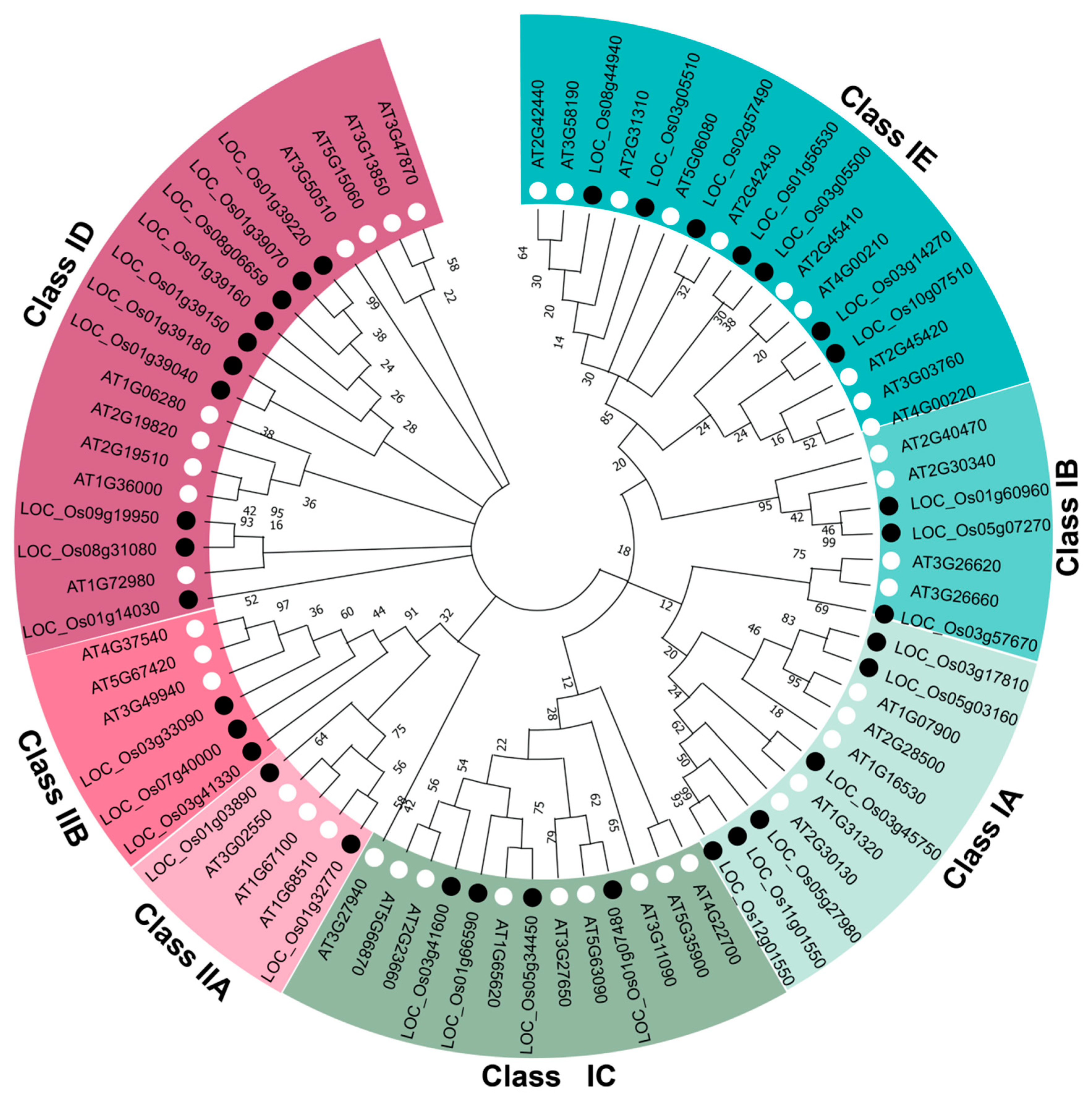
2.3. Phylogenetic Analysis and Evolutionary Relationships of OsLBDs
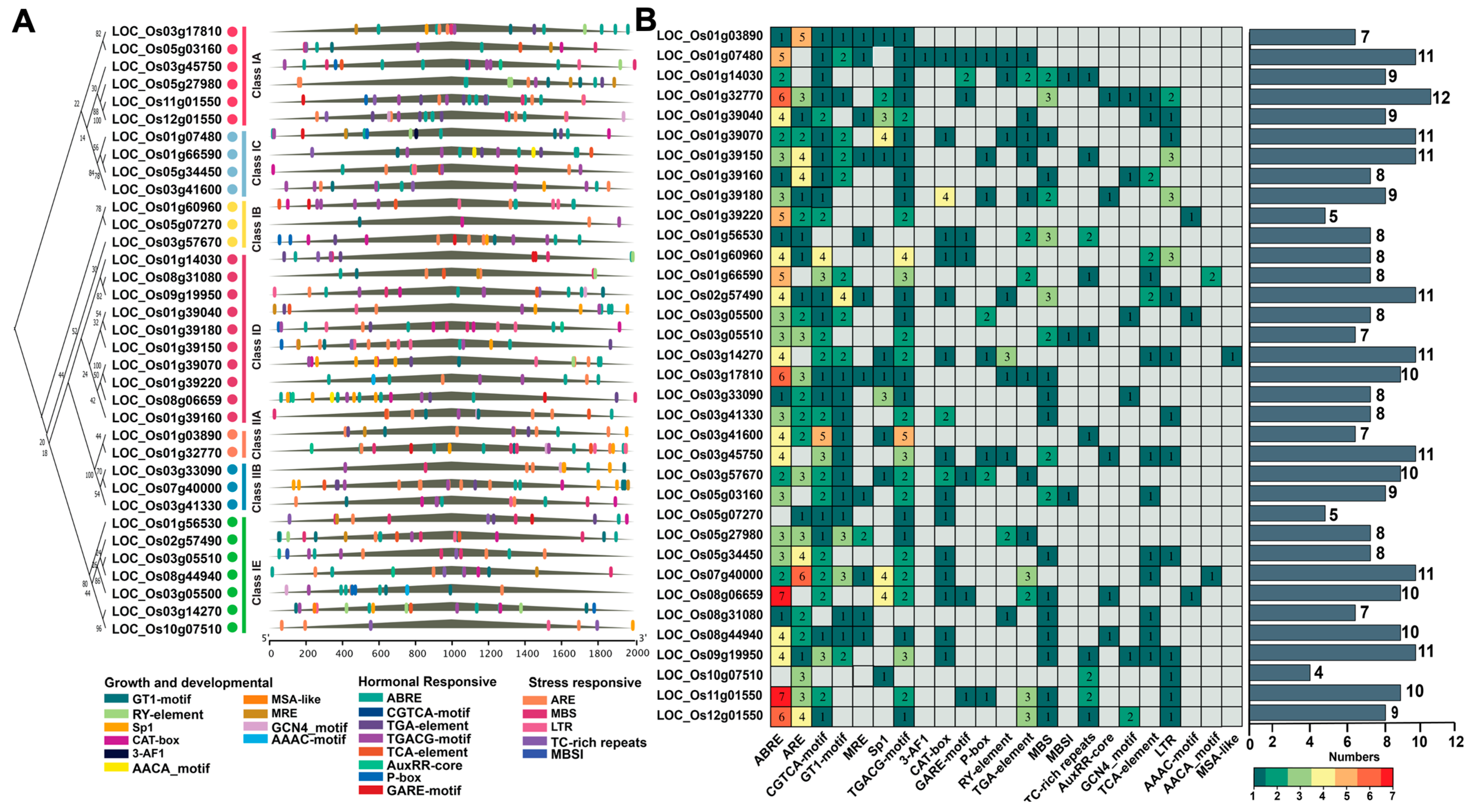
2.4. Gene Structure and Conserved Motif Analysis of the Rice OsLBD Gene Family
2.5. Cis-Regulatory Element Analysis in OsLBD Promoters
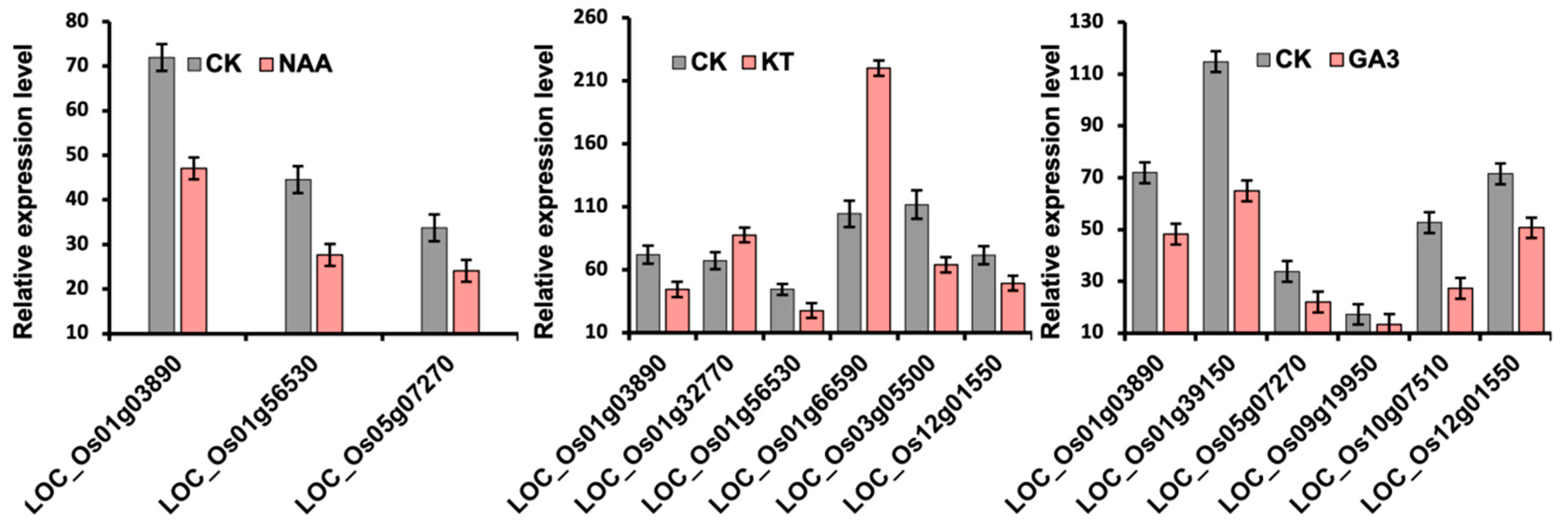
2.6. Expression of OsLBD Genes Under NAA, KT, and GA3 Hormonal Treatments
2.7. OsLBD Gene Duplication and Expression Divergence
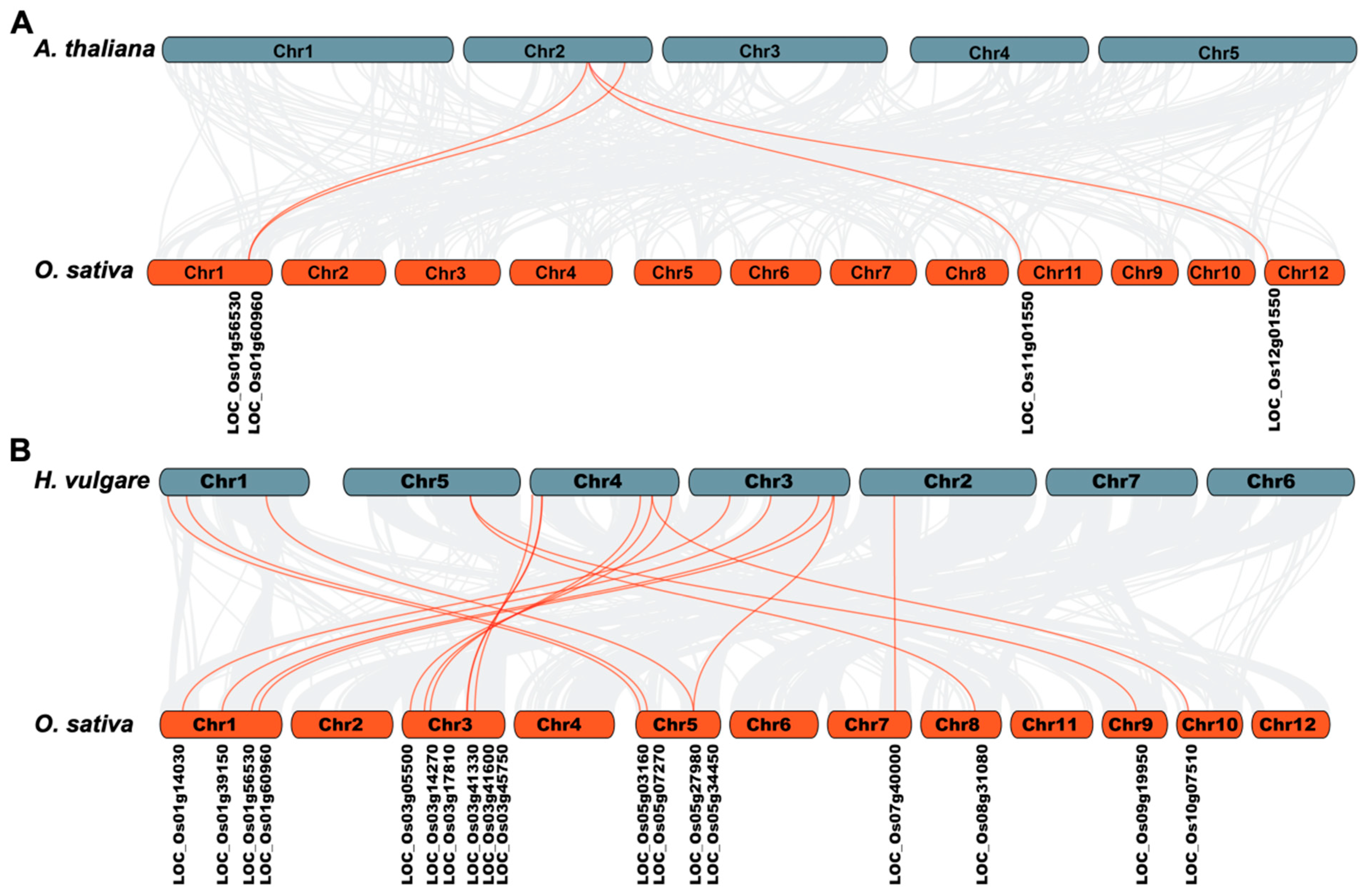
2.8. Collinearity and Evolution Analysis of OsLBD Genes
2.9. Expression Pattern of OsLBD Genes Throughout the Rice Life Cycle
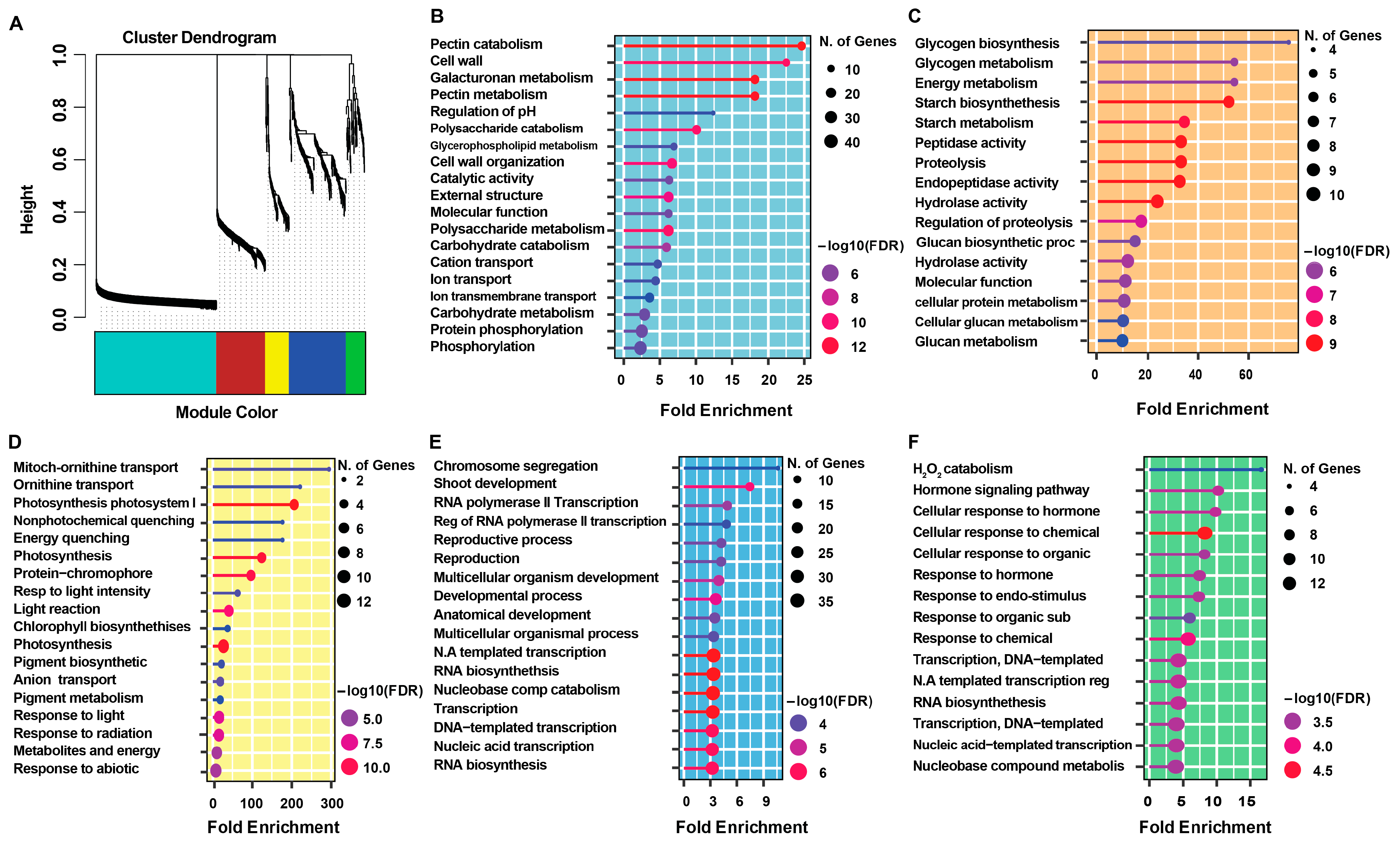
2.10. Identification of Co-Expressed Genes and Gene Enrichment Analysis
3. Materials and Methods
3.1. Data Acquisition and Sequence Analysis
3.2. Multiple Sequence Alignment and Phylogenetic Tree Construction
3.3. Cis-Regulatory Elements, Gene Structure, Conserved Motif, and Domain Analysis of OsLBD Genes
3.4. Chromosomal Distribution, Gene Duplication and Dual Synteny Analysis of OsLBD Genes
3.5. Expression Profiling and Identification of Co-Expressed of OsLBD Genes During Whole Life Cycle of Rice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhattacharya, K.R.; Ali, S.Z. On rice and the region of rice civilisation. Int. J. Sociol. Anthropol. 2016, 8, 65–75. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought resistance in rice from conventional to molecular breeding: A review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef] [PubMed]
- Sahebi, M.; Hanafi, M.M.; Rafii, M.Y.; Mahmud, T.M.M.; Azizi, P.; Osman, M.; Abiri, R.; Taheri, S.; Kalhori, N.; Shabanimofrad, M. Improvement of drought tolerance in rice (Oryza sativa L.): Genetics, genomic tools, and the WRKY gene family. BioMed Res. Int. 2018, 2018, 3158474. [Google Scholar] [CrossRef] [PubMed]
- Coudert, Y.; Dievart, A.; Droc, G.; Gantet, P. ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol. Biol. Evol. 2013, 30, 569–572. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.-R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Li, A.; Zhang, Y.; Wu, X.; Tang, W.; Wu, R.; Dai, Z.; Liu, G.; Zhang, H.; Wu, C.; Chen, G. DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol. Biol. 2008, 66, 491–502. [Google Scholar] [CrossRef]
- Liu, H.; Wang, S.; Yu, X.; Yu, J.; He, X.; Zhang, S.; Shou, H.; Wu, P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Evans, M.M. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009, 58, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-M.; Zhang, S.-Z.; Zheng, C.-C. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet. 2014, 93, 79–91. [Google Scholar] [CrossRef]
- Yu, J.; Xie, Q.; Li, C.; Dong, Y.; Zhu, S.; Chen, J. Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 2020, 251, 23. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, R.; Cheng, Y.; Lei, P.; Song, W.; Zheng, W.; Nie, X. Genome-wide identification, evolution, and expression analysis of LBD transcription factor family in bread wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 721253. [Google Scholar] [CrossRef]
- Yu, Q.; Hu, S.; Du, J.; Yang, Y.; Sun, X. Genome-wide identification and characterization of the lateral organ boundaries domain gene family in Brassica rapa var. rapa. Plant Divers. 2020, 42, 52–60. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, M.-J.; Park, M.Y.; Han, K.-H.; Kim, J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol. Plant 2013, 6, 1722–1725. [Google Scholar] [CrossRef]
- Chanderbali, A.S.; He, F.; Soltis, P.S.; Soltis, D.E. Out of the water: Origin and diversification of the LBD gene family. Mol. Biol. Evol. 2015, 32, 1996–2000. [Google Scholar] [CrossRef]
- Machida, Y.; Suzuki, T.; Sasabe, M.; Iwakawa, H.; Kojima, S.; Machida, C. Arabidopsis ASYMMETRIC LEAVES2 (AS2): Roles in plant morphogenesis, cell division, and pathogenesis. J. Plant Res. 2021, 134, 889–906. [Google Scholar] [CrossRef]
- Mangeon, A.; Bell, E.M.; Lin, W.; Jablonska, B.; Springer, P.S. Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J. Exp. Bot. 2011, 62, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, F.; Guo, J.; Zhang, X.S. Rice OsAS2 gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J. Plant Biol. 2009, 52, 374–381. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, D.; Xie, L.; Zhou, T.; Zhao, J.; Zhang, Q.; Yang, M.; Wu, W.; Lian, X. Rice transcription factors OsLBD37/38/39 regulate nitrate uptake by repressing OsNRT2.1/2.2/2.3 under high-nitrogen conditions. Crop J. 2022, 10, 1505–1515. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, P.; Chen, Z.; Zhang, L.; Wang, Y.; Xu, L. Genome-wide analysis of the LBD family in rice: Gene functions, structure and evolution. Comput. Biol. Med. 2023, 153, 106452. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Li, J.; Zhou, D.; Qiu, W.; Shi, Y.; Yang, J.-J.; Chen, S.; Wang, Q.; Pan, H. Application of weighted gene co-expression network analysis for data from paired design. Sci. Rep. 2018, 8, 622. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, L.; Xie, W.; Chen, Y.; Tang, W.; Yang, J.; Ye, R.; Liu, L.; Lin, Y.; Xu, C.; Xiao, J.; et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010, 61, 752–766. [Google Scholar] [CrossRef]
- Ouyang, Y.; Huang, X.; Lu, Z.; Yao, J. Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genom. 2012, 13, 100. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Han, B. Twenty years of rice genomics research: From sequencing and functional genomics to quantitative genomics. Mol. Plant 2022, 15, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, J.; Chen, L.; Huang, X.; Cheng, Z.; Han, B.; Zhang, Q.; Wu, C. Rice functional genomics research: Past decade and future. Mol. Plant 2018, 11, 359–380. [Google Scholar] [CrossRef]
- Bahar, N.H.A.; Lo, M.; Sanjaya, M.; Van Vianen, J.; Alexander, P.; Ickowitz, A.; Sunderland, T. Meeting the food security challenge for nine billion people in 2050: What impact on forests? Glob. Environ. Change 2020, 62, 102056. [Google Scholar] [CrossRef]
- Abbas, W.; Shalmani, A.; Zhang, J.; Sun, Q.; Zhang, C.; Li, W.; Cui, Y.; Xiong, M.; Li, Y. The GW5-WRKY53-SGW5 module regulates grain size variation in rice. New Phytol. 2024, 242, 2011–2025. [Google Scholar] [CrossRef]
- Abbas, W.; Sun, Q.; Cui, Y.; Shalmani, A.; Xu, P.; Fan, Y.; Zhang, D.; Wu, M.; Li, X.; Li, Y. The quantitative trait locus GWY10 controls rice grain width and yield. Plant Physiol. 2024, 196, 2286–2290. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenetics Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef]
- Guo, B.; Wang, J.; Lin, S.; Tian, Z.; Zhou, K.; Luan, H.; Lyu, C.; Zhang, X.; Xu, R. A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang Univ.-Sci. B 2016, 17, 763–774. [Google Scholar] [CrossRef]
- Sun, S.; Yi, J.; Gu, P.; Huang, Y.; Huang, X.; Li, H.; Fan, T.; Zhao, J.; Wang, R.; Gaballah, M.M.; et al. Comprehensive characterization and functional analysis of the lateral organ boundaries domain gene family in rice: Evolution, expression, and stress response. Int. J. Mol. Sci. 2025, 26, 3948. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Stark, T.L.; Liberles, D.A.; Holland, B.R.; O’Reilly, M.M. Analysis of a mechanistic Markov model for gene duplicates evolving under subfunctionalization. BMC Evol. Biol. 2017, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Crombez, E.; Van de Peer, Y.; Li, Z. The subordinate role of pseudogenization over recombinative deletion following polyploidization in angiosperms. Nat. Commun. 2025, 16, 6335. [Google Scholar] [CrossRef] [PubMed]
- Mangeon, A.; Lin, W.; Springer, P.S. Functional divergence in the Arabidopsis LOB-domain gene family. Plant Signal. Behav. 2012, 7, 1544–1547. [Google Scholar] [CrossRef]
- Copley, S.D. Evolution of new enzymes by gene duplication and divergence. FEBS J. 2020, 287, 1262–1283. [Google Scholar] [CrossRef]
- Vardhanabhuti, S.; Wang, J.; Hannenhalli, S. Position and distance specificity are important determinants of cis-regulatory motifs in addition to evolutionary conservation. Nucleic Acids Res. 2007, 35, 3203–3213. [Google Scholar] [CrossRef]
- Mitsis, T.; Efthimiadou, A.; Bacopoulou, F.; Vlachakis, D.; Chrousos, G.P.; Eliopoulos, E. Transcription factors and evolution: An integral part of gene expression (Review). World Acad. Sci. J. 2020, 2, 3–8. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, G.; Teixeira da Silva, J.A.; Zhao, C.; Duan, J. The methyl jasmonate-responsive transcription factor DobHLH4 promotes DoTPS10, which is involved in linalool biosynthesis in Dendrobium officinale during floral development. Plant Sci. 2021, 309, 110952. [Google Scholar] [CrossRef]
- Hobo, T.; Asada, M.; Kowyama, Y.; Hattori, T. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 1999, 19, 679–689. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Marand, A.P.; Eveland, A.L.; Kaufmann, K.; Springer, N.M. Cis-regulatory elements in plant development, adaptation, and evolution. Annu. Rev. Plant Biol. 2023, 74, 111–137. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, F.; Duan, D. Function analysis and stress-mediated cis-element identification in the promoter region of VqMYB15. Plant Signal. Behav. 2020, 15, 1773664. [Google Scholar] [CrossRef] [PubMed]
- Sosnowski, J.; Truba, M.; Vasileva, V. The impact of auxin and cytokinin on the growth and development of selected crops. Agriculture 2023, 13, 724. [Google Scholar] [CrossRef]
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Yang, H.; You, C.; Yang, S.; Zhang, Y.; Yang, F.; Li, X.; Chen, N.; Luo, Y.; Hu, X. The role of calcium/calcium-dependent protein kinases signal pathway in pollen tube growth. Front. Plant Sci. 2021, 12, 633293. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Emes, M.J. Starch biosynthesis in the developing endosperms of grasses and cereals. Agronomy 2017, 7, 81. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Cun, Z.; Chen, J.-W. Photosynthetic performance and photosynthesis-related gene expression coordinated in a shade-tolerant species Panax notoginseng under nitrogen regimes. BMC Plant Biol. 2020, 20, 273. [Google Scholar] [CrossRef]
- Simonini, S. Regulation of cell cycle in plant gametes: When is the right time to divide? Development 2025, 152, dev204217. [Google Scholar] [CrossRef]
- Su, J.; Liu, Y.; Han, F.; Gao, F.; Gan, F.; Huang, K.; Li, Z. ROS, an important plant growth regulator in root growth and development: Functional genes and mechanism. Biology 2024, 13, 1033. [Google Scholar] [CrossRef]
- Karasov, T.L.; Chae, E.; Herman, J.J.; Bergelson, J. Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 2017, 29, 666–680. [Google Scholar] [CrossRef]



| Gene IDs | AA Length | MW (Da) | pI | Negative Charges | Positive Charges | Instability Index | Stability | Aliphatic Index | GRAVY | Cellular Location |
|---|---|---|---|---|---|---|---|---|---|---|
| LOC_Os01g03890 | 307 | 32,379.55 | 6.89 | 30 | 29 | 52.62 | Unstable | 76.42 | −0.297 | Nucleus |
| LOC_Os01g07480 | 251 | 25,728.12 | 7.61 | 18 | 19 | 51.93 | Unstable | 78.65 | 0.003 | Nucleus |
| LOC_Os01g14030 | 249 | 26,091.37 | 9.72 | 18 | 31 | 45.7 | Unstable | 67.27 | −0.321 | Nucleus |
| LOC_Os01g32770 | 330 | 35,019.79 | 5.36 | 48 | 32 | 54.31 | Unstable | 77.27 | −0.399 | Nucleus |
| LOC_Os01g39040 | 356 | 38,510.31 | 4.9 | 53 | 38 | 77.89 | Unstable | 77.89 | −0.396 | Nucleus |
| LOC_Os01g39070 | 125 | 13,306.08 | 6.26 | 13 | 12 | 68.08 | Unstable | 68.16 | −0.425 | Nucleus |
| LOC_Os01g39150 | 251 | 26,857.04 | 5.69 | 30 | 23 | 75.7 | Unstable | 62.71 | −0.649 | Nucleus |
| LOC_Os01g39160 | 171 | 17,865.16 | 5.25 | 22 | 17 | 46.31 | Unstable | 72.75 | −0.127 | Nucleus |
| LOC_Os01g39180 | 321 | 35,174.2 | 9.45 | 29 | 39 | 64.75 | Unstable | 77.32 | −0.328 | Nucleus |
| LOC_Os01g39220 | 139 | 14,864.02 | 6.03 | 13 | 11 | 69.05 | Unstable | 77.41 | −0.192 | Nucleus |
| LOC_Os01g56530 | 245 | 25,730.04 | 7.72 | 22 | 23 | 50.79 | Unstable | 82.12 | −0.147 | Nucleus |
| LOC_Os01g60960 | 251 | 26,493.01 | 6.45 | 26 | 24 | 54.8 | Unstable | 76.29 | −0.173 | Nucleus |
| LOC_Os01g66590 | 269 | 27,621.93 | 8.17 | 17 | 19 | 39.38 | Stable | 69.89 | −0.08 | Nucleus |
| LOC_Os02g57490 | 216 | 22,457.64 | 8.09 | 15 | 17 | 53.54 | Unstable | 78.43 | 0.103 | Nucleus |
| LOC_Os03g05500 | 185 | 19,631.01 | 6.05 | 14 | 12 | 60.74 | Unstable | 66.65 | −0.186 | Nucleus |
| LOC_Os03g05510 | 259 | 26,736.57 | 5.96 | 21 | 14 | 43.6 | Unstable | 65.52 | −0.085 | Nucleus |
| LOC_Os03g14270 | 275 | 28,252.75 | 8.27 | 15 | 17 | 83.07 | Unstable | 67.53 | −0.321 | Nucleus |
| LOC_Os03g17810 | 239 | 25,556.79 | 6.06 | 23 | 20 | 62.94 | Unstable | 76.4 | −0.241 | Nucleus |
| LOC_Os03g33090 | 204 | 21,037.1 | 8.41 | 16 | 19 | 50.07 | Unstable | 81.37 | 0.042 | Nucleus |
| LOC_Os03g41330 | 232 | 23618.55 | 6.05 | 19 | 17 | 49.15 | Unstable | 81.34 | 0.023 | Nucleus |
| LOC_Os03g41600 | 307 | 33,077.8 | 6.65 | 25 | 20 | 58.81 | Unstable | 64.01 | −0.577 | Nucleus |
| LOC_Os03g45750 | 214 | 22,012.96 | 9.06 | 15 | 21 | 56.78 | Unstable | 70.75 | −0.131 | Nucleus |
| LOC_Os03g57670 | 171 | 18,322.4 | 7.63 | 17 | 18 | 65.46 | Unstable | 56.2 | −0.458 | Nucleus |
| LOC_Os05g03160 | 145 | 14,935.02 | 4.96 | 13 | 7 | 46.9 | Unstable | 85.86 | 0.166 | Nucleus |
| LOC_Os05g07270 | 207 | 21,567.58 | 8.98 | 12 | 17 | 63.58 | Unstable | 83.19 | 0.11 | Nucleus |
| LOC_Os05g27980 | 200 | 21,105.84 | 6.28 | 17 | 14 | 53.5 | Unstable | 78.2 | −0.145 | Nucleus |
| LOC_Os05g34450 | 279 | 28,044.35 | 8.23 | 16 | 18 | 41.59 | Unstable | 71.08 | −0.004 | Nucleus |
| LOC_Os07g40000 | 226 | 23,216.21 | 7.54 | 19 | 19 | 56.33 | Unstable | 69.16 | −0.115 | Nucleus |
| LOC_Os08g06659 | 196 | 21,283.44 | 4.62 | 36 | 18 | 81.05 | Unstable | 54.49 | −0.88 | Nucleus |
| LOC_Os08g31080 | 456 | 48,653.41 | 4.69 | 68 | 31 | 38.04 | Stable | 72.08 | −0.474 | Nucleus |
| LOC_Os08g44940 | 215 | 23,239.17 | 5.85 | 17 | 12 | 48.33 | Unstable | 67.26 | −0.16 | Nucleus |
| LOC_Os09g19950 | 362 | 37,637.55 | 5.27 | 37 | 22 | 46.48 | Unstable | 65.19 | −0.397 | Nucleus |
| LOC_Os10g07510 | 269 | 27,974.12 | 6.41 | 20 | 16 | 66.24 | Unstable | 67.55 | −0.336 | Nucleus |
| LOC_Os11g01550 | 156 | 17,404.87 | 6.42 | 14 | 12 | 57.14 | Unstable | 77.05 | −0.254 | Nucleus |
| LOC_Os12g01550 | 156 | 17,404.87 | 6.42 | 14 | 12 | 57.14 | Unstable | 77.05 | −0.254 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, W.; Shad, M.A.; Li, W.; Shalmani, A.; Zhang, J.; Iqbal, A.; Liu, L. A Comprehensive Profiling of the Rice LATERAL ORGAN BOUNDARIES DOMAIN (LBD) Gene Family: Structure, Evolution, and Expressional Dynamics. Plants 2025, 14, 3596. https://doi.org/10.3390/plants14233596
Abbas W, Shad MA, Li W, Shalmani A, Zhang J, Iqbal A, Liu L. A Comprehensive Profiling of the Rice LATERAL ORGAN BOUNDARIES DOMAIN (LBD) Gene Family: Structure, Evolution, and Expressional Dynamics. Plants. 2025; 14(23):3596. https://doi.org/10.3390/plants14233596
Chicago/Turabian StyleAbbas, Waseem, Munsif Ali Shad, Wei Li, Abdullah Shalmani, Jian Zhang, Adnan Iqbal, and Lin Liu. 2025. "A Comprehensive Profiling of the Rice LATERAL ORGAN BOUNDARIES DOMAIN (LBD) Gene Family: Structure, Evolution, and Expressional Dynamics" Plants 14, no. 23: 3596. https://doi.org/10.3390/plants14233596
APA StyleAbbas, W., Shad, M. A., Li, W., Shalmani, A., Zhang, J., Iqbal, A., & Liu, L. (2025). A Comprehensive Profiling of the Rice LATERAL ORGAN BOUNDARIES DOMAIN (LBD) Gene Family: Structure, Evolution, and Expressional Dynamics. Plants, 14(23), 3596. https://doi.org/10.3390/plants14233596








