The Central-Periphery Hypothesis Revisited: Implications for Long-Term Genetic Conservation
Abstract
1. Introduction
2. Results
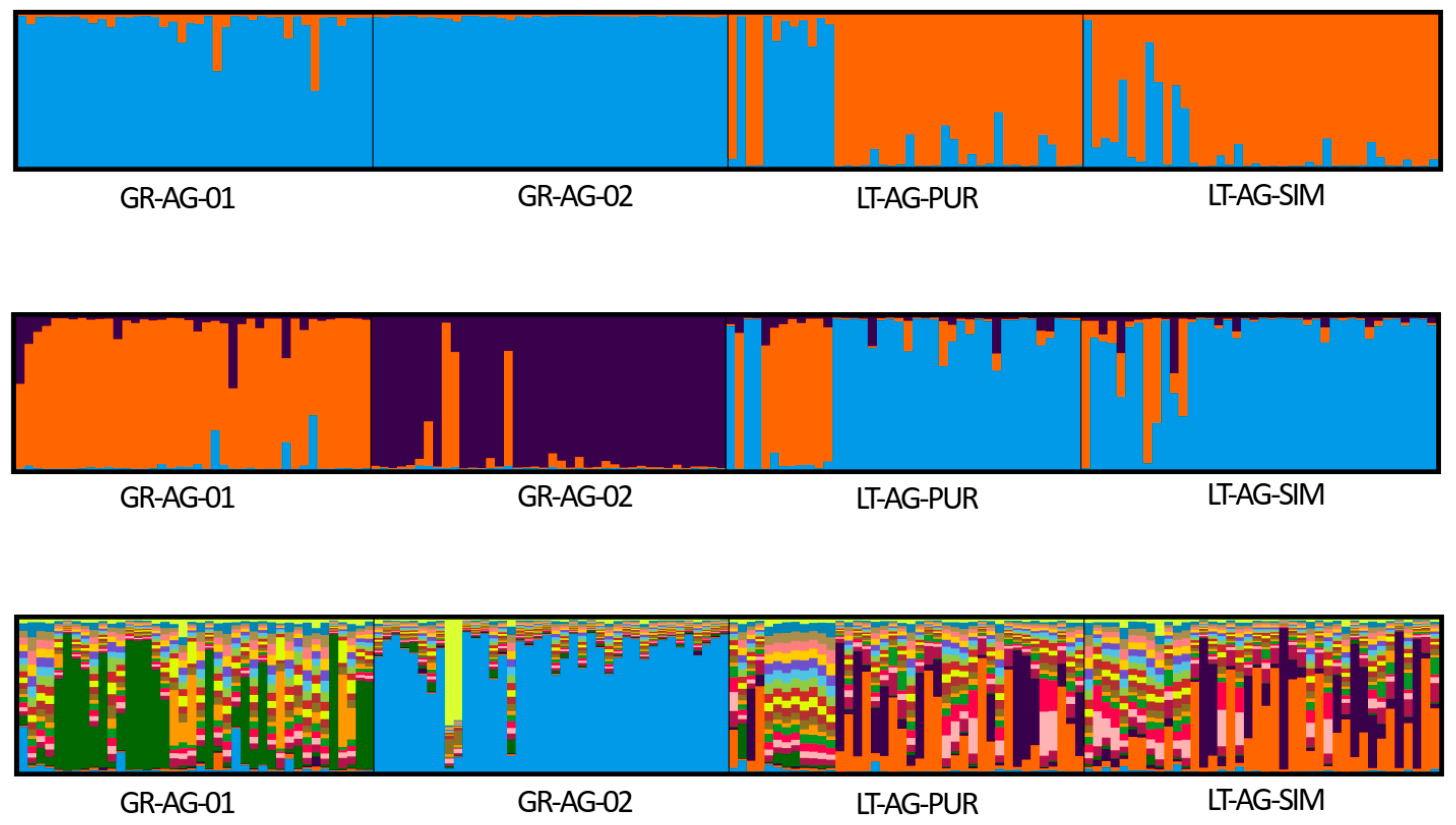
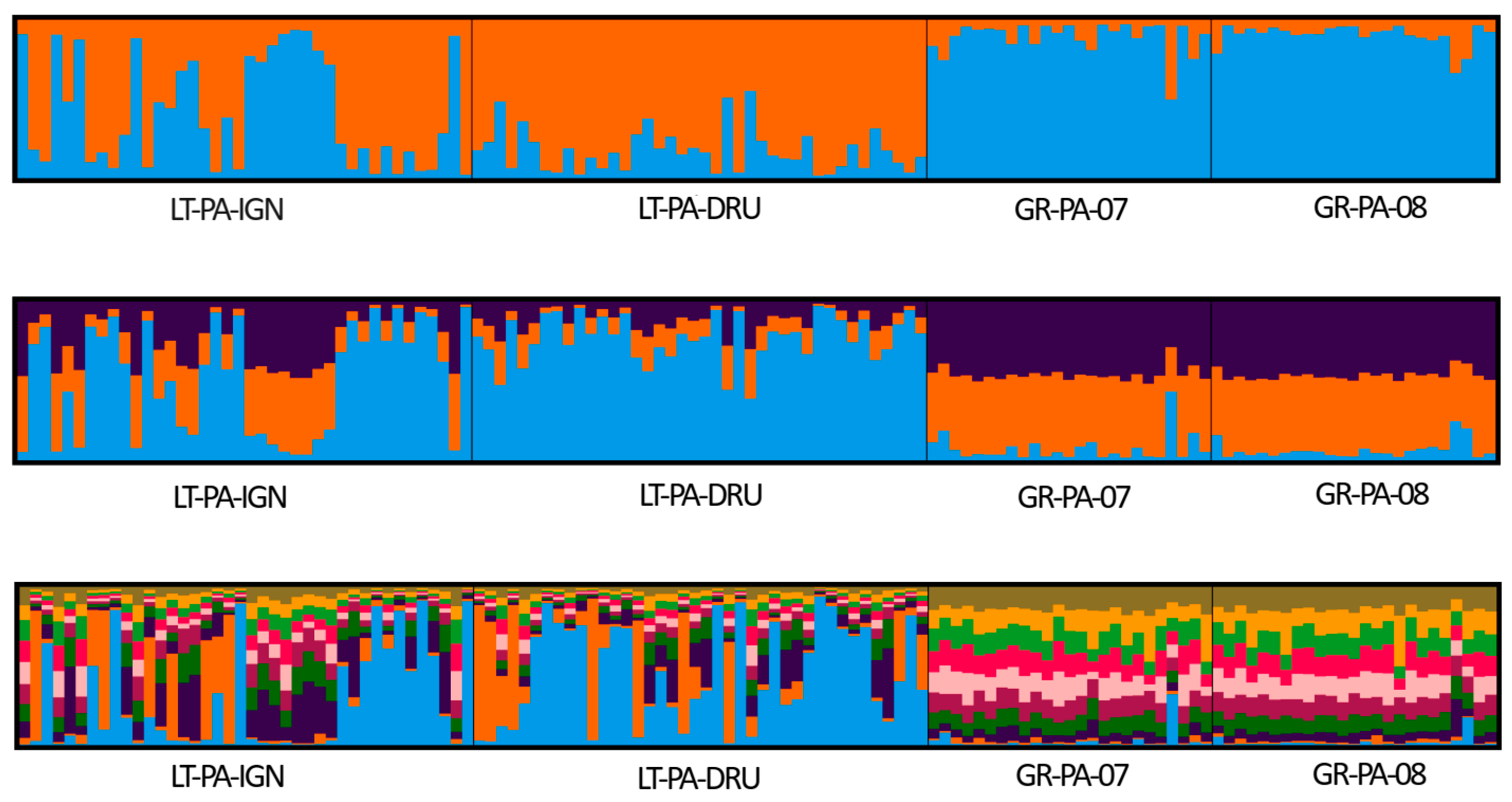
3. Discussion
4. Materials and Methods
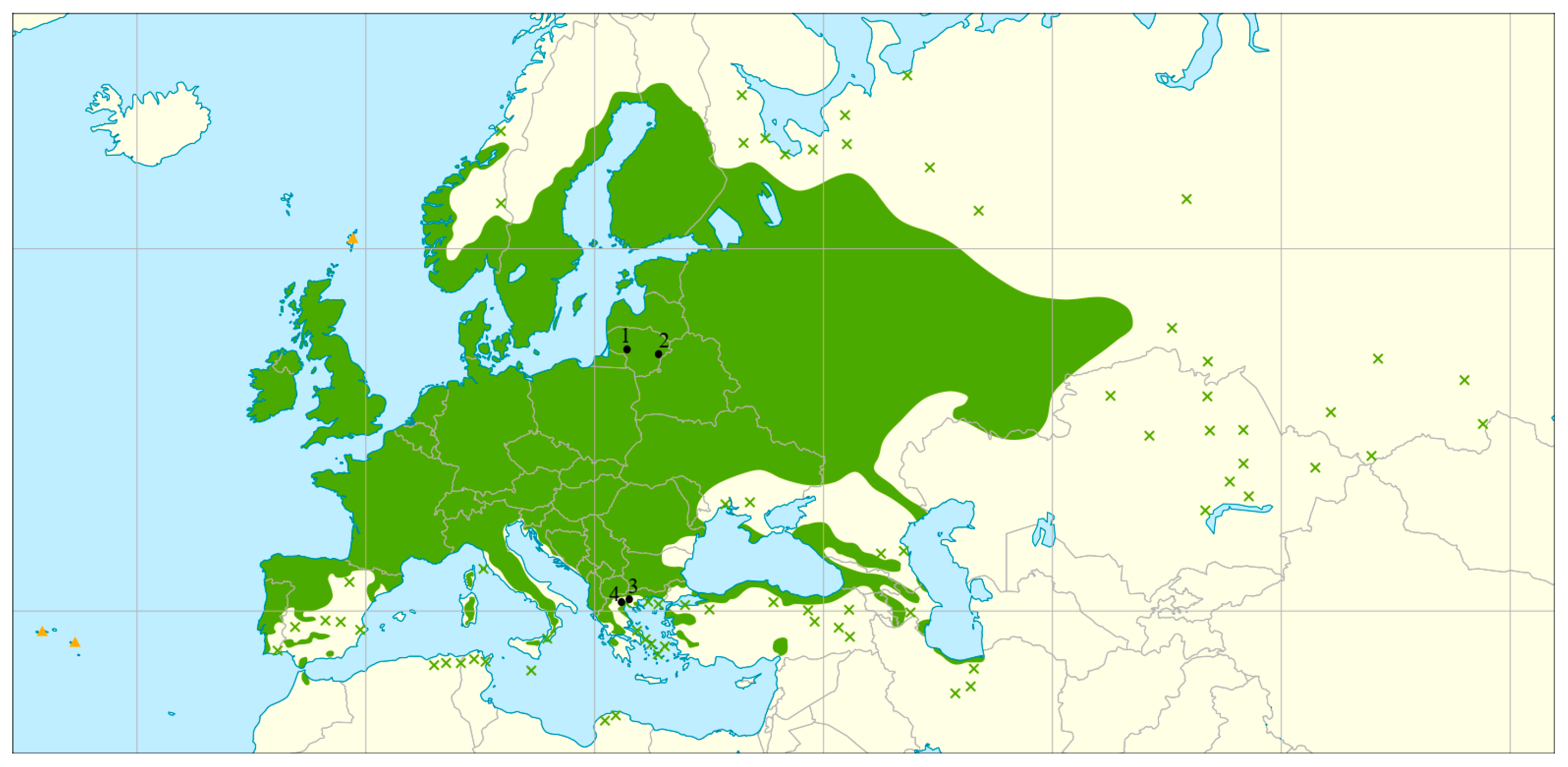
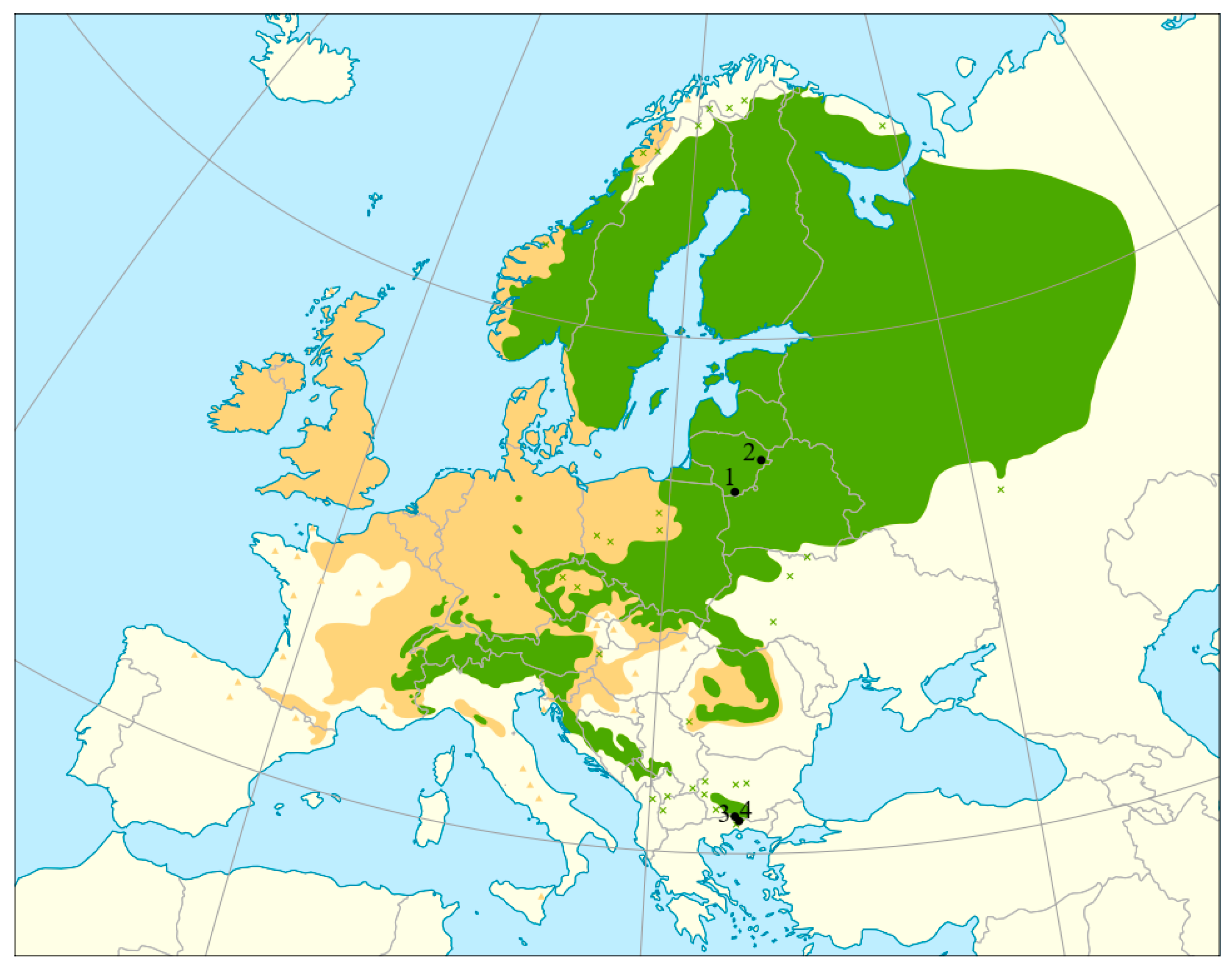
4.1. Study Sites and Sampling
4.2. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CPH | Central-periphery hypothesis |
| EST | Expressed sequence tag |
| GCU | Genetic conservation unit |
| PCoA | Principal coordinate analysis |
| PCR | Polymerase chain reaction |
| SSR | Simple sequence repeats |
References
- Kelleher, C.T.; De Vries, S.M.; Baliuckas, V.; Bozzano, M.; Frýdl, J.; Gonzalez Goicoechea, P.; Ivankovic, M.; Kandemir, G.; Koskela, J.; Kozioł, C.; et al. Approaches to the Conservation of Forest Genetic Resources in Europe in the Context of Climate Change; European Forest Genetic Resources Programme (EUFORGEN); Bioversity International: Rome, Italy, 2015; pp. xiv+46. [Google Scholar]
- Fady, B.; Aravanopoulos, F.A.; Alizoti, P.; Mátyás, C.; von Wühlisch, G.; Westergren, M.; Belletti, P.; Cvjetkovic, B.; Ducci, F.; Huber, G.; et al. Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. For. Ecol. Manag. 2016, 375, 66–75. [Google Scholar] [CrossRef]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Mol. Ecol. 2005, 8, 461–467. [Google Scholar] [CrossRef]
- Pörtner, H.O.; Roberts, D.C.; Tignor, M.; Poloczanska, E.S.; Mintenbeck, K.; Alegría, A.; Craig, M.; Langsdorf, S.; Löschke, S.; Möller, V.; et al. (Eds.) Climate Change 2022: Impacts, Adaptation, and Vulnerability. In Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2022; pp. 2233–2272. [Google Scholar] [CrossRef]
- Mayr, E. Populations, Species, and Evolution. An Abridgment of Animal Species and Evolution; Belknap Press: Cambridge, MA, USA, 1970. [Google Scholar]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The central-marginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef]
- Rajora, O.P.; DeVerno, L.; Mosseler, A.; Innes, D.J. Genetic diversity and population structure of disjunct Newfoundland and central Ontario populations of eastern white pine (Pinus strobus). Can. J. Bot. 1998, 76, 500–508. [Google Scholar] [CrossRef]
- Yang, A.; Dick, C.W.; Yao, X.; Huang, H. Impacts of biogeographic history and marginal population genetics on species range limits: A case study of Liriodendron chinense. Sci. Rep. 2016, 6, 25632. [Google Scholar] [CrossRef]
- Jaramillo-Correa, J.P.; Bagnoli, F.; Grivet, D.; Fady, B.; Aravanopoulos, F.A.; Vendramin, G.G.; González-Martínez, S.C. Evolutionary rate and genetic load in an emblematic Mediterranean tree following an ancient and prolonged population collapse. Mol. Ecol. 2020, 29, 4797–4811. [Google Scholar] [CrossRef]
- Vucetich, J.A.; Waite, T.A. Spatial patterns of demography and genetic processes across the species’ range: Null hypotheses forlandscape conservation genetics. Conserv. Genet. 2003, 4, 639–645. [Google Scholar] [CrossRef]
- Milesi, P.; Kastally, C.; Dauphin, B.; Cervantes, S.; Bagnoli, F.; Budde, K.B.; Cavers, S.; Fady, B.; Faivre-Rampant, P.; González-Martínez, S.C.; et al. Resilience of genetic diversity in forest trees over the Quaternary. Nat. Commun. 2024, 15, 8538. [Google Scholar] [CrossRef]
- Ha, Y.H.; An, J.B.; Chung, J.; Yoon, J.W.; Gil, H.Y. Genotyping-by-sequencing reveals low genetic diversity and peripheral isolation in Southern populations of Sophora koreensis, a Korean endemic shrub. Sci. Rep. 2025, 15, 17996. [Google Scholar] [CrossRef] [PubMed]
- Svenning, J.C.; Skov, F. Could the tree diversity pattern in Europe be generated by postglacial dispersal limitation? Ecol. Lett. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Feurdean, A.; Bhagwat, S.A.; Willis, K.J.; Birks, H.J.B.; Lischke, H.; Hickler, T. Tree migration-rates: Narrowing the gap between inferred post-glacial rates and projected rates. PLoS ONE 2013, 8, 71797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Chen, Q.; Wang, Y.; Wang, S.; Wang, G.; Li, P.; Liu, H.; Li, P.; Xu, C.; et al. Phylogeography and Population Demography of Parrotia subaequalis, a Hamamelidaceous Tertiary Relict ‘Living Fossil’Tree Endemic to East Asia Refugia: Implications from Molecular Data and Ecological Niche Modeling. Plants 2025, 14, 1754. [Google Scholar] [CrossRef]
- Stamford, M.D.; Taylor, E.B. Phylogeographic lineages of Arctic grayling (Thymallus arcticus) in North America, divergence, origins and affinities with Eurasian Thymallus. Mol. Ecol. 2004, 13, 1533–1549. [Google Scholar] [CrossRef]
- Muller, M.H.; Leppala, J.; Savolainen, O. Genome-wide effects of postglacial colonization in Arabidopsis lyrata. Heredity 2008, 100, 47–58. [Google Scholar] [CrossRef]
- Havrdová, A.; Douda, J.; Krak, K.; Vít, P.; Hadincová, V.; Zákravský, P.; Mandák, B. Higher genetic diversity in recolonized areas than in refugia of Alnus glutinosa triggered by continent—Wide lineage admixture. Mol. Ecol. 2015, 24, 4759–4777. [Google Scholar] [CrossRef]
- McVean, D.N. Biological flora of the British Isles: Alnus Mill. Alnus glutinosa (L.) Gaertn. (A. rotundifolia Stokes). J. Ecol. 1953, 41, 447–466. [Google Scholar]
- Douda, J.; Čejková, A.; Douda, K.; Kochánková, J. Development of alder carr after the abandonment of wet grasslands during the last 70 years. Ann. For. Sci. 2009, 66, 712. [Google Scholar] [CrossRef]
- Douda, J. The role of landscape configuration in plant composition of floodplain forests across different physiographic areas. J. Veg. Sci. 2010, 21, 1110–1124. [Google Scholar] [CrossRef]
- Cox, K.; Vanden Broeck, A.; Van Calster, H.; Mergeay, J. Temperature—Related natural selection in a wind—Pollinated tree across regional and continental scales. Mol. Ecol. 2011, 20, 2724–2738. [Google Scholar] [CrossRef]
- Lepais, O.; Muller, S.D.; Ben Saad-Limam, S.; Benslama, M.; Rhazi, L.; Belouahem-Abed, D.; Daoud-Bouattour, A.; Gammar, A.M.; Ghrabi-Gammar, Z.; Bacles, C.F.E. High genetic diversity and distinctiveness of rear-edge climate relicts maintained by ancient tetraploidisation for Alnus glutinosa. PLoS ONE 2013, 8, 75029. [Google Scholar] [CrossRef]
- Beatty, G.E.; Montgomery, W.I.; Tosh, D.G.; Provan, J. Genetic provenance and best practice woodland management: A case study in native alder (Alnus glutinosa). Tree Genet. Genomes 2015, 11, 92. [Google Scholar] [CrossRef]
- Mandák, B.; Vít, P.; Krak, K.; Trávníček, P.; Havrdová, A.; Hadincová, V.; Zákravský, P.; Jarolímová, V.; Bacles, C.F.E.; Douda, J. Flow cytometry, microsatellites and niche models reveal the origins and geographical structure of Alnus glutinosa populations in Europe. Ann. Bot. 2016, 117, 107–120. [Google Scholar] [CrossRef]
- Mingeot, D.; Husson, C.; Mertens, P.; Watillon, B.; Bertin, P.; Druart, P. Genetic diversity and genetic structure of black alder (Alnus glutinosa [L.] Gaertn) in the Belgium-Luxembourg-France cross-border area. Tree Genet. Genomes 2016, 12, 24. [Google Scholar] [CrossRef]
- Verbylaitė, R.; Aravanopoulos, F.A.; Baliuckas, V.; Juškauskaitė, A. Genetic monitoring of Alnus glutinosa natural populations using two generation cohorts. Forests 2023, 14, 330. [Google Scholar] [CrossRef]
- Caudullo, G.; Tinner, W.; De Rigo, D. Picea abies in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Publ. Off. EU: Reims, Luxembourg, 2016; p. e012300+. Available online: https://ies-ows.jrc.ec.europa.eu/efdac/download/Atlas/pdf/Picea_abies.pdf (accessed on 20 May 2025).
- Jalas, J.; Suominen, J. (Eds.) Atlas Florae Europaeae: Distribution of Vascular Plants in Europe—Vol. 2: Gymnospermae (Pinaceae To Ephedraceae); Committee for Mapping the Flora of Europe/Societas Biologica Fennica Vanamo: Helsinki, Finland, 1973; p. 40. [Google Scholar]
- Farjon, A.; Filer, D. An Atlas of the World’s Conifers: An Analysis of Their Distribution, Biogeography, Diversity and Conservation Status; Brill: Leiden, The Netherlands, 2013; pp. 139–170. [Google Scholar] [CrossRef]
- Horgan, T.; Keane, M.; McCarthy, R.; Lally, M.; Thompson, D.; O’Carroll, J. (Eds.) A Guide to Forest Tree Species Selection and Silviculture in IRELAND; COFORD: Dublin, Ireland, 2003; pp. x+255. Available online: https://www.coford.ie/media/coford/content/publications/projectreports/speciesmanual.pdf (accessed on 3 May 2025).
- Tjoelker, M.G.; Boratynski, A.; Bugala, W. (Eds.) Biology and Ecology of Norway Spruce, 1st ed.; Springer Science & Business Media: Dordrecht, The Netherlands, 2007; Volume 78, pp. 195–220. [Google Scholar] [CrossRef]
- Caré, O.; Müller, M.; Vornam, B.; Höltken, A.M.; Kahlert, K.; Krutovsky, K.V.; Gailing, O.; Leinemann, L. High morphological differentiation in crown architecture contrasts with low population genetic structure of German Norway spruce stands. Forests 2018, 9, 752. [Google Scholar] [CrossRef]
- Westergren, M.; Bozic, G.; Kraigher, H. Genetic diversity of core vs. peripheral Norway spruce native populations at a local scale in Slovenia. IForest 2018, 11, 104. [Google Scholar] [CrossRef]
- Stojnić, S.; Avramidou, E.V.; Fussi, B.; Westergren, M.; Orlović, S.; Matović, B.; Trudić, B.; Kraigher, H.; Aravanopoulos, F.A.; Konnert, M. Assessment of genetic diversity and population genetic structure of Norway spruce (Picea abies (L.) Karsten) at its southern lineage in Europe. Implications for conservation of forest genetic resources. Forests 2019, 10, 258. [Google Scholar] [CrossRef]
- Wang, X.; Bernhardsson, C.; Ingvarsson, P.K. Demography and natural selection have shaped genetic variation in the widely distributed conifer Norway spruce (Picea abies). Genome Biol. Evol. 2020, 12, 3803–3817. [Google Scholar] [CrossRef]
- Unger, G.M.; Konrad, H.; Geburek, T. Does spatial genetic structure increase with altitude? An answer from Picea abies in Tyrol, Austria. Plant Syst. Evol. 2011, 292, 133–141. [Google Scholar] [CrossRef]
- Verbylaitė, R.; Pliūra, A.; Lygis, V.; Suchockas, V.; Jankauskienė, J.; Labokas, J. Genetic diversity and its spatial distribution in self-regenerating Norway spruce and Scots pine stands. Forests 2017, 8, 470. [Google Scholar] [CrossRef]
- Máchová, P.; Trčková, O.; Cvrčková, H. Use of nuclear microsatellite loci for evaluating genetic diversity of selected populations of Picea abies (L.) Karsten in the Czech Republic. Forests 2018, 9, 92. [Google Scholar] [CrossRef]
- Bínová, Z.; Korecký, J.; Dvořák, J.; Bílý, J.; Zádrapová, D.; Jansa, V.; Lstibůrek, M. Genetic structure of Norway spruce ecotypes studied by SSR markers. Forests 2020, 11, 110. [Google Scholar] [CrossRef]
- Tollefsrud, M.M.; Kissling, R.O.Y.; Gugerli, F.; Johnsen, Ø.; Skrøppa, T.; Cheddadi, R.; VAN DER Knaap, W.O.; Latałowa, M.; Terhürne-Berson, R.; Litt, T.; et al. Genetic consequences of glacial survival and postglacial colonization in Norway spruce: Combined analysis of mitochondrial DNA and fossil pollen. Mol. Ecol. 2008, 17, 4134–4150. [Google Scholar] [CrossRef]
- Evanno, A.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Konnert, M.; Fady, B.; Gömöry, D.; A’Hara, S.; Wolter, F.; Ducci, F.; Koskela, J.; Bozzano, M.; Maaten, T.; Kowalczyk, J. European Forest Genetic Resources Programme (EUFORGEN): Use and Transfer of Forest Reproductive Material in Europe in the Context of Climate Change; European Forest Genetic Resources Programme (EUFORGEN); Bioversity International: Rome, Italy, 2015; pp. xvi+75. [Google Scholar]
- Cvjetkovic, B.; Konnert, M.; Fussi, B.; Mataruga, M.; Sijacic-Nikolic, M.; Danicic, V.; Lucic, A. Norway spruce (Picea abies Karst.) variability in progeny tests in Bosnia and Herzegovina. Genetika 2017, 49, 259–272. [Google Scholar] [CrossRef]
- Whitlock, M.C. G’ST and D do not replace FST. Mol. Ecol. 2011, 20, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J. Bottleneck: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Aravanopoulos, F.A.; Argiriou, A.; Kalivas, A.; Tsaftaris, A. Is the genetic diversity of small scattered forest tree populations at the southern limits of their range more prone to stochastic events? A wild cherry case study by microsatellite-based markers. Tree Genet. Genomes 2011, 7, 1299–1313. [Google Scholar] [CrossRef]
- Picard, N.; Marchi, M.; Serra-Varela, M.J.; Westergren, M.; Cavers, S.; Notivol, E.; Piotti, A.; Alizoti, P.; Bozzano, M.; González-Martínez, S.C.; et al. Marginality indices for biodiversity conservation in forest trees. Ecol. Indic. 2022, 143, 109367. [Google Scholar] [CrossRef]
- Avtzis, D.N.; Aravanopoulos, F.A. Host tree and insect genetic diversity on the borderline of natural distribution: A case study of Picea abies and Pityogenes chalcographus (Coleoptera, Scolytinae) in Greece. Silva Fenn. 2011, 45, 157–164. [Google Scholar] [CrossRef]
- Koskela, J.; Lefèvre, F.; Schueler, S.; Kraigher, H.; Olrik, D.C.; Hubert, J.; Longauer, R.; Bozzano, M.; Yrjänä, L.; Alizoti, P.; et al. Translating conservation genetics into management: Pan-European minimum requirements for dynamic conservation units of forest tree genetic diversity. Biol. Conserv. 2013, 157, 39–49. [Google Scholar] [CrossRef]
- De Vries, S.M.; Alan, M.; Bozzano, M.; Burianek, V.; Collin, E.; Cottrell, J.; Ivankovic, M.; Kelleher, C.T.; Koskela, J.; Rotach, P.; et al. Pan-European strategy for Genetic Conservation of Forest Trees and Establishment of a Core Network of Dynamic Conservation Units; European Forest Genetic Resources Programme (EUFORGEN); Bioversity International: Rome, Italy, 2015; pp. xii+40. [Google Scholar]
- Distribution Map of Black Alder (Alnus glutinosa) EUFORGEN. 2009. Available online: https://www.euforgen.org/fileadmin/templates/euforgen.org/upload/Documents/Maps/PDF/Alnus_glutinosa.pdf (accessed on 6 September 2023).
- Caudullo, G.; Welk, E.; San-Miguel-Ayanz, J. Distribution map of Norway spruce (Picea abies). Chorological maps for the main European woody species. Data Brief 2017, 12, 662–666. [Google Scholar] [CrossRef]
- Wu, B.; Lian, C.; Hogetsu, T. Development of microsatellite markers in white birch (Betula platyphylla var. japonica). Mol. Ecol. 2002, 3, 413–415. [Google Scholar] [CrossRef]
- Gürcan, K.; Mehlenbacher, S.A.; Botta, R.; Boccacci, P. Development, characterization, segregation, and mapping of microsatellite markers for European hazelnut (Corylus avellana L.) from enriched genomic libraries and usefulness in genetic diversity studies. Tree Genet. Genomes 2010, 6, 513–531. [Google Scholar] [CrossRef]
- Kulju, K.K.; Pekkinen, M.; Varvio, S. Twenty-three microsatellite primer pairs for Betula pendula (Betulaceae). Mol. Ecol. Notes 2004, 4, 471–473. [Google Scholar] [CrossRef]
- Tsuda, Y.; Ueno, S.; Ide, Y.; Tsumura, Y. Development of 14 EST-SSRs for Betula maximowicziana and their applicability to related species. Conserv. Genet. 2009, 10, 661–664. [Google Scholar] [CrossRef]
- Ogyu, K.; Tsuda, Y.; Sugaya, T.; Yoshimaru, H.; Ide, Y. Identification and characterization of microsatellite loci in Betula maximowicziana Regel. Mol. Ecol. Notes 2003, 3, 268–269. [Google Scholar] [CrossRef]
- Tsuda, Y.; Ueno, S.; Ranta, J.; Salminen, K.; Ide, Y.; Shinohara, K.; Tsumura, Y. Development of 11 EST-SSRs for Japanese white birch, Betula platyphylla var. japonica and their transferability to related species. Conserv. Genet. 2009, 10, 1385–1388. [Google Scholar] [CrossRef]
- Lepais, O.; Bacles, C.F. De novo discovery and multiplexed amplification of microsatellite markers for black alder (Alnus glutinosa) and related species using SSR-enriched shotgun pyrosequencing. J. Hered. 2011, 102, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Drašnarová, A.; Krak, K.; Vít, P.; Doudová, J.; Douda, J.; Hadincová, V.; Zákravský, P.; Mandák, B. Cross-amplification and multiplexing of SSR markers for Alnus glutinosa and A. incana. Tree Genet. Genomes 2014, 10, 865–873. [Google Scholar] [CrossRef]
- Besnard, G.; Acheré, V.; Faivre Rampant, P.; Favre, J.M.; Jeandroz, S. A set of cross—Species amplifying microsatellite markers developed from DNA sequence databanks in Picea (Pinaceae). Mol. Ecol. Notes 2003, 3, 380–383. [Google Scholar] [CrossRef]
- Schubert, R.; Mueller-Starck, G.; Riegel, R. Development of EST-PCR markers and monitoring their intrapopulational genetic variation in Picea abies (L.) Karst. Theor. Appl. Genet. 2001, 103, 1223–1231. [Google Scholar] [CrossRef]
- Rungis, D.; Bérubé, Y.; Zhang, J.; Ralph, S.; Ritland, C.E.; Ellis, B.E.; Douglas, C.; Ritland, K.; Bérubé, Y.; Bohlmann, J. Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theor. Appl. Genet. 2004, 109, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, A.; Olivieri, A.M.; Morgante, M. Identification and characterization of microsatellites in Norway spruce (Picea abies K.). Genome 1997, 40, 411–419. [Google Scholar] [CrossRef]
- Hodgetts, R.B.; Aleksiuk, M.A.; Brown, A.; Clarke, C.; Macdonald, E.; Nadeem, S.; Khasa, D. Development of microsatellite markers for white spruce (Picea glauca) and related species. Theor. Appl. Genet. 2001, 102, 1252–1258. [Google Scholar] [CrossRef]
- Scotti, I.; Paglia, G.; Magni, F.; Morgante, M. Efficient development of dinucleotide microsatellite markers in Norway spruce (Picea abies Karst.) through dot-blot selection. Theor. Appl. Genet. 2002, 104, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Scotti, I.; Magni, F.; Paglia, G.; Morgante, M. Trinucleotide microsatellites in Norway spruce (Picea abies): Their features and the development of molecular markers. Theor. Appl. Genet. 2002, 106, 40–50. [Google Scholar] [CrossRef]
- Scotti, I.; Magni, F.; Fink, R.; Powell, W.; Binelli, G.; Hedley, P.E. Microsatellite repeats are not randomly distributed within Norway spruce (Picea abies K.) expressed sequences. Genome 2000, 43, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Rajora, O.P.; Rahman, M.H.; Dayanandan, S.; Mosseler, A. Isolation, characterization, inheritance and linkage of microsatellite DNA markers in white spruce (Picea glauca) and their usefulness in other spruce species. Mol. Genet. Genom. 2001, 264, 871–882. [Google Scholar] [CrossRef]
- Scotti, I.; Paglia, G.; Magni, F.; Morgante, M. Population genetics of Norway spruce (Picea abies Karst.) at regional scale: Sensitivity of different microsatellite motif classes in detecting differentiation. Ann. For. Sci. 2006, 63, 485–491. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. Micro—Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.O.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.O.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Smouse, P.E.; Banks, S.C.; Peakall, R. Converting quadratic entropy to diversity: Both animals and alleles are diverse, but some are more diverse than others. PLoS ONE 2017, 12, e0185499. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C.H. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2010, 3, 244–262. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Matthew, S.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Liu, J.X. StructureSelector: A web based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Aravanopoulos, F.A. Conservation and monitoring of tree genetic resources in temperate forests. Curr. For. Rep. 2016, 2, 119–129. [Google Scholar] [CrossRef]
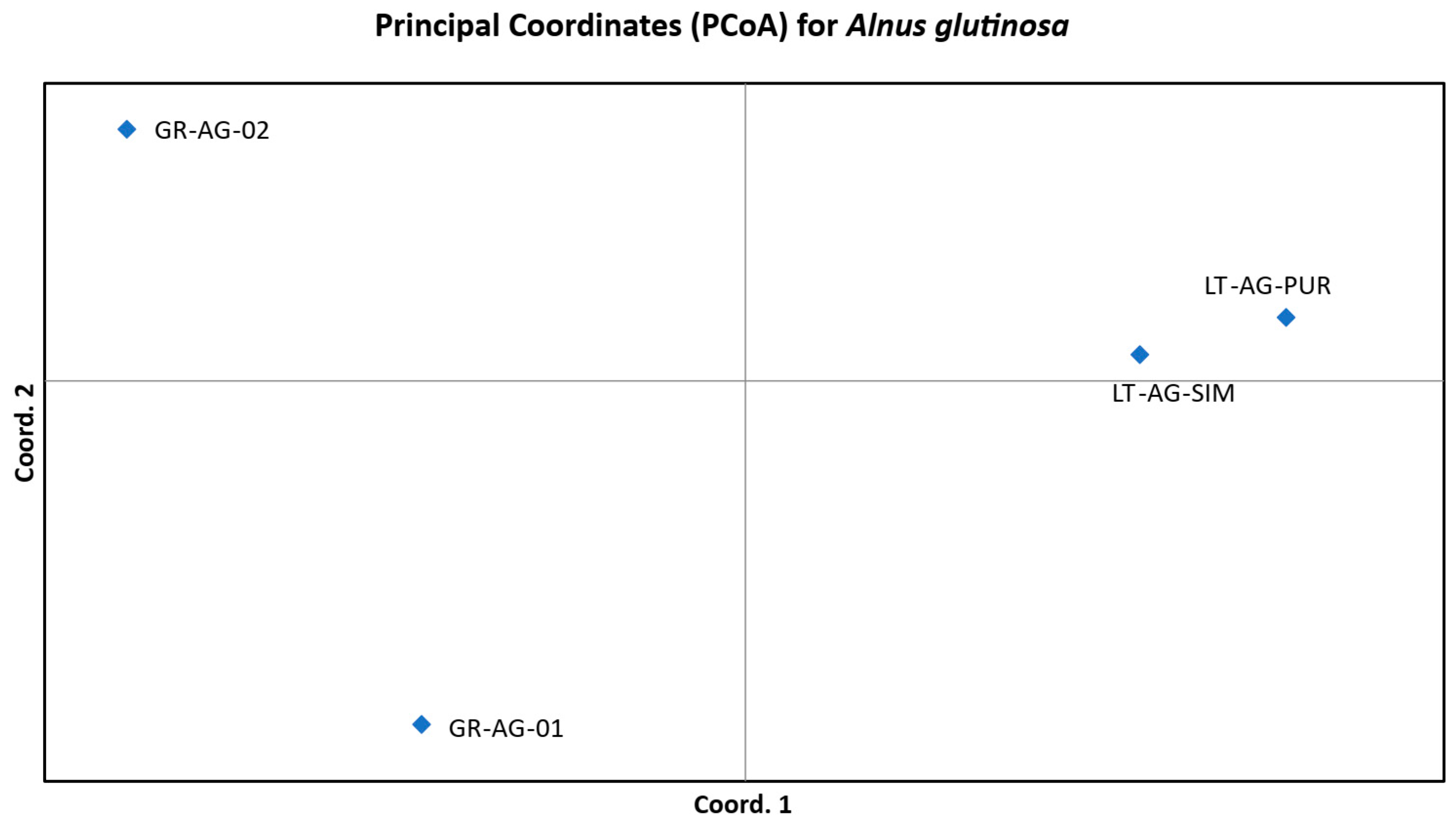
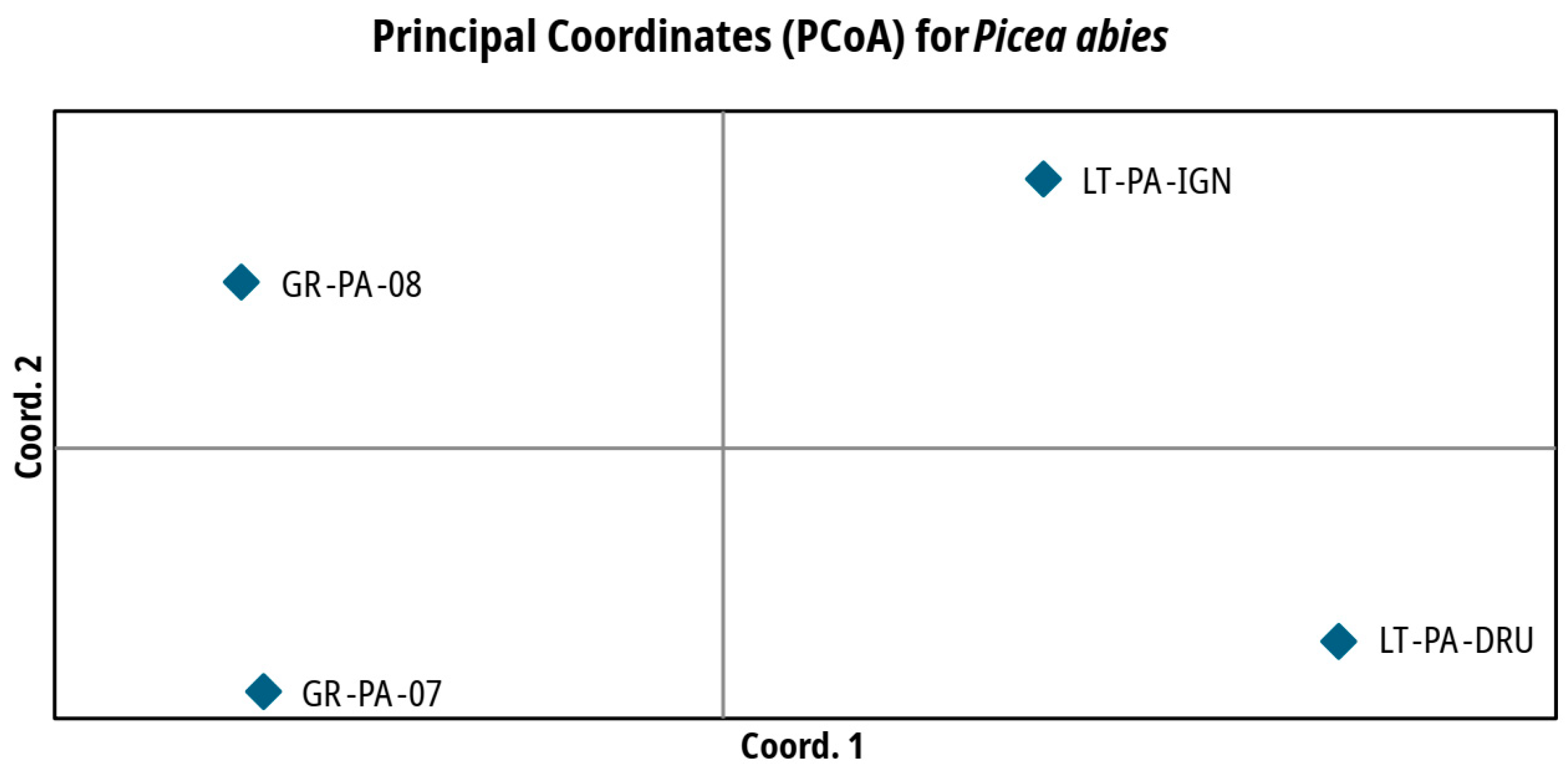
| Population | |||||||
|---|---|---|---|---|---|---|---|
| GR-AG-01 | GR-AG-02 | Mean, Peripheral | LT-AG-PUR | LT-AG-SIM | Mean, Core | Grand Mean | |
| Na | 9.78 ± 0.85 | 7.22 ± 0.60 | 8.50 ± 0.55 | 9.83 ± 0.74 | 10.50 ± 0.63 | 10.16 ± 0.47 | 9.33 ± 0.38 |
| Ae | 4.92 ± 0.53 | 3.92 ± 0.35 | 4.42 ± 0.32 | 4.71 ± 0.48 | 5.12 ± 0.54 | 4.91 ± 0.35 | 4.67 ± 0.24 |
| Ar | 9.15 ± 0.79 | 6.83 ± 0.55 | 7.99 ± 0.52 | 9.08 ± 0.64 | 9.70 ± 0.57 | 9.39 ± 0.43 | 9.59 ± 0.59 |
| No. of rare alleles | 85 | 50 | 117 * | 84 | 85 | 118 * | 149 ** |
| No. of private alleles | 24 | 10 | 37 * | 11 | 11 | 45 * | – |
| Ho | 0.78 ± 0.05 | 0.82 ± 0.04 | 0.80 ± 0.03 | 0.71 ± 0.04 | 0.73 ± 0.04 | 0.72 ± 0.03 | 0.76 ± 0.02 |
| He | 0.73 ± 0.05 | 0.71 ± 0.03 | 0.72 ± 0.03 | 0.72 ± 0.04 | 0.75 ± 0.04 | 0.73 ± 0.03 | 0.72 ± 0.02 |
| FIS | −0.06 ± 0.04 | −0.15 ± 0.07 | −0.08 ± 0.05 | 0.03 ± 0.04 | 0.04 ± 0.03 | 0.03 ± 0.03 | −0.04 ± 0.04 |
| Nm | 3.86 ± 0.42 | 3.24 ± 0.61 | 12.44 ± 2.00 | 44.16 ± 33.94 | 44.66 ± 33.74 | 50.54 ± 9.19 | 6.14 ± 0.77 |
| Ne (CI) | 87.6 (70.0–115.1) | 48.8 (40.3–60.6) | 67.5 (61.7–74.2) | 180.6 (127.2–301.2) | 119.7 (94.3–161.3) | 162.2 (139.3–192.6) | – |
| Population | |||||||
|---|---|---|---|---|---|---|---|
| GR-PA-07 | GR-PA-08 | Mean, Peripheral | LT-PA-IGN | LT-PA-DRU | Mean, Core | Grand Mean | |
| Na | 13.33 ± 1.61 | 13.61 ± 1.60 | 13.47 ± 1.12 | 13.56 ± 1.25 | 11.94 ± 1.46 | 12.75 ± 0.96 | 13.11 ± 0.73 |
| Ae | 7.14 ± 1.25 | 6.84 ± 1.07 | 6.98 ± 0.81 | 5.69 ± 0.65 | 5.00 ± 0.80 | 5.34 ± 0.51 | 6.17 ± 0.49 |
| Ar | 12.22 ± 1.37 | 12.32 ± 1.37 | 12.27 ± 0.97 | 10.44 ± 0.84 | 9.03 ± 0.98 | 9.74 ± 0.66 | 11.57 ± 1.12 |
| No. of rare alleles | 151 | 162 | 249 * | 167 | 142 | 223 * | 330 ** |
| No. of private alleles | 44 | 43 | 117 * | 29 | 21 | 83 * | – |
| Ho | 0.84 ± 0.04 | 0.85 ± 0.04 | 0.84 ± 0.03 | 0.83 ± 0.04 | 0.82 ± 0.05 | 0.83 ± 0.03 | 0.83 ± 0.02 |
| He | 0.78 ± 0.03 | 0.78 ± 0.04 | 0.78 ± 0.02 | 0.78 ± 0.03 | 0.73 ± 0.04 | 0.75 ± 0.02 | 0.77 ± 0.02 |
| FIS | –0.05 ± 0.05 | –0.07 ± 0.05 | –0.05 ± 0.05 | –0.05 ± 0.04 | –0.12 ± 0.07 | –0.10 ± 0.05 | –0.10 ± 0.05 |
| Nm | 17.48 ± 9.22 | 17.35 ± 9.27 | 38.41 ± 8.92 | 13.32 ± 4.43 | 10.98 ± 5.38 | 32.10 ± 5.19 | 10.24 ± 1.42 |
| Ne (CI) | ∞ (805.5–∞) | ∞ (147,375–∞) | 50,069 (1100.8–∞) | 103.0 (87.5–124.3) | 458.2 (253.2–2076.7) | 163.8 (147.3–183.9) | – |
| Population | LT-AG-PUR | LT-AG-SIM | GR-AG-01 | GR-AG-02 |
|---|---|---|---|---|
| LT-AG-PUR | 0.000 | 0.008 (0.165) | 0.286 (0.001) | 0.357 (0.001) |
| LT-AG-SIM | 0.008 (0.165) | 0.000 | 0.234 (0.001) | 0.307 (0.001) |
| GR-AG-01 | 0.046 (0.001) | 0.037 (0.001) | 0.000 | 0.178 (0.001) |
| GR-AG-02 | 0.058 (0.001) | 0.049 (0.001) | 0.031 (0.001) | 0.000 |
| Population | LT-PA-IGN | LT-PA-DRU | GR-PA-07 | GR-PA-08 |
|---|---|---|---|---|
| LT-PA-IGN | 0.000 | 0.043 (0.001) | 0.144 (0.001) | 0.149 (0.001) |
| LT-PA-DRU | 0.011 (0.001) | 0.000 | 0.221 (0.001) | 0.230 (0.001) |
| GR-PA-07 | 0.023 (0.001) | 0.034 (0.001) | 0.000 | 0.030 (0.020) |
| GR-PA-08 | 0.024 (0.001) | 0.035 (0.001) | 0.013 (0.020) | 0.000 |
| Sampling Locations and Population Codes | Type of Protected Area | No. of Trees Sampled; Total (Mature and Juvenile) | Population Type | Location Coordinates | |
|---|---|---|---|---|---|
| Latitude | Longitude | ||||
| Šimkaičiai, LT-AG-SIM | Genetic reserve | 40 (28, 12) | Core | 55.19456 | 22.83602 |
| Purviniškės, LT-AG-PUR | Seed stand | 40 (28, 12) | Core | 55.01534 | 25.62753 |
| Mouries, GR-AG-01 | Natura 2000 | 40 (27, 13) | Peripheral | 41.24923 | 22.77066 |
| Lake Chimaditis, GR-AG-02 | Natura 2000 | 40 (32, 8) | Peripheral | 40.60419 | 21.54777 |
| Druskininkai, LT-PA-DRU | Genetic reserve | 40 (28, 12) | Core | 53.96849 | 24.33219 |
| Ignalina, LT-PA-IGN | Genetic reserve | 40 (28, 12) | Core | 55.36613 | 26.19626 |
| Elatia, GR-PA-08 | Natura 2000 | 25 (25, 0) | Peripheral | 41.4850 | 24.3336 |
| Tsakalos, GR-PA-07 | Natura 2000 | 25 (25, 0) | Peripheral | 41.5302 | 24.2835 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verbylaitė, R.; Aravanopoulos, F.A.; Baliuckas, V.; Tourvas, N.; Farsakoglou, A.-M.; Kotina, V.-M.; Lyrou, F.G.; Juškauskaitė, A.; Petrokas, R.; Lygis, V. The Central-Periphery Hypothesis Revisited: Implications for Long-Term Genetic Conservation. Plants 2025, 14, 3563. https://doi.org/10.3390/plants14233563
Verbylaitė R, Aravanopoulos FA, Baliuckas V, Tourvas N, Farsakoglou A-M, Kotina V-M, Lyrou FG, Juškauskaitė A, Petrokas R, Lygis V. The Central-Periphery Hypothesis Revisited: Implications for Long-Term Genetic Conservation. Plants. 2025; 14(23):3563. https://doi.org/10.3390/plants14233563
Chicago/Turabian StyleVerbylaitė, Rita, Filippos A. Aravanopoulos, Virgilijus Baliuckas, Nikolaos Tourvas, Anna-Maria Farsakoglou, Vasiliki-Maria Kotina, Fani G. Lyrou, Aušra Juškauskaitė, Raimundas Petrokas, and Vaidotas Lygis. 2025. "The Central-Periphery Hypothesis Revisited: Implications for Long-Term Genetic Conservation" Plants 14, no. 23: 3563. https://doi.org/10.3390/plants14233563
APA StyleVerbylaitė, R., Aravanopoulos, F. A., Baliuckas, V., Tourvas, N., Farsakoglou, A.-M., Kotina, V.-M., Lyrou, F. G., Juškauskaitė, A., Petrokas, R., & Lygis, V. (2025). The Central-Periphery Hypothesis Revisited: Implications for Long-Term Genetic Conservation. Plants, 14(23), 3563. https://doi.org/10.3390/plants14233563








