Transcriptome-Based miR156-Mediated Expression Dynamics of SPL Transcription Factors During Vegetative to Reproductive Transition in Spinach
Abstract
1. Introduction
2. Results
2.1. Classification of Spinach Plant Growth Stages
2.2. Identification and Protein Physicochemical Properties of SPL Members in Spinach
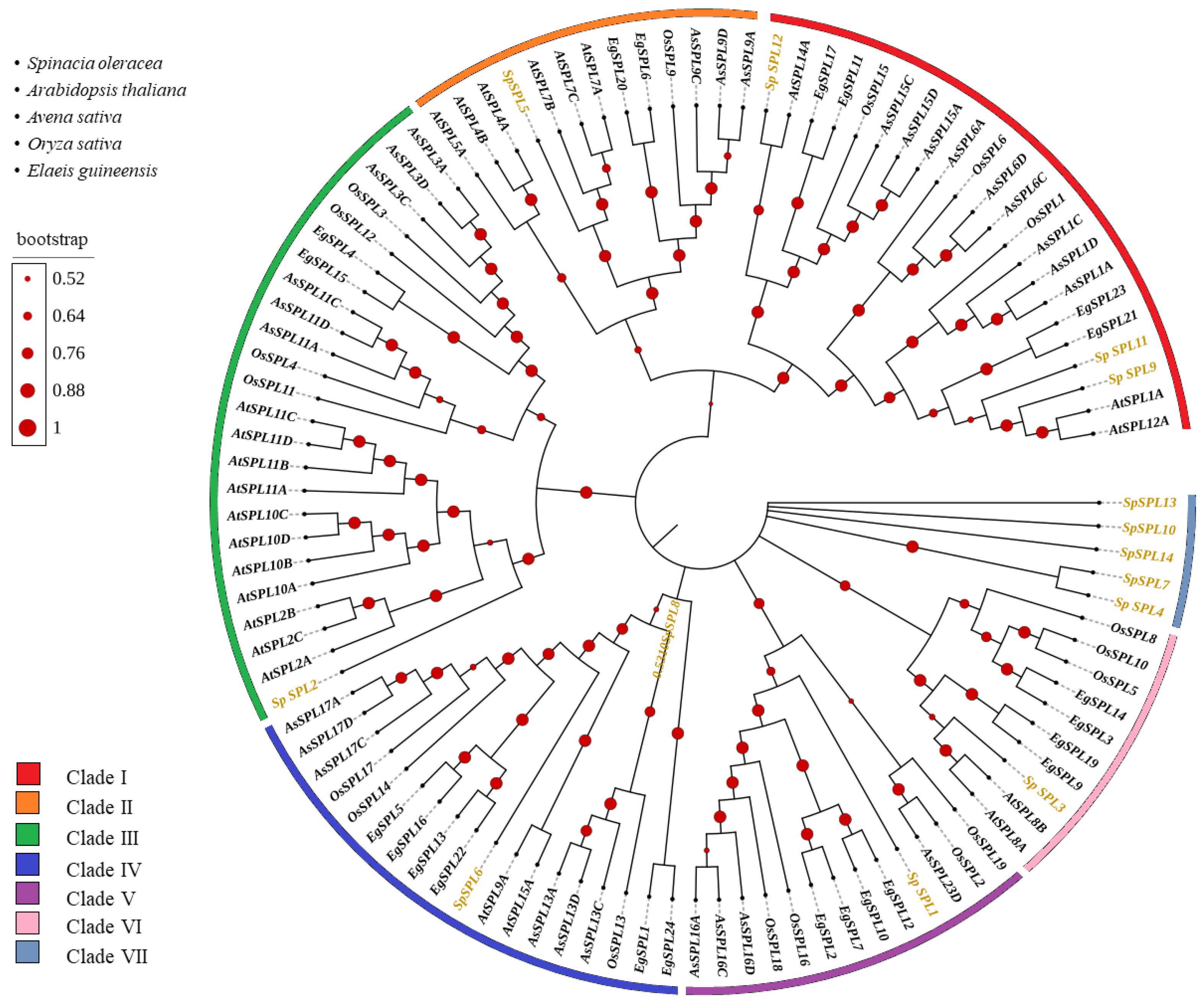
2.3. Phylogenetic Relationship of SPL Gene Members
2.4. Multiple Protein Sequence Alignment, Domain Confirmation, and Conserved Motif Analysis
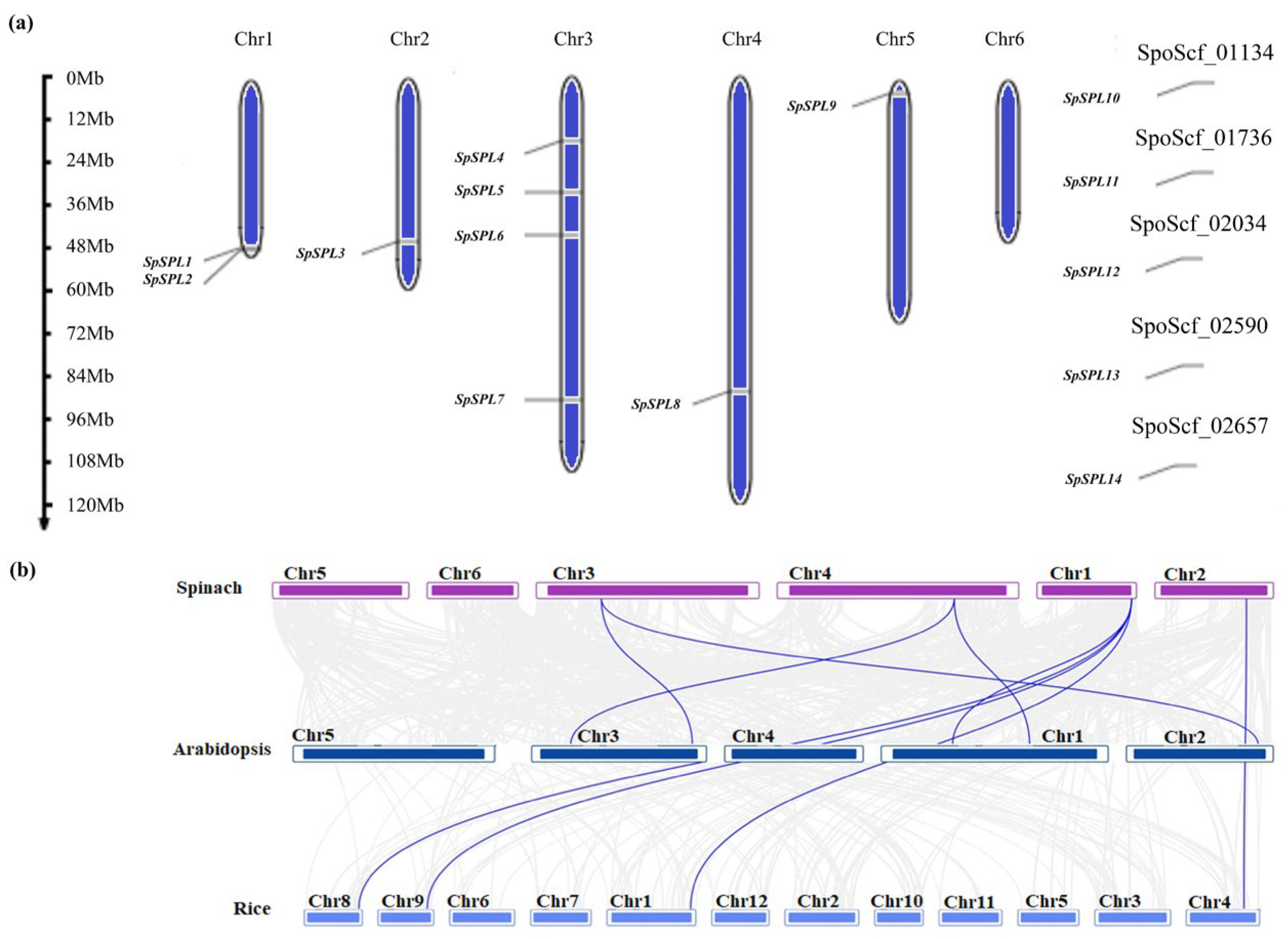
2.5. Chromosome Localization, Gene Duplication, and Collinearity Analysis
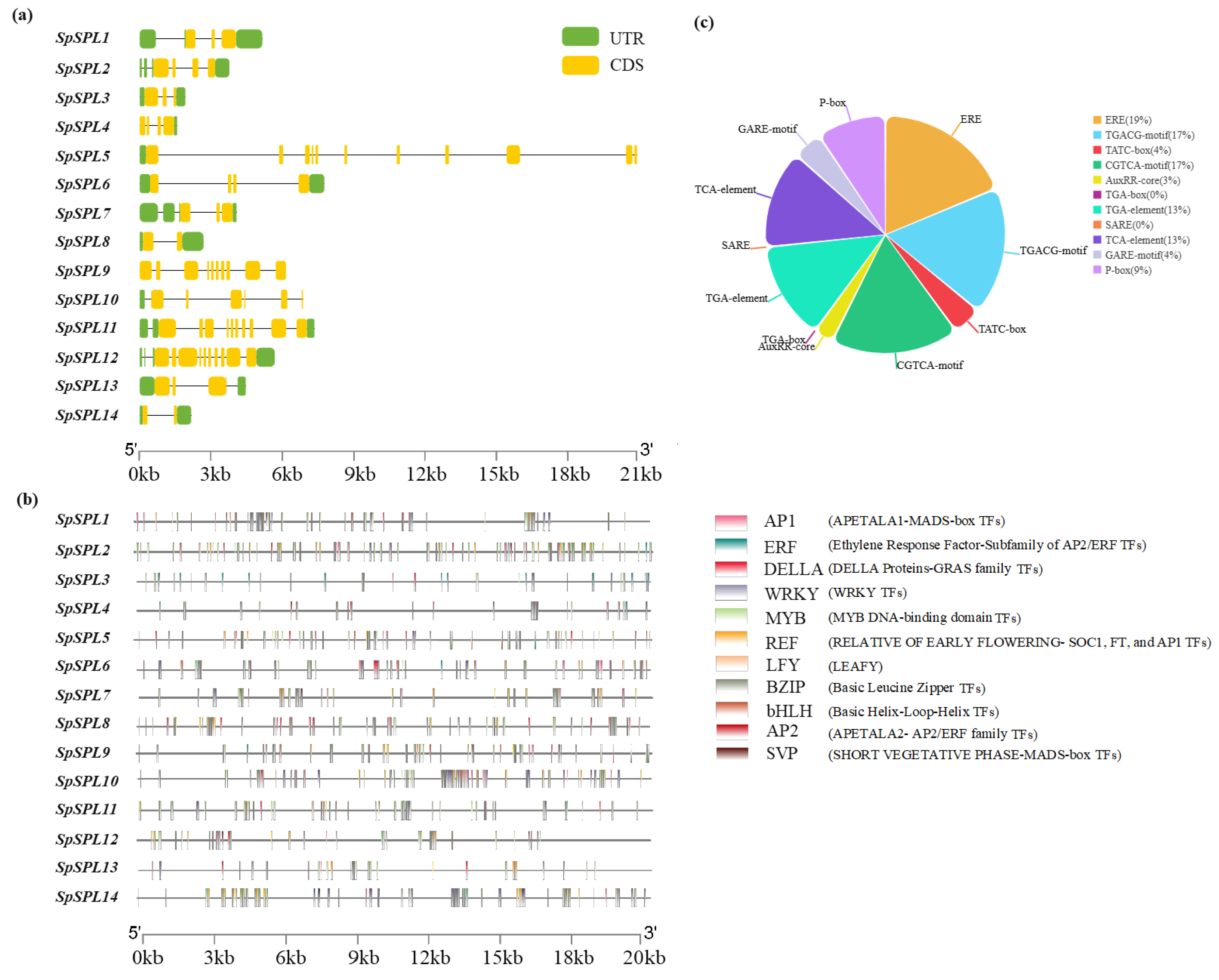
2.6. Gene Structure Distribution, Cis-Acting Elements, and Transcription Factor Binding Sites in the Promoter Region of SpSPLgene Members
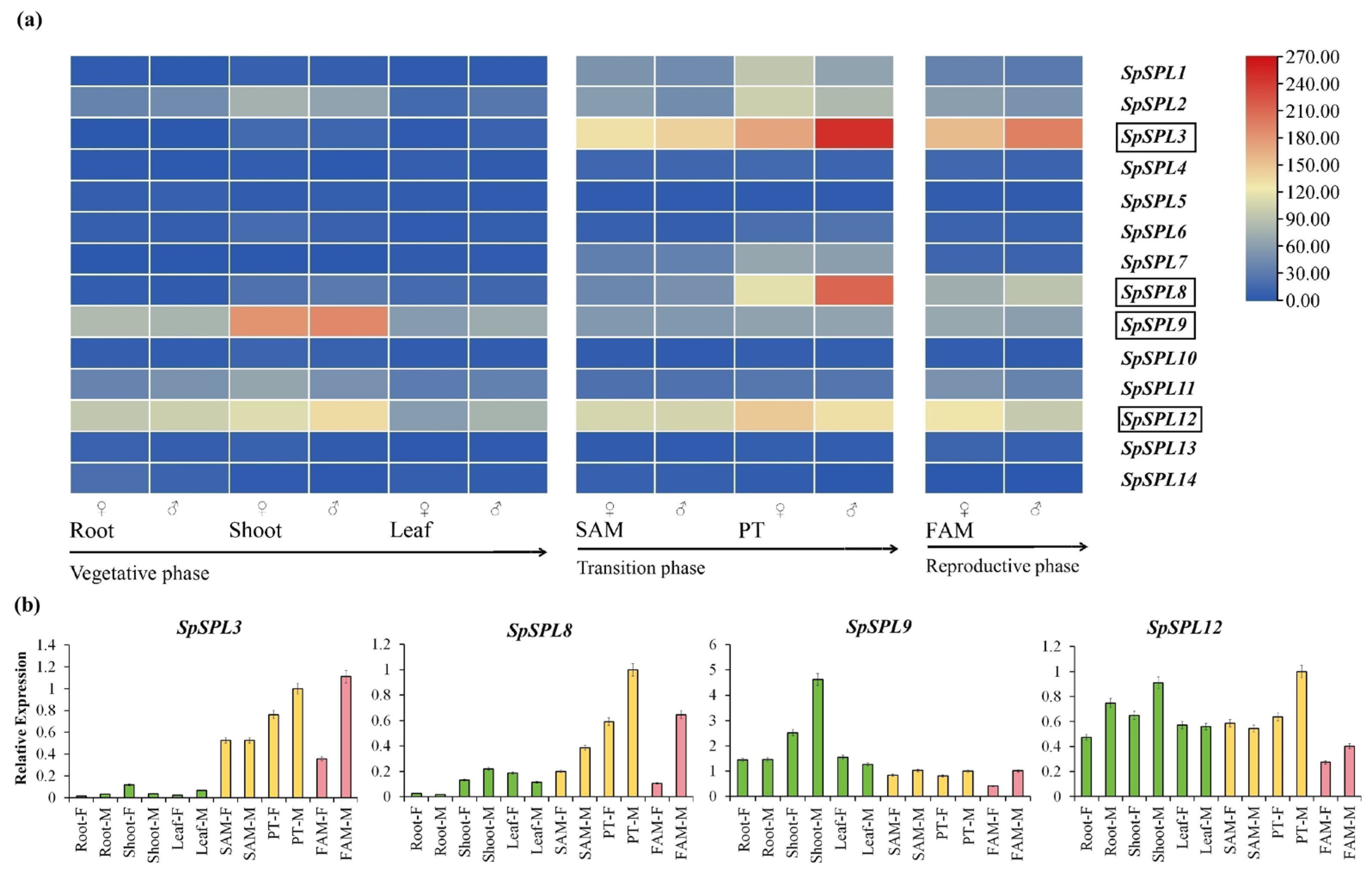
2.7. Expression Profiling of SpSPL Gene Members
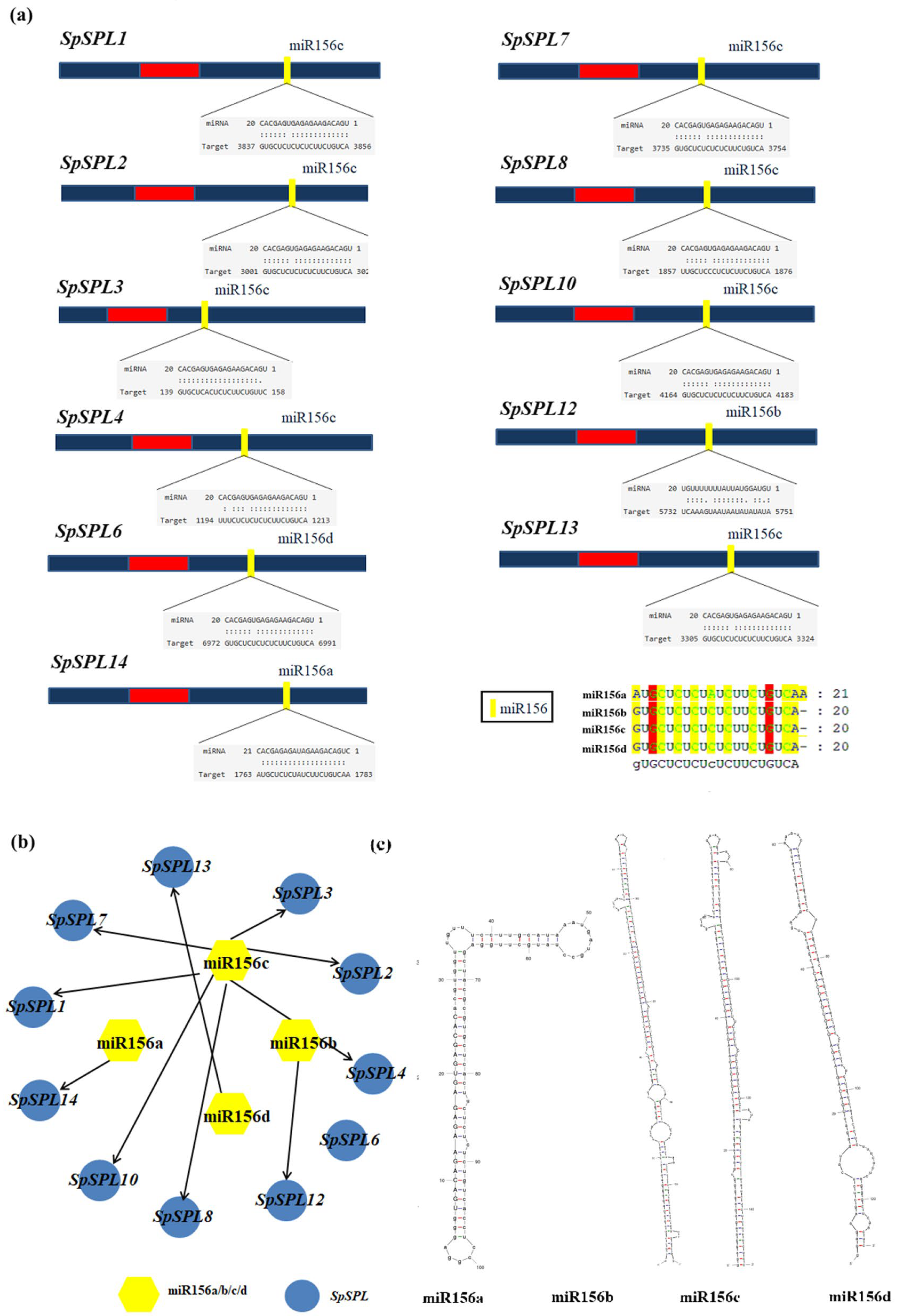
2.8. Identification of miR156 and Network Analysis in Spinach
2.9. Gene Ontology (GO) Enrichment Analysis
2.10. Proposed Model of miR156-Mediated Regulation of SpSPL Genes
3. Discussion
3.1. An Overview of SPL Gene Family Members
3.2. SpmiR156-SpSPL Module in Phase Transition
3.3. Functional Implication in Dioecious Spinach
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction, Library Construction, and Sequencing
4.3. RNA-Seq Data Analysis
4.4. Identification and Physicochemical Properties Analysis of SpSPL Gene Members
4.5. Phylogenetic and Sequence Analysis of SpSPL Gene Members
4.6. Chromosome Distribution, Collinearity Analysis, and Gene Duplication of SpSPL Gene Members
4.7. Multi-Sequence Alignment and Domain Confirmation of SpSPL Gene Members
4.8. Gene Structure and Cis-Element and Transcription Factor Binding Sites in Promoter Region
4.9. MicroRNAs Identification and Analysis of SpSPL Gene Members
4.10. qRT-PCR Validation of the Expression of Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| Sp | Spinacia olaracea |
| SPL | SQUAMOSA Promoter-Binding Protein-Like |
| miRNA | MicroRNA |
| TFs | Transcription Factors |
| GO | Gene Ontology |
| qRT-PCR | Quantitative Real-Time Polymerase Chain Reaction |
| SAM | Shoot Apical Meristem |
| FAM | Flower Apical Meristem |
| IM | Inflorescence Meristem |
| PT | Phase Transition Meristem |
| F | Female |
| M | Male |
| DEGs | Differentially Expressed Genes |
| CDs | Coding Sequences |
| IPs | Predicted Isoelectric |
| GRAVY | Grand Average of Hydropathy |
| TPM | Transcripts per Million |
| GSDS | Gene Structure Display Server |
| CDD | Conserved Domain Database |
| PWM | Position Weight Matrix |
References
- Huijser, P.; Schmid, M. The Control of Developmental Phase Transitions in Plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef]
- Tanaka, N.; Itoh, H.; Sentoku, N.; Kojima, M.; Sakakibara, H.; Izawa, T.; Itoh, J.-I.; Nagato, Y. The COP1 Ortholog PPS Regulates the Juvenile–Adult and Vegetative–Reproductive Phase Changes in Rice. Plant Cell 2011, 23, 2143–2154. [Google Scholar] [CrossRef]
- Luo, Y.; Guo, Z.; Li, L. Evolutionary Conservation of MicroRNA Regulatory Programs in Plant Flower Development. Dev. Biol. 2013, 380, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Aung, B.; Gruber, M.Y.; Amyot, L.; Omari, K.; Bertrand, A.; Hannoufa, A. Micro RNA 156 as a Promising Tool for Alfalfa Improvement. Plant Biotechnol. J. 2015, 13, 779–790. [Google Scholar] [CrossRef]
- Ahsan, M.U.; Hayward, A.; Irihimovitch, V.; Fletcher, S.; Tanurdzic, M.; Pocock, A.; Beveridge, C.A.; Mitter, N. Juvenility and Vegetative Phase Transition in Tropical/Subtropical Tree Crops. Front. Plant Sci. 2019, 10, 729. [Google Scholar] [CrossRef]
- He, J.; Xu, M.; Willmann, M.R.; McCormick, K.; Hu, T.; Yang, L.; Starker, C.G.; Voytas, D.F.; Meyers, B.C.; Poethig, R.S. Threshold-Dependent Repression of SPL Gene Expression by MiR156/MiR157 Controls Vegetative Phase Change in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007337. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, L.; Wu, G. Epigenetic Regulation of Juvenile-to-Adult Transition in Plants. Front. Plant Sci. 2018, 9, 1048. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, P.; Liu, S.; Yang, Q.; Guo, H. The Control of Developmental Phase Transitions by MicroRNAs and Their Targets in Seed Plants. Int. J. Mol. Sci. 2020, 21, 1971. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Bartel, D.P. Antiquity of MicroRNAs and Their Targets in Land Plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar] [CrossRef]
- Poethig, R.S. Vegetative Phase Change and Shoot Maturation in Plants. Curr. Top. Dev. Biol. 2013, 105, 125–152. [Google Scholar]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of MiR156-Regulated squamosa promoter binding protein-like (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Wei, X.; Ke, H.; Wen, A.; Gao, B.; Shi, J.; Feng, Y. Structural Basis of MicroRNA Processing by Dicer-like 1. Nat. Plants 2021, 7, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of Plant MicroRNA Targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef]
- Gai, Y.-P.; Zhao, H.-N.; Zhao, Y.-N.; Zhu, B.-S.; Yuan, S.-S.; Li, S.; Guo, F.-Y.; Ji, X.-L. MiRNA-Seq-Based Profiles of MiRNAs in Mulberry Phloem Sap Provide Insight into the Pathogenic Mechanisms of Mulberry Yellow Dwarf Disease. Sci. Rep. 2018, 8, 812. [Google Scholar] [CrossRef]
- Nozawa, M.; Miura, S.; Nei, M. Origins and Evolution of MicroRNA Genes in Plant Species. Genome Biol. Evol. 2012, 4, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Q.; Zhao, Y.; Li, X.; Ma, Q. Comparative Genome Analysis of the SPL Gene Family Reveals Novel Evolutionary Features in Maize. Genet. Mol. Biol. 2019, 394, 380–394. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, Z.; Zhao, X.; Cao, H.; Wang, L.; Liu, S.; Wang, C.; Liu, M.; Wang, L.; Liu, Z. Superstar MicroRNA, MiR156, Involved in Plant Biological Processes and Stress Response: A Review. Sci. Hortic. 2023, 316, 112010. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal Regulation of Shoot Development in Arabidopsis thaliana by MiR156 and Its Target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A New Family of DNA Binding Proteins Includes Putative Transcriptional Regulators of the Antirrhinum majus Floral Meristem Identity Gene SQUAMOSA. Mol. Gen. Genet. MGG 1996, 250, 7–16. [Google Scholar]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E. A Novel Zinc-Binding Motif Revealed by Solution Structures of DNA-Binding Domains of Arabidopsis SBP-Family Transcription Factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, S.; Kumar, S.; Kumar Sharma, M.; Kumar Sinha, M. Hermetic Storage: A Technology for Reducing Grains Losses during Storage. Int. J. Chem. Stud. 2019, 6, 763–768. [Google Scholar]
- Cardon, G.; Höhmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular Characterisation of the Arabidopsis SBP-Genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative Study of SBP-Gene Family in Arabidopsis and Rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Li, Y.; Deng, Y.; Qin, D.; An, X. International Journal of Biological Macromolecules Study of the SPL Gene Family and MiR156- SPL Module in Populus tomentosa: Potential Roles in Juvenile-to-Adult Phase Transition and Reproductive Phase. Int. J. Biol. Macromol. 2025, 296, 139547. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target Mimicry Provides a New Mechanism for Regulation of MicroRNA Activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X. Regulation of OsSPL14 by OsmiR156 Defines Ideal Plant Architecture in Rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.-J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 Promotes Panicle Branching and Higher Grain Productivity in Rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Luo, L.; Li, W.; Miura, K.; Ashikari, M.; Kyozuka, J. Control of Tiller Growth of Rice by OsSPL14 and Strigolactones, Which Work in Two Independent Pathways. Plant Cell Physiol. 2012, 53, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Shen, G.; Peng, K.; Huang, Z.; Tong, J.; Kabir, M.H.; Wang, J.; Zhang, J.; Qin, G.; Xiao, L. The Alteration in the Architecture of a T-DNA Insertion Rice Mutant Osmtd1 Is Caused by Up-regulation of MicroRNA156f. J. Integr. Plant Biol. 2015, 57, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Wu, C.; Xiong, L. Genomic Organization, Differential Expression, and Interaction of SQUAMOSA Promoter-Binding-like Transcription Factors and MicroRNA156 in Rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Wang, J.-W.; Park, M.Y.; Wang, L.-J.; Koo, Y.; Chen, X.-Y.; Weigel, D.; Poethig, R.S. MiRNA Control of Vegetative Phase Change in Trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Hirakawa, H.; Toyoda, A.; Itoh, T.; Suzuki, Y.; Nagano, A.J.; Sugiyama, S.; Onodera, Y. A Spinach Genome Assembly with Remarkable Completeness, and Its Use for Rapid Identification of Candidate Genes for Agronomic Traits. DNA Res. 2021, 28, dsab004. [Google Scholar] [CrossRef]
- Li, L.; Xu, J.B.; Zhu, Z.W.; Ma, R.; Wu, X.Z.; Geng, Y.K. Genome-Wide Identification and Expression Analysis of the SPL Transcription Factor Family and Its Response to Abiotic Stress in Pisum sativum L. BMC Genom. 2024, 25, 539. [Google Scholar]
- Zhou, L.; Yarra, R. Genome-Wide Analysis of SPL/MiR156 Module and Its Expression Analysis in Vegetative and Reproductive Organs of Oil Palm (Elaeis guineensis). Int. J. Mol. Sci. 2023, 24, 13658. [Google Scholar] [CrossRef] [PubMed]
- Fan, E.; Liu, C.; Wang, Z.; Wang, S.; Ma, W.; Lu, N.; Liu, Y.; Fu, P.; Wang, R.; Lv, S. Genome-Wide Identification and Expression Analysis of the SQUAMOSA Promoter-Binding Protein-like (SPL) Transcription Factor Family in Catalpa bungei. Int. J. Mol. Sci. 2023, 25, 97. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Liu, J.; Huang, J.; Wang, C.; Zhang, L.; Feng, S. Genome-Wide Identification of MiR156 and SPL Family Genes and Phenotypic Analysis of Vegetative Phase Change in Pepper (Capsicum annuum L.). Gene 2023, 877, 147542. [Google Scholar] [CrossRef]
- Hou, H.; Li, J.; Gao, M.; Singer, S.D.; Wang, H.; Mao, L.; Fei, Z.; Wang, X. Genomic Organization, Phylogenetic Comparison and Differential Expression of the SBP-Box Family Genes in Grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef]
- Li, B.; Zhao, Y.; Wang, S.; Zhang, X.; Wang, Y.; Shen, Y.; Yuan, Z. Genome-Wide Identification, Gene Cloning, Subcellular Location and Expression Analysis of SPL Gene Family in P. granatum L. BMC Plant Biol. 2021, 21, 400. [Google Scholar] [CrossRef]
- Song, M.; Wang, R.; Zhou, F.; Wang, R.; Zhang, S.; Li, D.; Song, J.; Yang, S.; Yang, Y. SPLs-Mediated Flowering Regulation and Hormone Biosynthesis and Signaling Accompany Juvenile-Adult Phase Transition in Pyrus. Sci. Hortic. 2020, 272, 109584. [Google Scholar] [CrossRef]
- Guo, A.-Y.; Zhu, Q.-H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-Wide Identification and Evolutionary Analysis of the Plant Specific SBP-Box Transcription Factor Family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Fan, Y.; Xue, G.; He, A.; Yang, H.; He, C.; Li, Y.; Ruan, J.; Yan, J.; Cheng, J. Genome-Wide Identification and Characterization of the SPL Gene Family and Its Expression in the Various Developmental Stages and Stress Conditions in Foxtail Millet (Setaria italica). BMC Genom. 2022, 23, 389. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Z.; Xu, Y.; Kong, L.; Shi, J.; Liu, Y.; Fu, C.; Wang, X.; Wang, Z.-Y.; Zhou, C. Genome-Wide Characterization of SPL Family in Medicago truncatula Reveals the Novel Roles of MiR156/SPL Module in Spiky Pod Development. BMC Genom. 2019, 20, 552. [Google Scholar] [CrossRef]
- Ren, Y.; Ma, R.; Fan, Y.; Zhao, B.; Cheng, P.; Fan, Y.; Wang, B. Genome-Wide Identification and Expression Analysis of the SPL Transcription Factor Family and Its Response to Abiotic Stress in Quinoa (Chenopodium quinoa). BMC Genom. 2022, 23, 773. [Google Scholar] [CrossRef]
- Wang, Y.; Ruan, Q.; Zhu, X.; Wang, B.; Wei, B.; Wei, X. Identification of Alfalfa SPL Gene Family and Expression Analysis under Biotic and Abiotic Stresses. Sci. Rep. 2023, 13, 84. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H. The MiR156/SPL Module, a Regulatory Hub and Versatile Toolbox, Gears up Crops for Enhanced Agronomic Traits. Mol. Plant 2015, 8, 677–688. [Google Scholar] [CrossRef]
- Yu, N.; Yang, J.-C.; Yin, G.-T.; Li, R.-S.; Zou, W.-T. Genome-Wide Characterization of the SPL Gene Family Involved in the Age Development of Jatropha curcas. BMC Genom. 2020, 21, 368. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.K.; Goel, R.; Kumari, S.; Dahuja, A. Genomic Organization, Phylogenetic Comparison, and Expression Profiles of the SPL Family Genes and Their Regulation in Soybean. Dev. Genes Evol. 2017, 227, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Peng, H.; Chen, F.; Liu, Y.; Chen, B.; Li, W. Genome-Wide Identification and Characterization, Phylogenetic Comparison and Expression Profiles of SPL Transcription Factor Family in B. juncea (Cruciferae). PLoS ONE 2019, 14, e0224704. [Google Scholar] [CrossRef]
- Lawrence, E.H.; Leichty, A.R.; Doody, E.E.; Ma, C.; Strauss, S.H.; Poethig, R.S. Vegetative Phase Change in Populus Tremula× Alba. New Phytol. 2021, 231, 351–364. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Zhang, J.; Zhang, Y.; Han, Q.; Hu, T.; Xu, X.; Liu, H.; Li, H.; Ye, Z. Over-expression of Sly-miR156a in Tomato Results in Multiple Vegetative and Reproductive Trait Alterations and Partial Phenocopy of the Sft Mutant. FEBS Lett. 2011, 585, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banerjee, A.K. MicroRNA156: A Potential Graft-Transmissible MicroRNA That Modulates Plant Architecture and Tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014, 164, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Aung, B.; Gao, R.; Gruber, M.Y.; Yuan, Z.-C.; Sumarah, M.; Hannoufa, A. MsmiR156 Affects Global Gene Expression and Promotes Root Regenerative Capacity and Nitrogen Fixation Activity in Alfalfa. Transgenic Res. 2017, 26, 541–557. [Google Scholar] [CrossRef]
- You, C.; He, W.; Hang, R.; Zhang, C.; Cao, X.; Guo, H.; Chen, X.; Cui, J.; Mo, B. FIERY1 Promotes MicroRNA Accumulation by Suppressing RRNA-Derived Small Interfering RNAs in Arabidopsis. Nat. Commun. 2019, 10, 4424. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Wu, M.-F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The MicroRNA-Regulated SBP-Box Transcription Factor SPL3 Is a Direct Upstream Activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef]
- Feng, C.; Zhang, X.; Du, B.; Xiao, Y.; Wang, Y.; Sun, Y.; Zhou, X.; Wang, C.; Liu, Y.; Li, T.-H. MicroRNA156ab Regulates Apple Plant Growth and Drought Tolerance by Targeting Transcription Factor MsSPL13. Plant Physiol. 2023, 192, 1836–1857. [Google Scholar] [CrossRef]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-Wide Identification and Analysis of the SBP-Box Family Genes in Apple (Malus × Domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Xiong, Z.; Zhu, X.; Feng, P.; Hu, Z.; Fang, R.; Zhang, Y.; Liu, Q. RcSPL1—RcTAF15b Regulates the f Lowering Time of Rose (Rosa chinensis). Hortic. Res. 2023, 10, uhad083. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.S.; Ning, C.; Li, Z.; Hu, C.; Zhang, J. Floral Regulation: The Significant Virtue of Horticultural Flowering Plants. Horticulturae 2025, 11, 102. [Google Scholar] [CrossRef]
- Davoudpour, Y.; Schmidt, M.; Calabrese, F.; Richnow, H.H.; Musat, N. High Resolution Microscopy to Evaluate the Efficiency of Surface Sterilization of Zea Mays Seeds. PLoS ONE 2020, 15, e0242247. [Google Scholar] [CrossRef]
- Manuela, D.; Xu, M. Juvenile Leaves or Adult Leaves: Determinants for Vegetative Phase Change in Flowering Plants. Int. J. Mol. Sci. 2020, 21, 9753. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Sci. 2000, 290, 1151–1155. [Google Scholar] [CrossRef]



| Gene IDs | Putative IDs | CHR | Type | CDS Length | Protein Length | Start | End | Strand | Introns | Exons | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spo04944 | SpSPL1 | chr1 | gene | 1185 | 394 | 48,064,777 | 48,070,072 | + | 2 | 3 | nucleus |
| Spo04935 | SpSPL2 | chr1 | gene | 1362 | 453 | 48,289,996 | 48,293,867 | + | 3 | 4 | nucleus |
| Spo08630 | SpSPL3 | chr2 | gene | 858 | 285 | 46,729,688 | 46,731,659 | + | 2 | 3 | nucleus |
| Spo01383 | SpSPL4 | chr3 | gene | 948 | 315 | 18,256,316 | 18,257,931 | + | 4 | 3 | nucleus |
| Spo06804 | SpSPL5 | Chr3 | gene | 2469 | 822 | 19,484,558 | 19,506,030 | + | 10 | 11 | nucleus |
| Spo16305 | SpSPL6 | chr3 | gene | 1092 | 363 | 33,003,448 | 33,011,420 | + | 3 | 4 | nucleus |
| Spo06968 | SpSPL7 | chr3 | gene | 1050 | 349 | 92,766,316 | 92,770,496 | + | 2 | 3 | nucleus |
| Spo26325 | SpSPL8 | chr4 | gene | 675 | 224 | 90,130,430 | 90,133,186 | + | 1 | 2 | nucleus |
| Spo02184 | SpSPL9 | chr5 | gene | 2984 | 994 | 3,852,598 | 3,858,915 | − | 9 | 10 | plasma membrane |
| Spo06850 | SpSPL10 | SpoScf_01134 | gene | 1482 | 493 | 112,527 | 119,578 | + | 5 | 6 | nucleus |
| Spo14961 | SpSPL11 | SpoScf_01736 | gene | 2970 | 989 | 5796 | 13,340 | + | 9 | 10 | endomembrane |
| Spo24998 | SpSPL12 | SpoScf_02034 | gene | 3237 | 1078 | 4996 | 10,827 | + | 9 | 10 | plasma membrane |
| Spo16283 | SpSPL13 | SpoScf_02590 | gene | 1563 | 520 | 28,195 | 32,774 | + | 2 | 3 | nucleus |
| Spo22151 | SpSPL14 | SpoScf_02657 | gene | 330 | 109 | 7708 | 9930 | + | 1 | 2 | nucleus |
| Gene IDs | Putative IDs | Formulas | Molecular Weight (Da) | Theoretical pI | Instability Index | Aliphatic Index | Gravy |
|---|---|---|---|---|---|---|---|
| Spo04944 | SpSPL1 | C1862H2921N565O595S14 | 43,190.94 | 9.23 | 46.65 | 58.65 | −0.738 |
| Spo04935 | SpSPL2 | C2139H3358N630O694S18 | 49,581.08 | 8.49 | 56.9 | 60.93 | −0.616 |
| Spo08630 | SpSPL3 | C1391H2160N438O431S14 | 32,363.97 | 9.3 | 75.8 | 56.49 | −0.833 |
| Spo01383 | SpSPL4 | C1495H2358N454O481S17 | 34,932.94 | 8.57 | 70.59 | 63.46 | −0.609 |
| Spo06804 | SpSPL5 | C4027H6371N1137O1225S53 | 92,013.95 | 6.61 | 47.93 | 79.29 | −0.353 |
| Spo16305 | SpSPL6 | C1631H2533N501O548S20 | 38,569.28 | 7.11 | 62.46 | 52.59 | −0.647 |
| Spo06968 | SpSPL7 | C1632H2555N493O523S29 | 38,523.96 | 7.93 | 57.67 | 51.92 | −0.64 |
| Spo26325 | SpSPL8 | C1035H1698N338O344S15 | 24,861.83 | 9.59 | 66.92 | 45.85 | −0.983 |
| Spo02184 | SpSPL9 | C4821H7583N1389O1510S47 | 110,669.46 | 5.67 | 50.26 | 79.94 | −0.419 |
| Spo06850 | SpSPL10 | C2406H3791N67O768S26 | 55,183.14 | 6.15 | 54.53 | 74.14 | −0.468 |
| Spo14961 | SpSPL11 | C4797H7634N1378O1468S49 | 109,670.67 | 7.51 | 51.33 | 81.31 | −0.391 |
| Spo24998 | SpSPL12 | C5119H8107N1497O1620S57 | 118,370.16 | 8.13 | 57.63 | 71.45 | −0.438 |
| Spo16283 | SpSPL13 | C2452H3850N712O823S24 | 57,241.26 | 5.99 | 53.83 | 65.79 | −0.582 |
| Spo22151 | SpSPL14 | C428H818N174O172S8 | 12,611.85 | 7.68 | 67.45 | 42.02 | −1.093 |
| Duplicate Gene Pairs Putative IDs | Ka | Ks | Ka/Ks | Duplication | Type of Mutation/Evolution |
|---|---|---|---|---|---|
| SpSPL10/SpSPL13 | 0.598279 | 1.604524 | 0.37286998 | SD | Negitive mutation/Purifing |
| SpSPL1/SpSPL2 | 0.635706 | 2.991455 | 0.212507439 | TD | Negitive mutation/Purifing |
| SpSPL4/SpSPL7 | 0.166343 | 0.348476 | 0.477344591 | TD | Negitive mutation/Purifing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, E.; Zheng, Y.; Wang, T.; Cai, L.; Ming, R. Transcriptome-Based miR156-Mediated Expression Dynamics of SPL Transcription Factors During Vegetative to Reproductive Transition in Spinach. Plants 2025, 14, 3543. https://doi.org/10.3390/plants14223543
Khalid E, Zheng Y, Wang T, Cai L, Ming R. Transcriptome-Based miR156-Mediated Expression Dynamics of SPL Transcription Factors During Vegetative to Reproductive Transition in Spinach. Plants. 2025; 14(22):3543. https://doi.org/10.3390/plants14223543
Chicago/Turabian StyleKhalid, Ehsan, Yutong Zheng, Tengqi Wang, Lingmin Cai, and Ray Ming. 2025. "Transcriptome-Based miR156-Mediated Expression Dynamics of SPL Transcription Factors During Vegetative to Reproductive Transition in Spinach" Plants 14, no. 22: 3543. https://doi.org/10.3390/plants14223543
APA StyleKhalid, E., Zheng, Y., Wang, T., Cai, L., & Ming, R. (2025). Transcriptome-Based miR156-Mediated Expression Dynamics of SPL Transcription Factors During Vegetative to Reproductive Transition in Spinach. Plants, 14(22), 3543. https://doi.org/10.3390/plants14223543





