Integrative Roles of miRNAs and circRNAs in Plant Antiviral Gene Regulation and Autophagy
Abstract
1. Introduction
2. Gene Regulation by Small RNAs in Plants
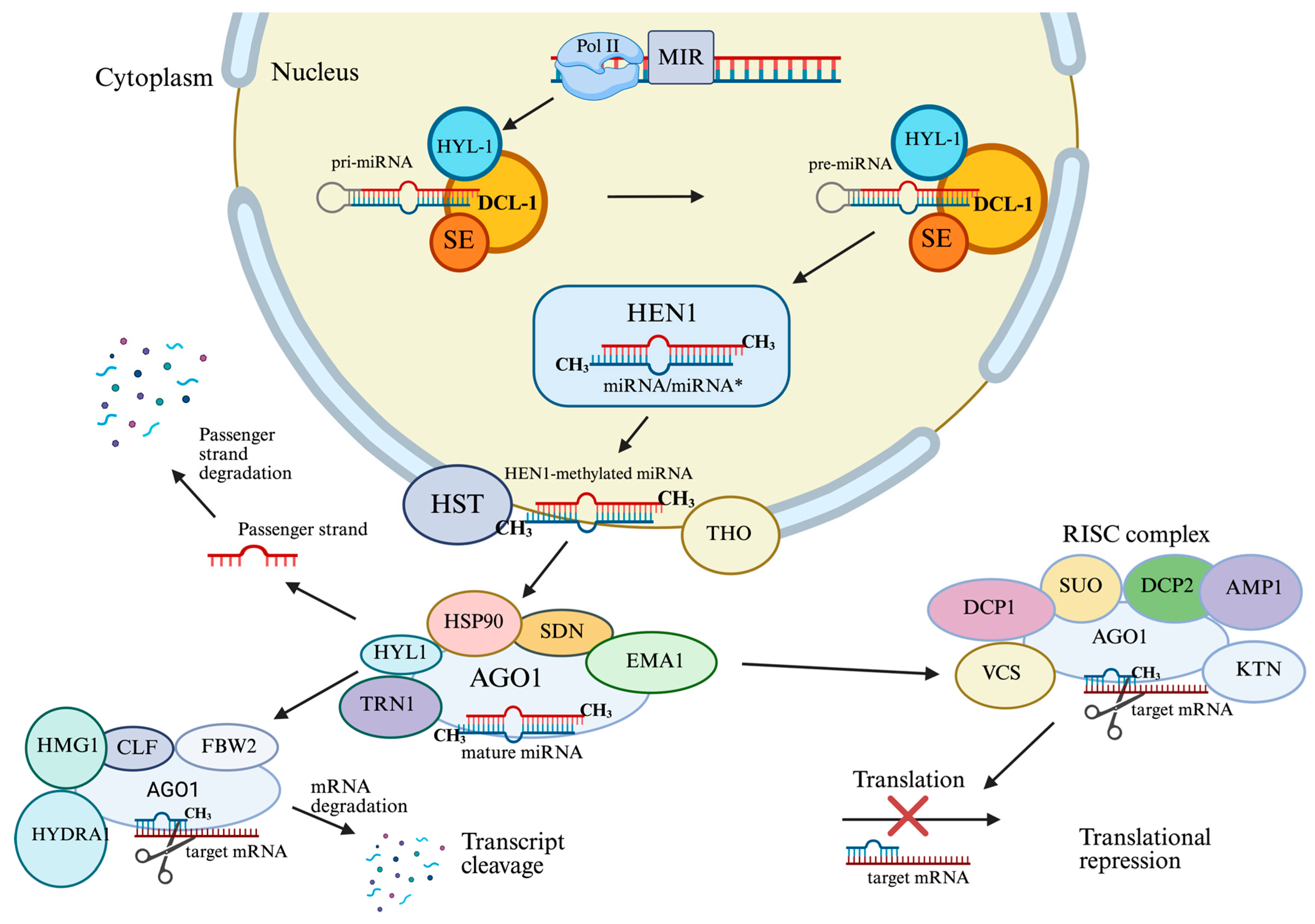
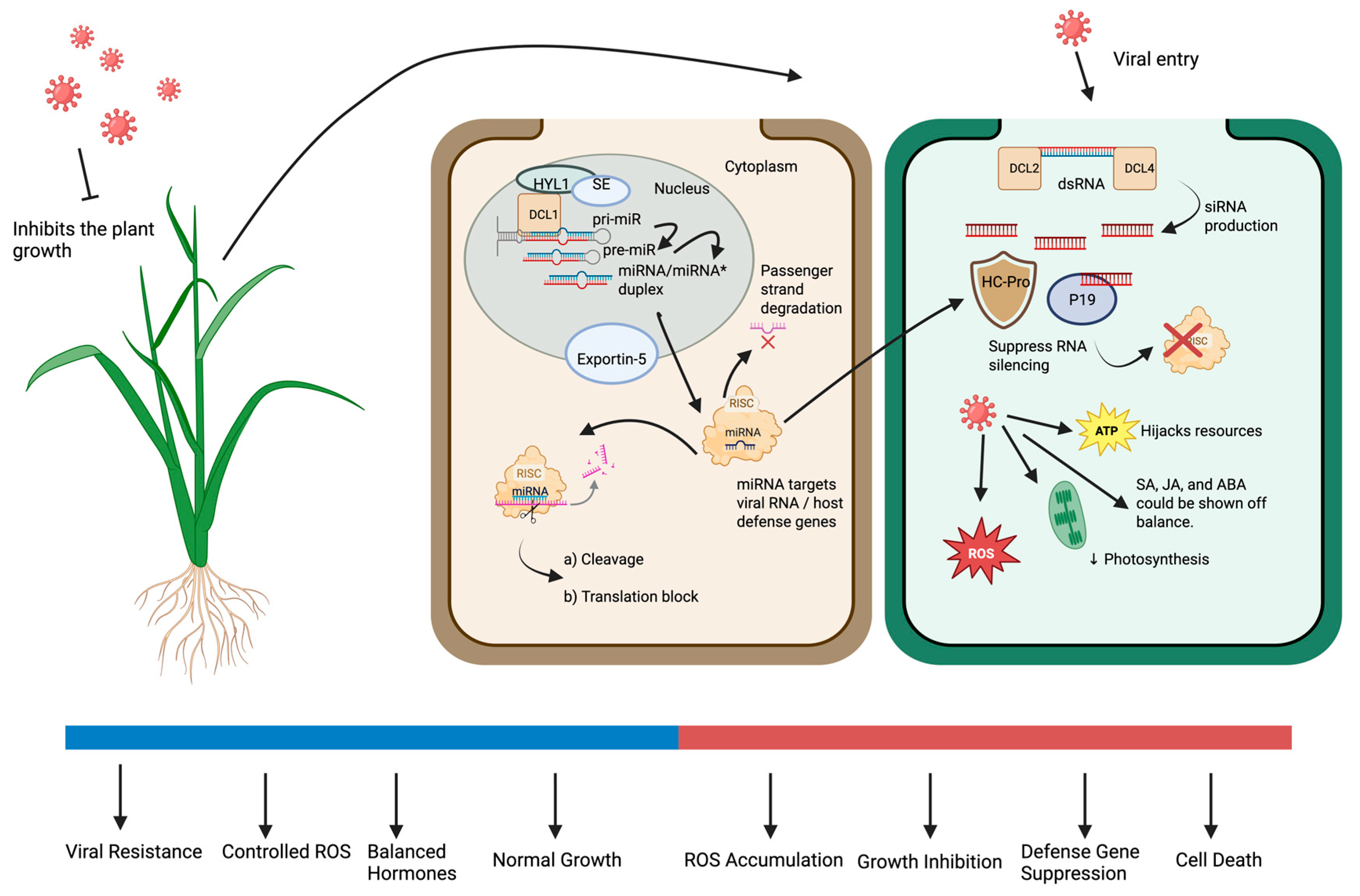
2.1. Plant miRNA Biogenesis
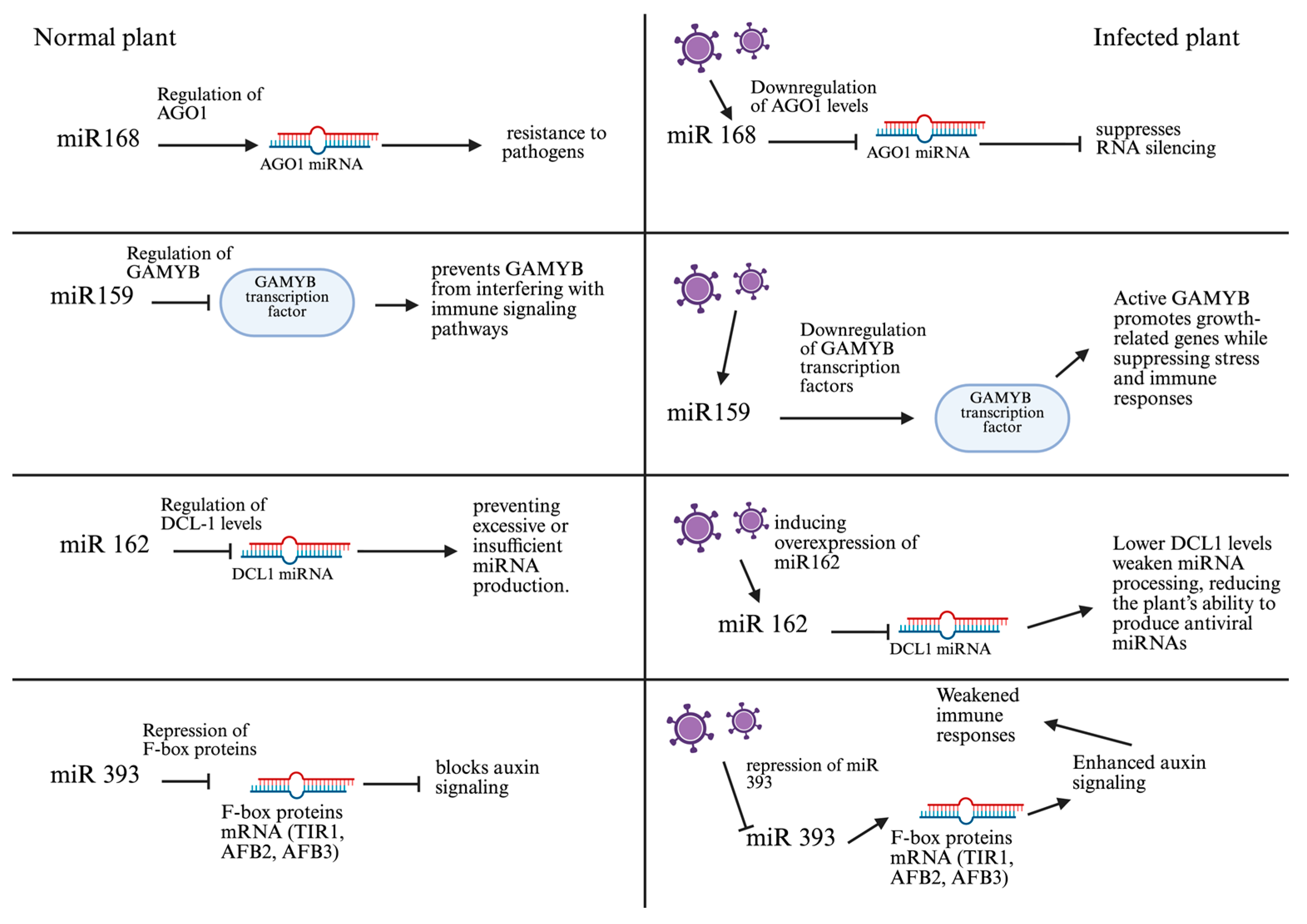
2.2. Functional Roles of microRNAs in Plant Antiviral Immunity
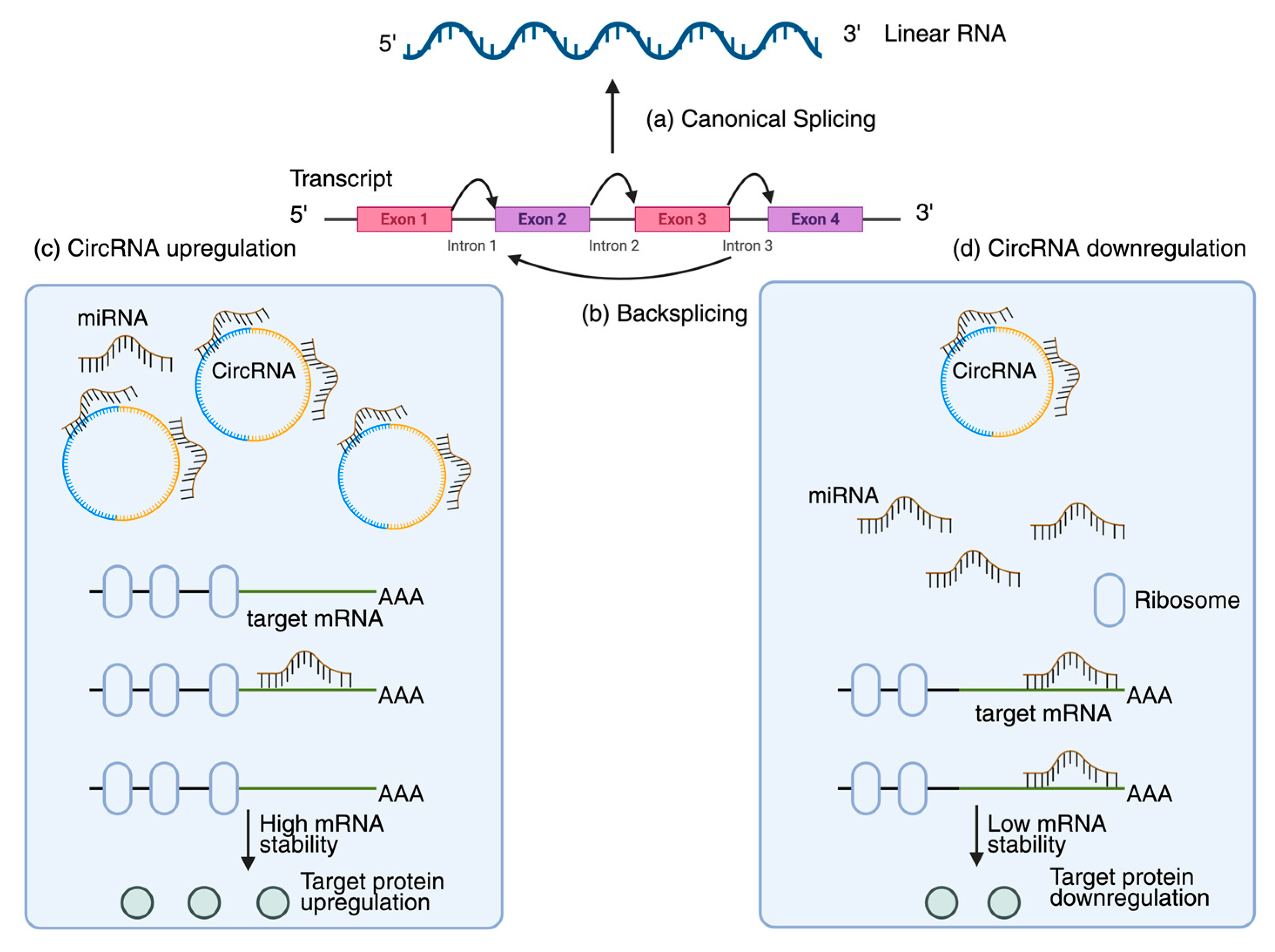
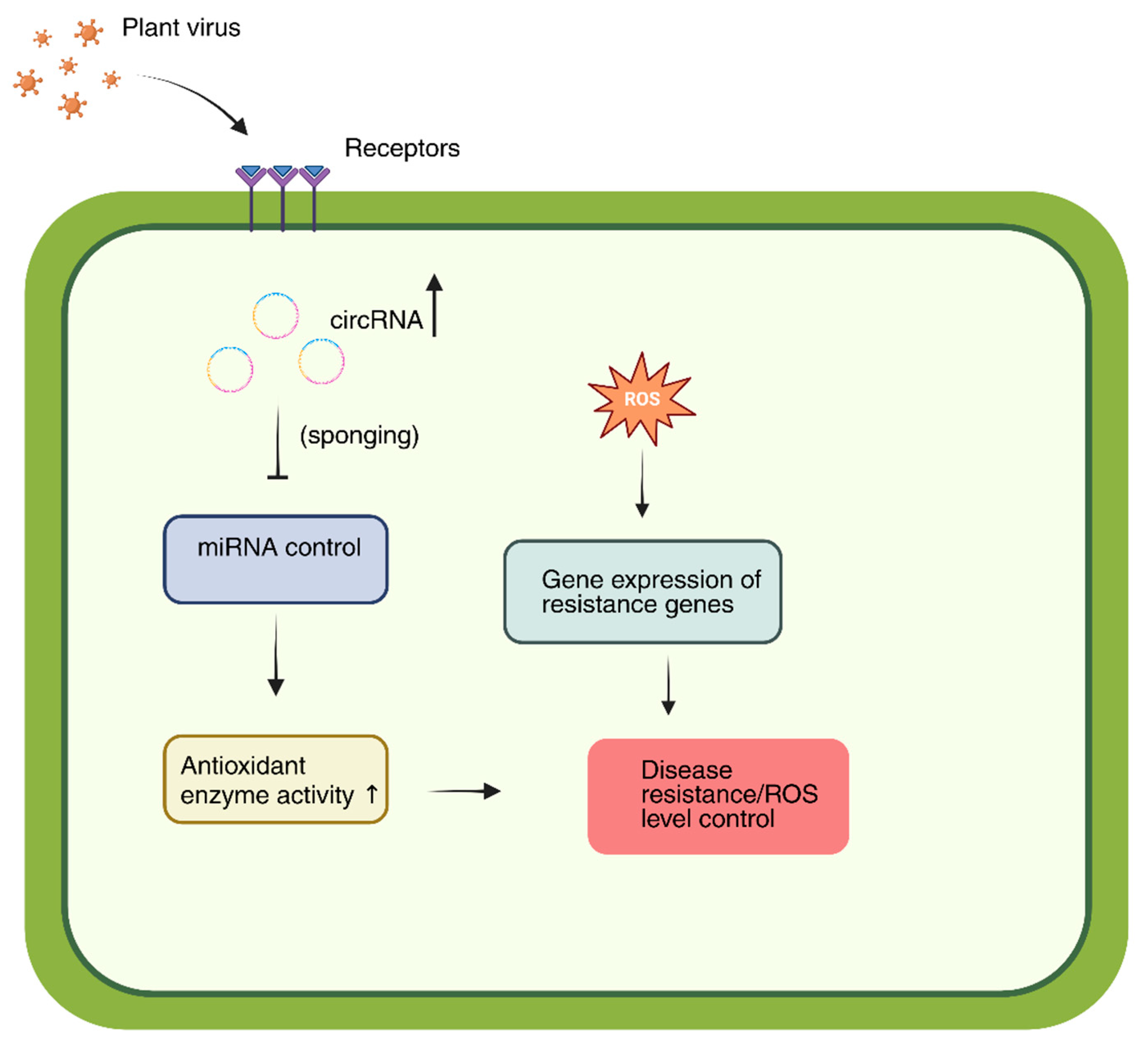
2.3. Roles of Plant Circular RNAs in Regulating Programmed Cell Death During Viral Infection
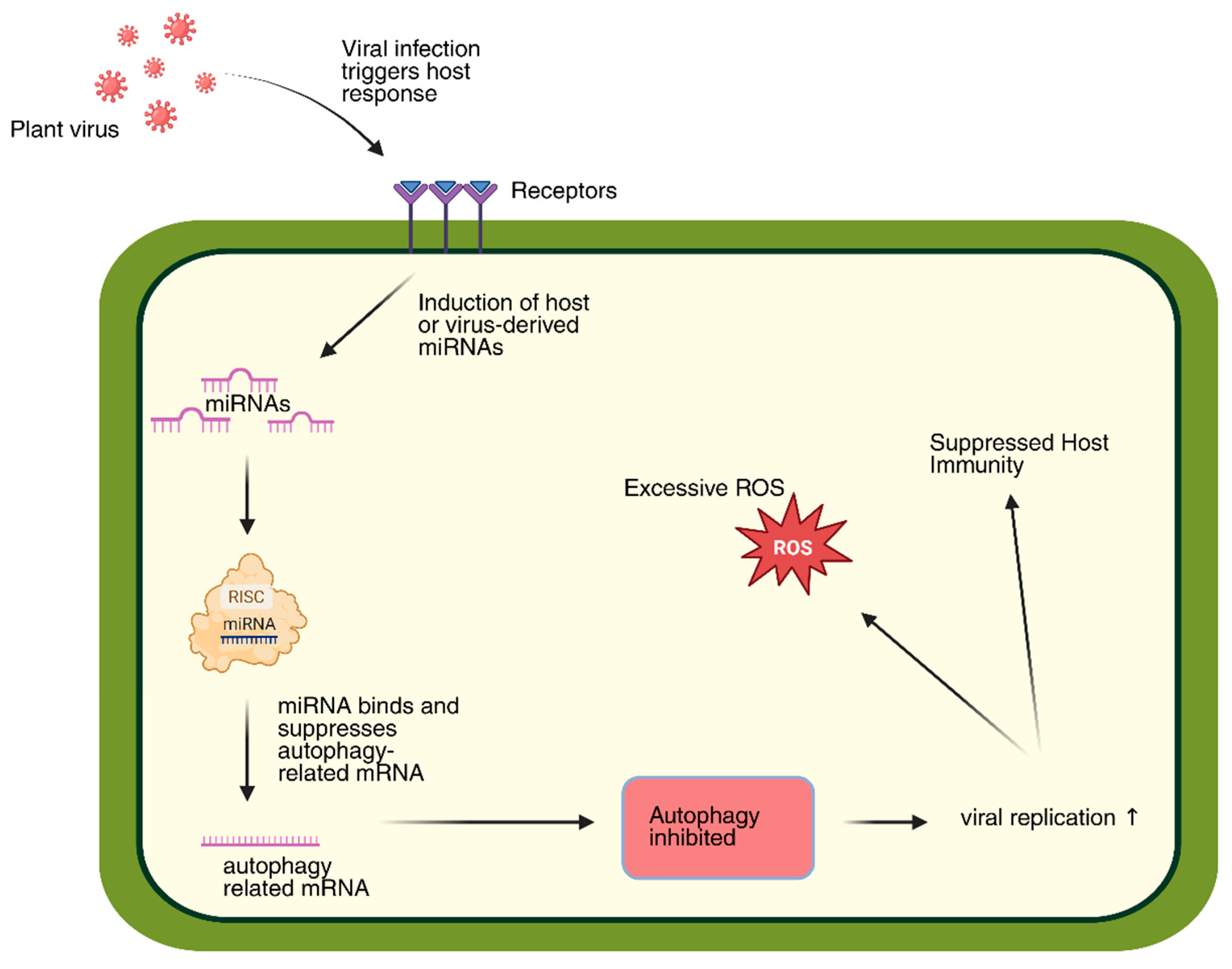
2.4. Complex Interaction of Viruses and Autophagy
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| sRNA | small RNA |
| miR | microRNA |
| circRNA | circular RNA |
| SAR | systemic acquired resistance |
| RISC | RNA-induced silencing complex |
| PCD | programmed cell death |
| ROS | reactive oxygen species |
| VSR | viral suppressor of RNA silencing |
| HR | hypersensitive response |
References
- Lu, Y.; Gan, Q.; Chi, X.; Qin, S. Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep. 2008, 27, 571–1579. [Google Scholar] [CrossRef]
- Ding, T.; Li, W.; Li, F.; Ren, M.; Wang, W. microRNAs: Key Regulators in Plant Responses to Abiotic and Biotic Stresses via Endogenous and Cross-Kingdom Mechanisms. Int. J. Mol. Sci. 2024, 25, 1154. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Q. MicroRNA-Based Biotechnology for Plant Improvement. J. Cell Physiol. 2015, 230, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.-F. PTI-ETI Crosstalk: An Integrative View of Plant Immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Gouveia, B.C.; Calil, I.P.; Machado, J.P.B.; Santos, A.A.; Fontes, E.P.B. Immune Receptors and Co-Receptors in Antiviral Innate Immunity in Plants. Front. Microbiol. 2017, 7, 2139. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef]
- Kawasaki, T.; Yamada, K.; Yoshimura, S.; Yamaguchi, K. Chitin Receptor-Mediated Activation of MAP Kinases and ROS Production in Rice and Arabidopsis. Plant Signal. Behav. 2017, 12, e1361076. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.H.; Chen, S.; Zhou, J.-M. Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Wang, W.; Feng, B.; Zhou, J.-M.; Tang, D. Plant Immune Signaling: Advancing on Two Frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef]
- Martin, R.; Qi, T.; Zhang, H.; Liu, F.; King, M.; Toth, C.; Nogales, E.; Staskawicz, B.J. Structure of the Activated ROQ1 Resistosome Directly Recognizing the Pathogen Effector XopQ. Science 2020, 370, eabd9993. [Google Scholar] [CrossRef]
- Nguyen, Q.-M.; Iswanto, A.B.B.; Son, G.H.; Kim, S.H. Recent Advances in Effector-Triggered Immunity in Plants: New Pieces in the Puzzle Create a Different Paradigm. Int. J. Mol. Sci. 2021, 22, 4709. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.; Li, Y.; Tung, J.; Deng, Y.; Baker, B.; Dinesh-Kumar, S.P.; Li, F. Conserved Transcription Factors NRZ1 and NRM1 Regulate NLR Receptor-Mediated Immunity. Plant Physiol. 2024, 195, 832–849. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-Triggered Immunity: From Pathogen Perception to Robust Defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Kontra, L.; Csorba, T.; Tavazza, M.; Lucioli, A.; Tavazza, R.; Moxon, S.; Tisza, V.; Medzihradszky, A.; Turina, M.; Burgyán, J. Distinct Effects of P19 RNA Silencing Suppressor on Small RNA Mediated Pathways in Plants. PLoS Pathog. 2016, 12, e1005935. [Google Scholar] [CrossRef]
- Atabekova, A.K.; Solovieva, A.D.; Chergintsev, D.A.; Solovyev, A.G.; Morozov, S.Y. Role of Plant Virus Movement Proteins in Suppression of Host RNAi Defense. Int. J. Mol. Sci. 2023, 24, 9049. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.J.; Prokhnevsky, A.I.; Gopinath, K.; Dolja, V.V.; Carrington, J.C. Viral RNA Silencing Suppressors Inhibit the microRNA Pathway at an Intermediate Step. Genes Dev. 2004, 18, 1179–1186. [Google Scholar] [CrossRef]
- Yu, X.-Q.; Niu, H.-Q.; Liu, C.; Wang, H.-L.; Yin, W.; Xia, X. PTI-ETI Synergistic Signal Mechanisms in Plant Immunity. Plant Biotechnol. J. 2024, 22, 2113–2128. [Google Scholar] [CrossRef]
- Mengistu, A.; Tenkegna, T. The role of miRNA in plant–virus interaction: A review. Mol. Biol. Rep. 2021, 48, 2853–2861. [Google Scholar] [CrossRef]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef]
- Várallyay, É.; Havelda, Z. Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol. Plant Pathol. 2013, 14, 567–575. [Google Scholar] [CrossRef]
- Hafrén, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef]
- Hofius, D.; Li, L.; Hafrén, A.; Coll, N.S. Autophagy as an emerging arena for plant–pathogen interactions. Curr. Opin. Plant Biol. 2017, 38, 117–123. [Google Scholar] [CrossRef]
- Belter, A.; Popenda, M.; Sajek, M.; Woźniak, T.; Naskręt-Barciszewska, M.Z.; Szachniuk, M.; Jurga, S.; Barciszewski, J. A new molecular mechanism of RNA circularization and the microRNA sponge formation. J. Biomol. Struct. Dyn. 2020, 40, 3038–3045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Dai, M. CircRNA: A rising star in plant biology. J. Genet. Genom. 2022, 49, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.Y.; Chen, L.; Liu, C.; Zhu, Q.H.; Fan, L. Widespread noncoding circular RNA s in plants. New Phytol. 2015, 208, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yuan, M.; Zhao, Y.; Quan, Q.; Yu, D.; Yang, H.; Tang, X.; Xin, X.; Cai, G.; Qian, Q.; et al. Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol. J. 2021, 19, 1240–1252. [Google Scholar] [CrossRef]
- Sha, A.; Zhao, J.; Yin, K.; Tang, Y.; Wang, Y.; Wei, X.; Hong, Y.; Liu, Y. Virus-Based MicroRNA Silencing in Plants. Plant Physiol. 2014, 164, 36–47. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef]
- Pérez-Quintero, Á.L.; Neme, R.; Zapata, A.; López, C. Plant microRNAs and their role in defense against viruses: A bioinformatics approach. BMC Plant Biol. 2010, 10, 138. [Google Scholar] [CrossRef]
- Hake, S. MicroRNAs: A Role in Plant Development. Curr. Biol. 2003, 13, R851–R852. [Google Scholar] [CrossRef]
- Bajczyk, M.; Jarmolowski, A.; Jozwiak, M.; Pacak, A.; Pietrykowska, H.; Sierocka, I.; Swida-Barteczka, A.; Szewc, L.; Szweykowska-Kulinska, Z. Recent Insights into Plant miRNA Biogenesis: Multiple Layers of miRNA Level Regulation. Plants 2023, 12, 342. [Google Scholar] [CrossRef]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In Response to Abiotic Stress, DNA Methylation Confers EpiGenetic Changes in Plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef]
- Omarov, R.; Sparks, K.; Smith, L.; Zindovic, J.; Scholthof, H.B. Biological relevance of a stable biochemical interaction between the tombusvirus-encoded P19 and short interfering RNAs. J. Virol. 2006, 80, 3000–3008. [Google Scholar] [CrossRef]
- Liu, S.-R.; Zhou, J.-J.; Hu, C.-G.; Wei, C.-L.; Zhang, J.-Z. MicroRNA-Mediated Gene Silencing in Plant Defense and Viral Counter-Defense. Front. Microbiol. 2017, 8, 1801. [Google Scholar] [CrossRef]
- Omarov, R.T.; Ciomperlik, J.J.; Scholthof, H.B. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc. Natl. Acad. Sci. USA 2007, 104, 1714–1719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiang, Y.; Chen, S.; Shi, M.; Jiang, X.; He, Z.; Gao, S. Mechanisms of MicroRNA Biogenesis and Stability Control in Plants. Front. Plant Sci. 2022, 13, 844149. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Xue, L.; An, L. Functional diversity of miRNA in plants. Plant Sci. 2007, 172, 423–432. [Google Scholar] [CrossRef]
- Pontes, O.; Pikaard, C.S. siRNA and miRNA processing: New functions for Cajal bodies. Curr. Opin. Genet. Dev. 2008, 18, 197–203. [Google Scholar] [CrossRef]
- Li, Q.; Liu, Y.; Zhang, X. Biomolecular condensates in plant RNA silencing: Insights into formation, function, and stress responses. Plant Cell 2024, 36, 227–245. [Google Scholar] [CrossRef]
- Dong, Q.; Hu, B.; Zhang, C. microRNAs and their roles in plant development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Tomczyk, K.; Mickiewicz, A.; Sarzynska, J.; Szachniuk, M. Bioinformatics Study of Structural Patterns in Plant MicroRNA Precursors. Biomed Res. Int. 2017, 2017, 6783010. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, X. microRNA biogenesis and stabilization in plants. Fundam. Res. 2023, 3, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 2021, 17, e1009242. [Google Scholar] [CrossRef] [PubMed]
- Iksat, N.; Madirov, A.; Artykbayeva, D.; Shevchenko, O.; Zhanassova, K.; Baikarayev, Z.; Masalimov, Z. Heat Stress Induces Partial Resistance to Tomato Bushy Stunt Virus in Nicotiana benthamiana Via Combined Stress Pathways. Viruses 2025, 17, 1250. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.V.; Ratnaparkhe, M.B.; Kumawat, G.; Gupta, G.K.; Husain, S.M. Plant miRNAome and antiviral resistance: A retrospective view and prospective challenges. Virus Genes 2014, 48, 1–14. [Google Scholar] [CrossRef]
- Tong, B.; Shi, Y.; Ntambiyukuri, A.; Li, X.; Zhan, J.; Wang, A.; Xiao, D.; He, L. Integration of Small RNA and degradome sequencing reveals the regulatory network of al-induced programmed cell death in peanut. Int. J. Mol. Sci. 2021, 23, 246. [Google Scholar] [CrossRef]
- Dalmadi, Á.; Miloro, F.; Bálint, J.; Várallyay, É.; Havelda, Z. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021, 49, 12912–12928. [Google Scholar] [CrossRef]
- Vaucheret, H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE 2009, 4, e6442. [Google Scholar] [CrossRef]
- Li, W.; Cui, X.; Meng, Z.; Huang, X.; Xie, Q.; Wu, H.; Jin, H.; Zhang, D.; Liang, W. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012, 158, 1279–1292. [Google Scholar] [CrossRef]
- Vaucheret, H.; Mallory, A.C.; Bartel, D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 2006, 22, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhu, H.; Li, N.; Batley, J.; Wang, Y. The miR393-target module regulates plant development and responses to biotic and abiotic stresses. Int. J. Mol. Sci. 2022, 23, 9477. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Hao, K.; Lv, Z.; Yu, L.; Bu, Q.; Ren, J.; Zhang, H.; Chen, R.; Zhang, L. Profiling of phytohormone-specific microRNAs and characterization of the miR160-ARF1 module involved in glandular trichome development and artemisinin biosynthesis in Artemisia annua. Plant Biotechnol. J. 2023, 21, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Hao, K.; Wang, Y.; Zhu, Z.; Wu, Y.; Chen, R.; Zhang, L. miR160: An indispensable regulator in plant. Front. Plant Sci. 2022, 13, 833322. [Google Scholar] [CrossRef]
- Yin, X.; Wang, J.; Cheng, H.; Wang, X.; Yu, D. Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 2013, 237, 1213–1225. [Google Scholar] [CrossRef]
- Verma, S.; Sarkar, A.K. miRNA-mediated regulation of biotic and abiotic stress responses in plants. In Agricultural Biotechnology: Latest Research and Trends; Springer Nature: Singapore, 2022; pp. 463–492. [Google Scholar] [CrossRef]
- Couzigou, J.M.; Combier, J.P. Plant microRNA s: Key regulators of root architecture and biotic interactions. New Phytol. 2016, 212, 22–35. [Google Scholar] [CrossRef]
- Li, S.; Castillo-González, C.; Yu, B.; Zhang, X. The functions of plant small RNA s in development and in stress responses. Plant J. 2017, 90, 654–670. [Google Scholar] [CrossRef]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and function of miR159 in plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef]
- Imran, M.; Liu, T.; Wang, Z.; Wang, M.; Liu, S.; Gao, X.; Wang, A.; Liu, S.; Tian, Z.; Zhang, M. Evolutionary conservation of nested MIR159 structural microRNA genes and their promoter characterization in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 948751. [Google Scholar] [CrossRef]
- Jiao, B.; Peng, Q.; Wu, B.; Liu, S.; Zhou, J.; Yuan, B.; Lin, H.; Xi, D. The miR172/TOE3 module regulates resistance to tobacco mosaic virus in tobacco. Plant J. 2024, 119, 2672–2686. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Haq, Q.M.; Mukherjee, S.K. MicroRNA profiling of tomato leaf curl new delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol. J. 2010, 7, 281. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeon, H.S.; Kim, H.G.; Park, O.K. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 2017, 214, 343–360. [Google Scholar] [CrossRef]
- Jensen, M.K.; Hagedorn, P.H.; De Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkjaer, M.F. Transcriptional regulation by an NAC (NAM–ATAF1, 2–CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, S.; Zhang, F.; Chen, L.; Hao, X.; Pan, Q.; Fu, X.; Li, L.; Sun, X.; Tang, K. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 2016, 57, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T.; et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, C.; Zeng, J.; Yun, Z.; Liu, Y.; Qu, H.; Jiang, Y.; Duan, X.; Xia, R. Micro RNA 528, a hub regulator modulating ROS homeostasis via targeting of a diverse set of genes encoding copper-containing proteins in monocots. New Phytol. 2020, 225, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kumar, M.; Choudhary, D.; Aher, L.; Rane, J.; Singh, N.P. RNAi Approach: A Powerful Technique for Gene Function Studies and Enhancing Abiotic Stress Tolerance in Crop Plants. In Biotechnologies of Crop Improvement; Gosal, S., Wani, S., Eds.; Springer: Singapore, 2018; pp. 113–127. [Google Scholar] [CrossRef]
- Leng, X.; Wang, P.; Zhu, X.; Li, X.; Zheng, T.; Shangguan, L.; Fang, J. Ectopic expression of CSD1 and CSD2 targeting genes of miR398 in grapevine is associated with oxidative stress tolerance. Funct. Integr. Genom. 2017, 17, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, Q.; Zuo, Z.F.; Liu, L. MicroRNA398: A master regulator of plant development and stress responses. Int. J. Mol. Sci. 2022, 23, 10803. [Google Scholar] [CrossRef]
- Lin, K.Y.; Wu, S.Y.; Hsu, Y.H.; Lin, N.S. MiR398-regulated antioxidants contribute to Bamboo mosaic virus accumulation and symptom manifestation. Plant Physiol. 2022, 188, 593–607. [Google Scholar] [CrossRef]
- Lian, S.; Cho, W.K.; Kim, S.M.; Choi, H.; Kim, K.H. Time-course small RNA profiling reveals rice miRNAs and their target genes in response to rice stripe virus infection. PLoS ONE 2016, 11, e0162319. [Google Scholar] [CrossRef]
- Shamloo-Dashtpagerdi, R.; Shahriari, A.G.; Tahmasebi, A.; Vetukuri, R.R. Potential role of the regulatory miR1119-MYC2 module in wheat (Triticum aestivum L.) drought tolerance. Front. Plant Sci. 2023, 14, 1161245. [Google Scholar] [CrossRef]
- Chen, Y.; Dong, J.; Bennetzen, J.L.; Zhong, M.; Yang, J.; Zhang, J.; Li, S.; Hao, X.; Zhang, Z.; Wang, X. Integrating transcriptome and microRNA analysis identifies genes and microRNAs for AHO-induced systemic acquired resistance in N. tabacum. Sci. Rep. 2017, 7, 12504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, T.; Li, N.; Tang, G.; Tang, J. MicroRNA166: Old Players and New Insights into Crop Agronomic Traits Improvement. Genes 2024, 15, 944. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Gong, W.; Boscá, S.; Tucker, M.; Vaucheret, H.; Laux, T. Dose-dependent AGO1-mediated inhibition of the miRNA165/166 pathway modulates stem cell maintenance in Arabidopsis shoot apical meristem. Plant Commun. 2020, 1, 100002. [Google Scholar] [CrossRef]
- Li, Q.; Shen, H.; Yuan, S.; Dai, X.; Yang, C. miRNAs and lncRNAs in tomato: Roles in biotic and abiotic stress responses. Front. Plant Sci. 2023, 13, 1094459. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, C.; Ding, G.; Jin, Y. Evolution of MIR159/319 microRNA genes and their post-transcriptional regulatory link to siRNA pathways. BMC Evol. Biol. 2011, 11, 112. [Google Scholar] [CrossRef]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Shi, S.; Liu, X.; Zhao, S.; Yang, Z.; et al. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol. Plant 2016, 9, 1302–1314. [Google Scholar] [CrossRef]
- Permar, V.; Singh, A.; Pandey, V.; Alatar, A.A.; Faisal, M.; Jain, R.K.; Praveen, S. Tospo viral infection instigates necrosis and premature senescence by microRNA controlled programmed cell death in Vigna unguiculata. Physiol. Mol. Plant Pathol. 2014, 88, 77–84. [Google Scholar] [CrossRef]
- Maghuly, F.; Ramkat, R.C.; Laimer, M. Virus versus host plant microRNAs: Who determines the outcome of the interaction? PLoS ONE 2014, 9, e98263. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Gao, Z.; Wang, F.; Xu, T.; Qi, M.; Liu, Y.; Li, T. MicroRNA162 regulates stomatal conductance in response to low night temperature stress via abscisic acid signaling pathway in tomato. Front. Plant Sci. 2023, 14, 1045112. [Google Scholar] [CrossRef]
- Barciszewska-Pacak, M.; Knop, K.; Jarmołowski, A.; Szweykowska-Kulińska, Z. Arabidopsis thaliana microRNA162 level is posttranscriptionally regulated via splicing and polyadenylation site selection. Acta Biochim. Pol. 2016, 63, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Parent, J.S.; Van Ex, F.; Wolff, P.; Martínez, G.; Köhler, C.; Martienssen, R.A. Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 2018, 50, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, D.; Zhang, C.; Tan, H.; Li, Y.; Lv, S.; Hou, X.; Cui, X. Genome-wide identification of turnip mosaic virus-responsive microRNAs in non-heading Chinese cabbage by high-throughput sequencing. Gene 2015, 571, 178–187. [Google Scholar] [CrossRef]
- Tousi, N.; Eini, O.; Ahmadvand, R.; Carra, A.; Miozzi, L.; Noris, E.; Accotto, G.P. In silico prediction of miRNAs targeting ToLCV and their regulation in susceptible and resistant tomato plants. Australas. Plant Pathol. 2017, 46, 379–386. [Google Scholar] [CrossRef]
- Zhang, P.; Li, S.; Chen, M. Characterization and function of circular RNAs in plants. Front. Mol. Biosci. 2020, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Ding, Y.; Xu, X.; Ye, C.Y.; Zhu, Q.H.; Guo, L.; Fan, L. Recent origination of circular RNAs in plants. New Phytol. 2022, 233, 515–525. [Google Scholar] [CrossRef]
- Zhao, W.; Chu, S.; Jiao, Y. Present scenario of circular RNAs (circRNAs) in plants. Front. Plant Sci. 2019, 10, 379. [Google Scholar] [CrossRef]
- Alkan, A.H.; Akgül, B. Endogenous miRNA Sponges. Methods Mol. Biol. 2022, 2257, 91–104. [Google Scholar] [CrossRef]
- Zhang, P.; Meng, X.; Chen, H.; Liu, Y.; Xue, J.; Zhou, Y.; Chen, M. PlantCircNet: A database for plant circRNA–miRNA–mRNA regulatory networks. Database 2017, 2017, bax089. [Google Scholar] [CrossRef]
- Zhang, P.; Meng, Y.; Li, Y.; Wang, Q.; Zhang, Y. GreenCircRNA: A database for plant circRNAs that act as miRNA decoys. Nucleic Acids Res. 2020, 48, D1189–D1196. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef]
- Roenhorst, J.W.; Boonham, N.; Winter, S.; Menzel, W.; van der Vlugt, R.A.A. The plant viruses and viroids database and collections of Q-bank. EPPO Bull. 2013, 43, 238–243. [Google Scholar] [CrossRef]
- Satish, D.; Mukherjee, S.K.; Gupta, D. PAmiRDB: A Web Resource for Plant miRNAs Targeting Viruses. Sci. Rep. 2019, 9, 4627. [Google Scholar] [CrossRef]
- Adams, M.J.; Antoniw, J.F. DPVweb: A Comprehensive Database of Plant and Fungal Virus Genes and Genomes. Nucleic Acids Res. 2006, 34, D382–D385. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-H.; Wang, Y.-X.; Xiao, J.; Jia, Y.-F.; Liu, F.; Wang, W.-X.; Wei, Q.; Lai, F.-X.; Fu, Q.; Wan, P.-J. Defense regulatory network associated with circRNA in rice in response to brown planthopper infestation. Plants 2024, 13, 373. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Bazin, J.; Webb, S.; Crespi, M.; Zubieta, C. CircRNAs in plants. Plant Sci. 2018, 274, 101–106. [Google Scholar] [CrossRef]
- Pan, X.; Xu, S.; Cao, G.; Chen, S.; Zhang, T.; Yang, B.B.; Zhou, G.; Yang, X. A novel peptide encoded by a rice circular RNA confers broad-spectrum disease resistance in rice plants. New Phytol. 2025, 246, 689–701. [Google Scholar] [CrossRef]
- Ghorbani, A.; Izadpanah, K.; Peters, J.R.; Dietzgen, R.G.; Mitter, N. Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant Sci. 2018, 274, 402–409. [Google Scholar] [CrossRef]
- Hashim, G.M.; Haight, T.; Chen, X.; Zovoilis, A.; Venkataraman, S. Profiling Plant circRNAs Provides Insights into the Expression of Plant Genes Involved in Viral Infection. Life 2025, 15, 1143. [Google Scholar] [CrossRef]
- Chen, G.; Cui, J.; Wang, L.; Zhu, Y.; Lu, Z.; Jin, B. Genome-wide identification of circular RNAs in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1678. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.; Wu, W.; Dong, Y.; Wang, M.; Yi, D.; Zhou, Y.; Xu, Q. Systematic identification and functional analysis of circular RNAs during rice black-streaked dwarf virus infection in the Laodelphax striatellus (Fallén) midgut. Front. Microbiol. 2020, 11, 588009. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ma, Y.; Guo, T.; Li, G. Identification, biogenesis, function, and mechanism of action of circular RNAs in plants. Plant Commun. 2023, 4, 100430. [Google Scholar] [CrossRef]
- Wojciechowska, N.; Michalak, K.M.; Bagniewska-Zadworna, A. Autophagy-an underestimated coordinator of construction and destruction during plant root ontogeny. Planta 2021, 254, 15. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J.; Kang, Y. Novel exploitation of autophagy by tombusviruses. Virology 2024, 603, 110363. [Google Scholar] [CrossRef]
- Mao, J.; Lin, E.; He, L.; Yu, J.; Tan, P.; Zhou, Y. Autophagy and viral infection. In Autophagy Regulation of Innate Immunity; Springer: Singapore, 2019; pp. 55–78. [Google Scholar] [CrossRef]
- Li, F.; Zhang, C.; Tang, Z.; Zhang, L.; Dai, Z.; Lyu, S.; Li, Y.; Hou, X.; Bernards, M.; Wang, A. A plant RNA virus activates selective autophagy in a UPR-dependent manner to promote virus infection. New Phytol. 2020, 228, 622–639. [Google Scholar] [CrossRef] [PubMed]
- Šečić, E.; Kogel, K.H.; Ladera-Carmona, M.J. Biotic stress-associated microRNA families in plants. J. Plant Physiol. 2021, 263, 153451. [Google Scholar] [CrossRef]
- Černý, M.; Habanova, H.; Berka, M.; Luklova, M.; Brzobohatý, B. Hydrogen peroxide: Its role in plant biology and crosstalk with signalling networks. Int. J. Mol. Sci. 2018, 19, 2812. [Google Scholar] [CrossRef]
- Stephani, M.; Dagdas, Y. Plant selective autophagy-still an uncharted territory with a lot of hidden gems. J. Mol. Biol. 2020, 432, 63–79. [Google Scholar] [CrossRef]
- Wu, M.Y.; Li, Z.W.; Lu, J.H. Molecular modulators and receptors of selective autophagy: Disease implication and identification strategies. Int. J. Biol. Sci. 2024, 20, 751. [Google Scholar] [CrossRef]
- Paludan, S.R.; Pradeu, T.; Masters, S.L.; Mogensen, T.H. Constitutive immune mechanisms: Mediators of host defence and immune regulation. Nat. Rev. Immunol. 2021, 21, 137–150. [Google Scholar] [CrossRef]
- Jiang, L.; Lu, Y.; Zheng, X.; Yang, X.; Chen, Y.; Zhang, T.; Zhao, X.; Wang, S.; Zhao, X.; Song, X.; et al. The plant protein NbP3IP directs degradation of Rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy-related protein NbATG8. New Phytol. 2021, 229, 1036–1051. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Xie, X.; Yue, N.; Li, J.; Wang, X.B.; Han, C.; Yu, J.; Liu, Y.; Li, D. Barley stripe mosaic virus γb Protein Subverts Autophagy to Promote Viral Infection by Disrupting the ATG7-ATG8 Interaction. Plant Cell 2018, 30, 1582–1595. [Google Scholar] [CrossRef]
- Huang, X.; Chen, S.; Yang, X.; Yang, X.; Zhang, T.; Zhou, G. Friend or Enemy: A Dual Role of Autophagy in Plant Virus Infection. Front. Microbiol. 2020, 11, 736. [Google Scholar] [CrossRef]
- Wang, Y.; Gong, Q.; Liu, Y. Multifaceted roles of autophagy in plant-virus-insect interactions. New Phytol. 2025, 248, 1166–1170. [Google Scholar] [CrossRef]
- Meng, Y.; Ismayil, A.; Liu, Y. Autophagy in plant-virus interactions. Annu. Rev. Virol. 2020, 7, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.H.; Peng, X.B.; Sun, M.X. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.B.; Montenegro Alonso, A.P.; Chisholm, J.; Xiao, H.; Sanfaçon, H. Transcriptomic changes associated with infection of Nicotiana benthamiana plants with tomato ringspot virus (genus Nepovirus) during the acute symptomatic stage and after symptom recovery. PLoS ONE 2025, 20, e0328517. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.J.; Ho, T.H. An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol. 2008, 147, 1710–1722. [Google Scholar] [CrossRef]
- Wan, J.; Meng, S.; Wang, Q.; Zhao, J.; Qiu, X.; Wang, L.; Li, J.; Lin, Y.; Mu, L.; Dang, K.; et al. Suppression of microRNA168 enhances salt tolerance in rice (Oryza sativa L.). BMC Plant Biol. 2022, 22, 563. [Google Scholar] [CrossRef]
- Pertermann, R.; Tamilarasan, S.; Gursinsky, T.; Gambino, G.; Schuck, J.; Weinholdt, C.; Lilie, H.; Grosse, I.; Golbik, R.P.; Pantaleo, V.; et al. A viral suppressor modulates the plant immune response early in infection by regulating microRNA activity. mBio 2018, 9, e00419-18. [Google Scholar] [CrossRef] [PubMed]
- Iksat, N.; Masalimov, Z.; Omarov, R. Plant virus resistance biotechnological approaches: From genes to the CRISPR/Cas gene editing system. J. Water Land Dev. 2023, 57, 147–158. [Google Scholar] [CrossRef]
- Mahas, A.; Aman, R.; Mahfouz, M. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 2023, 20, 263. [Google Scholar] [CrossRef]
- Duan, C.G.; Wang, C.H.; Fang, R.X.; Guo, H.S. Artificial MicroRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 2008, 82, 11084–11095. [Google Scholar] [CrossRef]
- Kis, A.; Tholt, G.; Ivanics, M.; Várallyay, É.; Jenes, B.; Havelda, Z. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol. Plant Pathol. 2016, 17, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Gasparis, S.; Kała, M.; Przyborowski, M.; Orczyk, W.; Nadolska-Orczyk, A. Artificial MicroRNA-Based Specific Gene Silencing of Grain Hardness Genes in Polyploid Cereals Appeared to Be Not Stable Over Transgenic Plant Generations. Front. Plant Sci. 2017, 7, 2017. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, D.; Chen, S.L.; Gong, B.Q.; Guo, Y.; Xu, L.; Zhang, X.N.; Li, J.F. Engineering Artificial MicroRNAs for Multiplex Gene Silencing and Simplified Transgenic Screen. Plant Physiol. 2018, 178, 989–1001. [Google Scholar] [CrossRef]
- Mickiewicz, A.; Rybarczyk, A.; Sarzynska, J.; Figlerowicz, M.; Blazewicz, J. AmiRNA Designer—New method of artificial miRNA design. Acta Biochim. Pol. 2016, 63, 71–77. [Google Scholar] [CrossRef]
- Fahim, M.; Larkin, P.J. Designing effective amiRNA and multimeric amiRNA against plant viruses. Methods Mol. Biol. 2013, 942, 357–377. [Google Scholar] [CrossRef]

| miRNA | Targets | Related Mechanism Against Viruses | Viruses |
|---|---|---|---|
| miR168 | AGO1 | Maintains proper AGO1 levels to ensure stable antiviral RNA silencing activity and efficient degradation of viral RNAs | TBSV, TNV, CMV (Tombusvirus, Tobamovirus, Cucumovirus) |
| miR159 | R2R3 and MYB transcription factors | Regulates MYB transcription factors involved in gibberellin and abscisic acid (ABA) signaling pathways, thereby suppressing viral gene expression and limiting replication | CMV, TYLCV (Cucumovirus, Geminivirus) |
| miR393 | TIR1, AFB2, AFB3 | Suppresses expression of F-box proteins (TIR1, AFB2, AFB3), blocking the auxin signaling pathway and triggering systemic acquired resistance (SAR) through SA-dependent signal transduction | CMV, TuMV (Cucumovirus, Potyvirus) |
| miR166 | Class III Homeodomain- Leucine Zipper (HD-ZIP III) transcription factors | Regulates HD-ZIP III transcription factors associated with hormonal and vascular development; modulation of their expression enhances SA-dependent SAR signaling and promotes PR-gene expression under viral infection | CMV, BCTV (Cucumovirus, Curtovirus) |
| miR160 | ARF10, ARF16, ARF17 | Modulates ARF10/16/17 to balance auxin-mediated growth and SA-mediated defense responses, thereby enhancing immune signaling and restricting viral replication | TuMV, CMV (Potyvirus, Cucumovirus) |
| miR398 | CSD1, CSD2, CSD3 | Regulates antioxidant enzymes CSD1 and CSD2 to control reactive oxygen species (ROS) levels and alleviate virus-induced oxidative stress, stabilizing plant defense responses | CMV, TMV (Cucumovirus, Tobamoirus) |
| miR172 | APETALA2-like transcription factors | Suppresses APETALA2-like transcription factors, modulating ethylene-response factor (ERF) signaling to reduce viral load and enhance plant tolerance | TuMV, CMV (Potyvirus, Cucumovirus) |
| miR162 | DCL1 | Regulates DCL1 expression to prevent overaccumulation and maintain balanced RNA-silencing activity against viral RNA genomes | CMV. TMV (Cucumovirus, Tobamoirus) |
| Database | Focus | Main Features | Example Use |
|---|---|---|---|
| PlantCircNet (http://bis.zju.edu.cn/plantcircnet/index.php (accessed on 5 November 2025)) | ciRNA-miRNA-mRNA network visualization | Visualization of circRNA-miRNA regulatory interactions | Identify miRNA sponges in plants |
| Q-bank Plant Viruses & Viroids (https://qbank.eppo.int/ (accessed on 6 November 2025)) | Prioritizes viruses and viroids that are subject to plant health regulations | Provides taxonomic, biological, and regulatory information on plant viruses and viroids, including curated DNA barcodes and sequence data | Obtain DNA barcodes and validated protocols for rapid and accurate identification |
| PAmiRDB (https://bioinfo.icgeb.res.in/pamirdb/index.html (accessed on 10 November 2025)) | miRNAs and their predicted targets in virus genomes | Contains over 2600 plant miRNAs and their predicted targets across approximately 500 viral species | Identify plant miRNAs predicted to target the virus’s genes |
| DPVweb (http://www.dpvweb.net/ (accessed on 10 November 2025)) | Gives a curated information on complete or nearly complete sequences of plant, fungal, and protozoan viruses, viroids, and satellites, currently covering around 9000 entries | Each entry includes start and end positions of genes and non-coding regions, checked for accuracy, with standardized gene/protein nomenclature within genera and families | Retrieve all annotated gene and protein sequences for that virus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iksat, N.; Baikarayev, Z.; Shevchenko, O.; Zhanassova, K.; Bekturova, A.; Zhangazin, S.; Masalimov, Z. Integrative Roles of miRNAs and circRNAs in Plant Antiviral Gene Regulation and Autophagy. Plants 2025, 14, 3541. https://doi.org/10.3390/plants14223541
Iksat N, Baikarayev Z, Shevchenko O, Zhanassova K, Bekturova A, Zhangazin S, Masalimov Z. Integrative Roles of miRNAs and circRNAs in Plant Antiviral Gene Regulation and Autophagy. Plants. 2025; 14(22):3541. https://doi.org/10.3390/plants14223541
Chicago/Turabian StyleIksat, Nurgul, Zhaksat Baikarayev, Oleksiy Shevchenko, Kuralay Zhanassova, Assemgul Bekturova, Sayan Zhangazin, and Zhaksylyk Masalimov. 2025. "Integrative Roles of miRNAs and circRNAs in Plant Antiviral Gene Regulation and Autophagy" Plants 14, no. 22: 3541. https://doi.org/10.3390/plants14223541
APA StyleIksat, N., Baikarayev, Z., Shevchenko, O., Zhanassova, K., Bekturova, A., Zhangazin, S., & Masalimov, Z. (2025). Integrative Roles of miRNAs and circRNAs in Plant Antiviral Gene Regulation and Autophagy. Plants, 14(22), 3541. https://doi.org/10.3390/plants14223541







