Ectopic Over-Expression of BjuAGL9-2 Promotes Flowering and Pale-Yellow Phenotype in Arabidopsis
Abstract
1. Introduction
2. Results
2.1. BjuAGL9-2 Functions in Promoting Plant Flowering and Pale-Yellow Phenotype in A. thaliana
2.2. BjuAGL9-2 Interaction Protein Screening
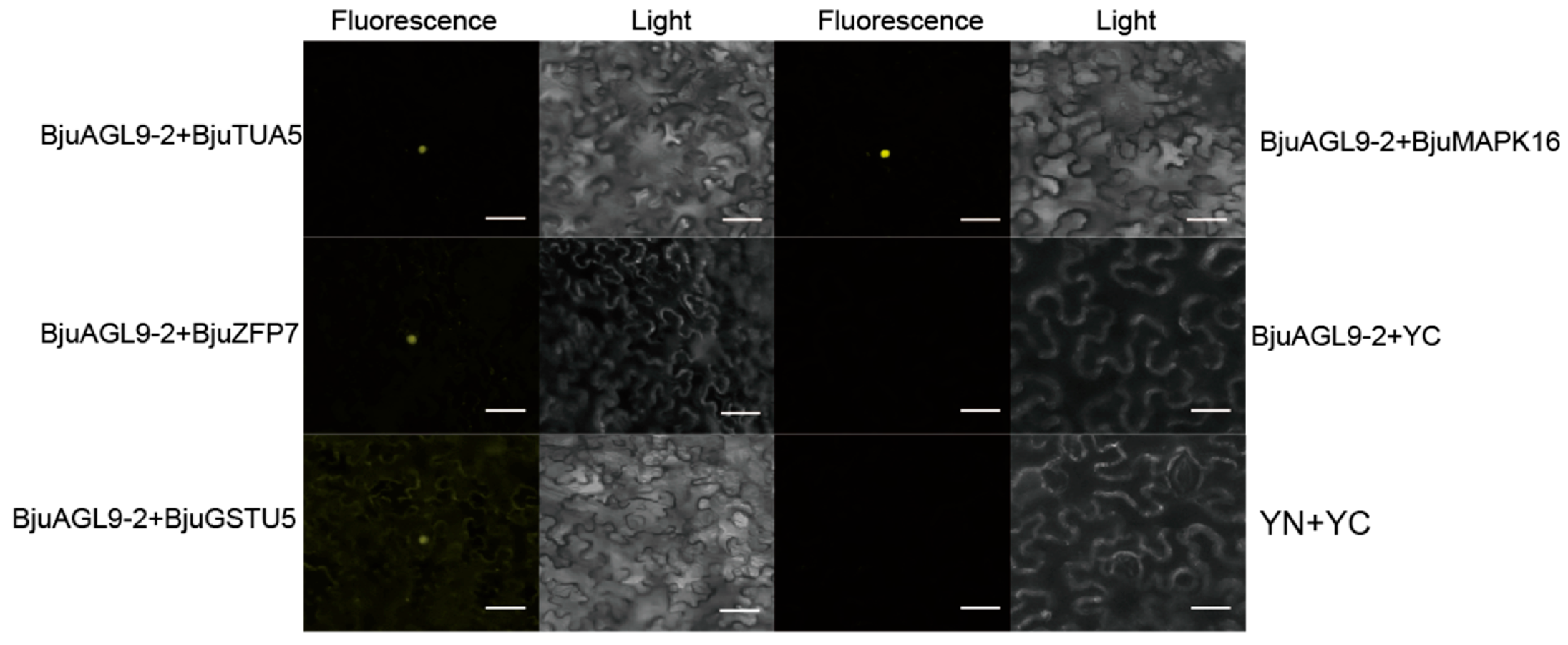
2.3. Interactive Analyses Between BjuAGL9-2 and Screened Proteins
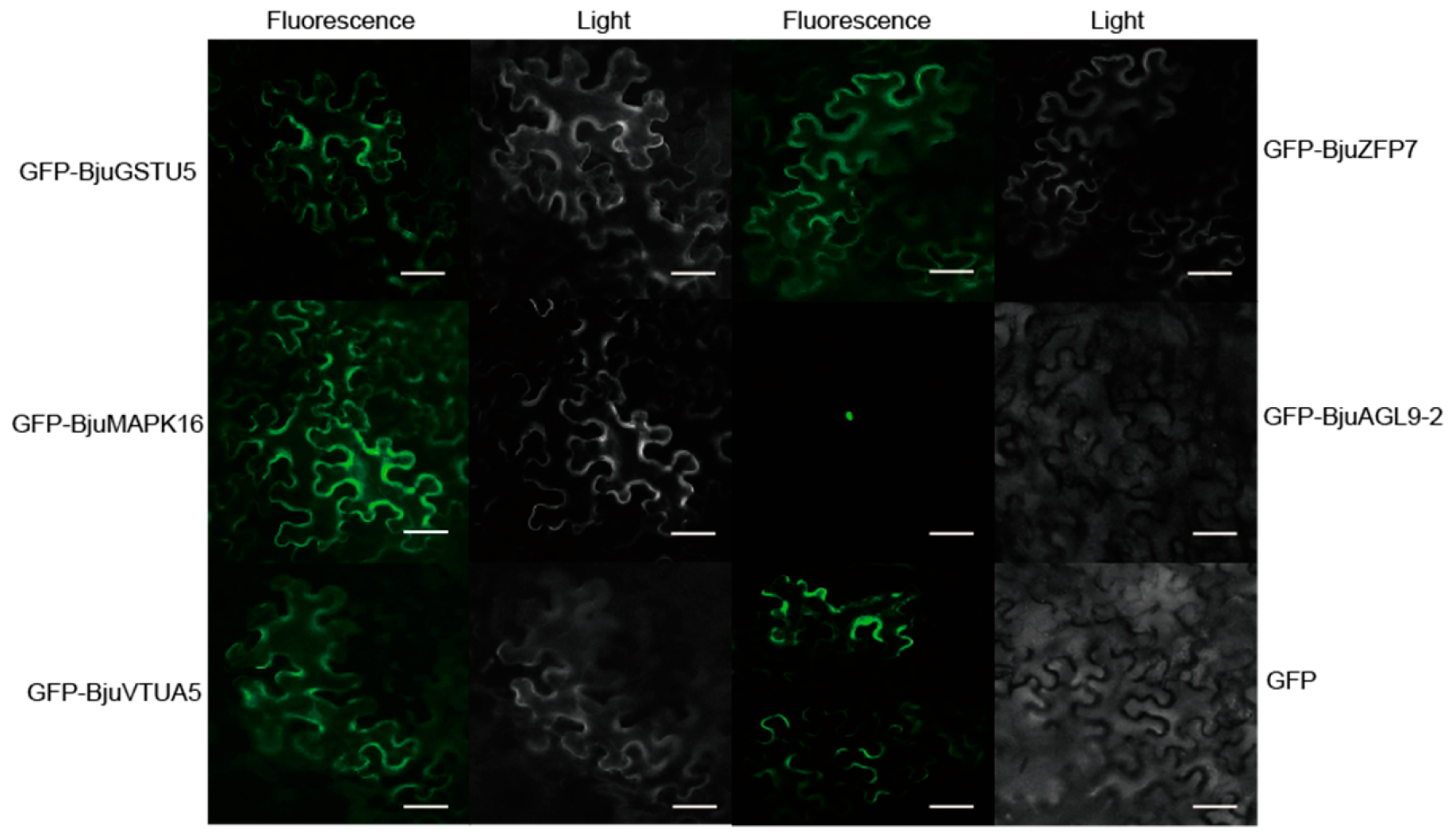
2.4. Sub-Cellular Location Assays
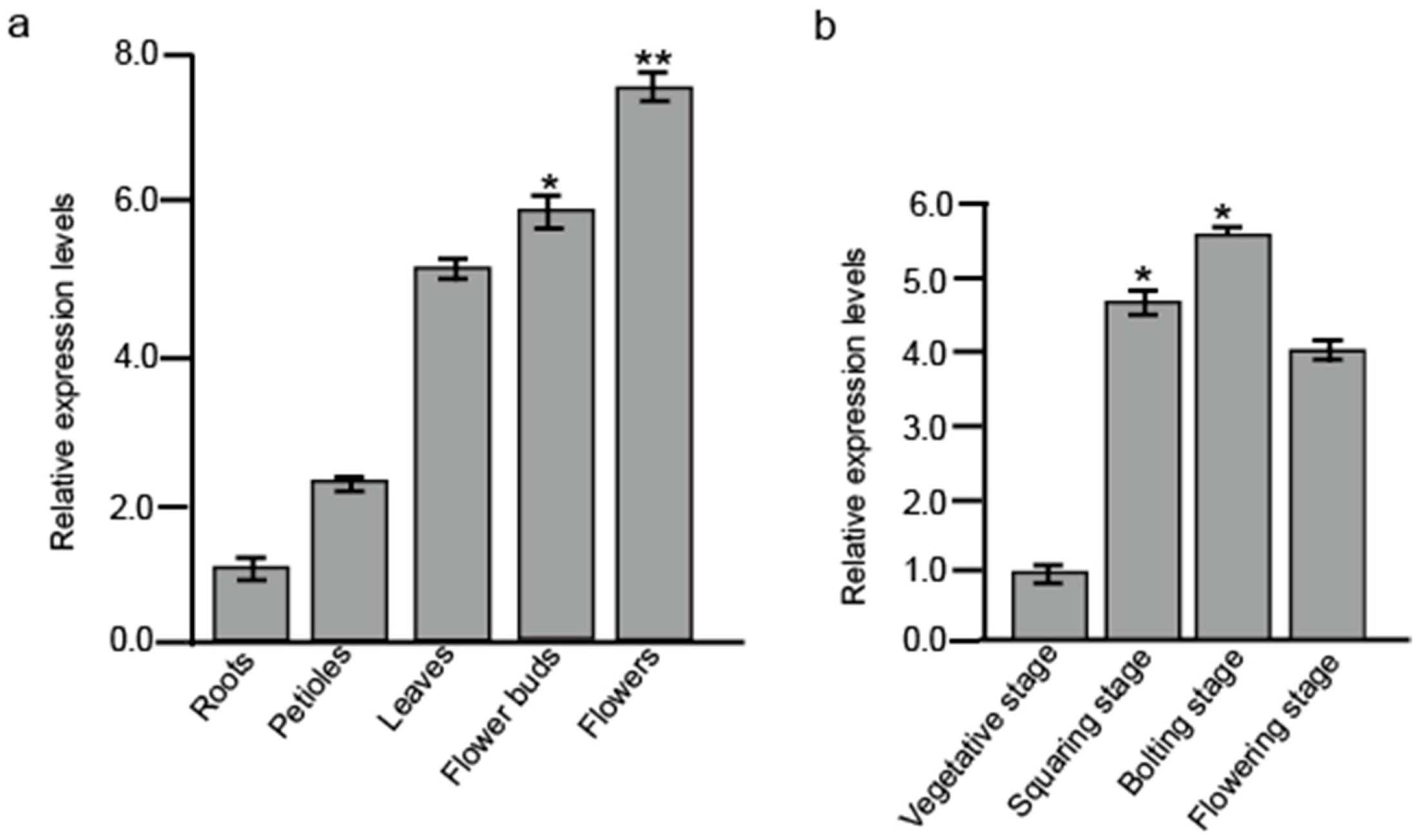
2.5. Interactive Protein-Encoding Gene Expression Analysis in Different Tissues and at Different Development Stages in B. juncea
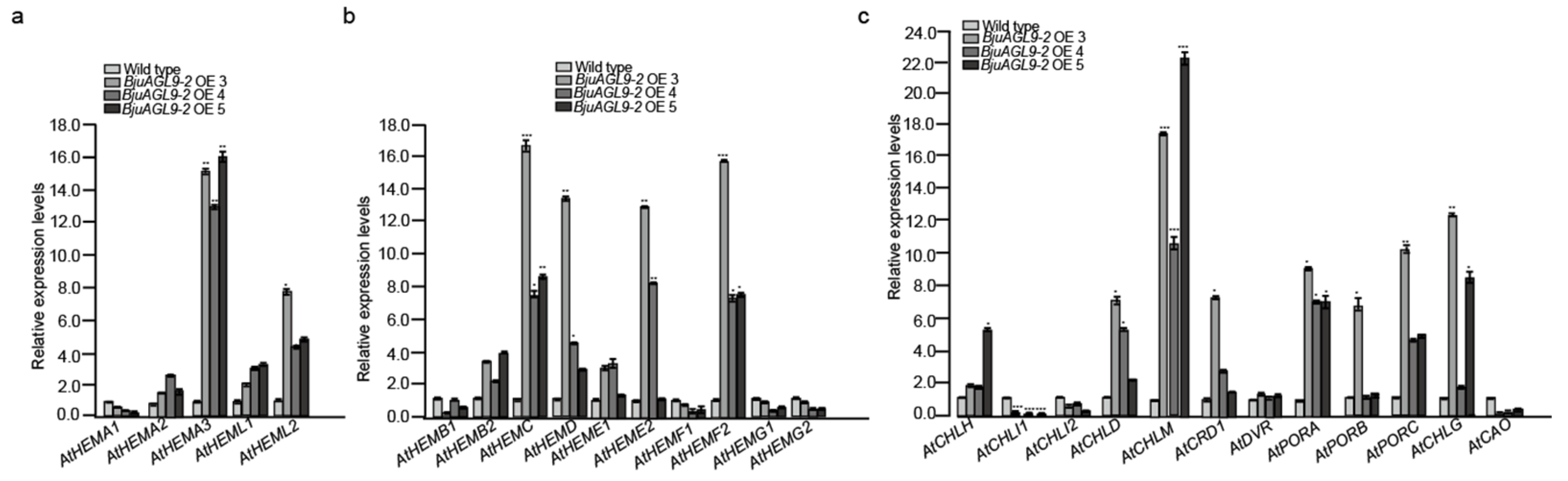
2.6. Chlorophyll Synthesis Gene Expression Levels in Arabidopsis OE Lines
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. BjuAGL9-2 Over-Expression Line Screening and Phenotype Observation
4.3. BjuAGL9-2 Interaction Protein Screening
4.4. The Interaction Analysis Between BjuAGL9-2 and Screened Proteins
4.5. Bimolecular Florescence (BiFC) Analysis and Confocal Microscopy
4.6. Sub-Cellular Localization Assays
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fornara, F.; De, M.A.; Coupland, G. Snapshot: Control of flowering in Arabidopsis. Cell 2010, 141, 5502. [Google Scholar] [CrossRef]
- Méndez-Vigo, B.; Martínez-Zapater, J.M.; Alonso-Blanco, C. The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. Plos Genet. 2013, 9, e1003289. [Google Scholar] [CrossRef]
- Weigel, D. Natural variation in Arabidopsis: From molecular genetics to ecological genomics. Plant Physiol. 2011, 158, 2–22. [Google Scholar] [CrossRef]
- Andres, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Benlloch, R.; Berbel, A.; Serrano-Mislata, A.; Madueno, F. Floral initiation and inflorescence architecture: A comparative view. Ann. Bot. 2007, 100, 659–676. [Google Scholar] [CrossRef]
- Parcy, F. Flowering: A time for integration. Int. J. Dev. Biol. 2005, 49, 585–593. [Google Scholar] [CrossRef]
- Serrano-Mislata, A.; Goslin, K.; Zheng, B.B.; Rae, L.; Wellmer, F.; Graciet, E.; Madueno, F. Regulatory interplay between leafy, apetala 1/cauliflower and terminal flower 1: New insights into an old relationship. Plant Signal Behav. 2017, 12, e1370164. [Google Scholar] [CrossRef]
- Kater, M.M.; Dreni, L.; Colombo, L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 2006, 57, 3433–3444. [Google Scholar] [CrossRef]
- Tanabe, Y.; Hasebe, M.; Sekimoto, H.; Nishiyama, T.; Kitani, M.; Henschel, K.; Munster, T.; Theissen, G.; Nozaki, H.; Ito, M. Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc. Natl. Acad. Sci. USA 2005, 102, 2436–2441. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Parenicova, L.; Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.K.; Song, X.M.; Liu, T.K.; Huang, Z.N.; Ren, J.; Hou, X.L.; Li, Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mollecular Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Daher, H.; Galien, A.; Hugouvieux, V.; Zubieta, C. Structural basis for plant MADS transcription factor oligomerization. Comput. Struct. Biotechnol. J. 2019, 17, 946–953. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Munster, T.; Theiben, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Wang, M.; Meyerowitz, E.M. DNA-binding properties of Arabidopsis MADS domain homeotic proteins apetala1, apetala3, Ppistillata and agamous. Nucleic Acids Res. 1996, 24, 3134–3141. [Google Scholar] [CrossRef]
- Hayes, T.E.; Sengupta, P.; Cochran, B.H. The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 1988, 2, 1713–1722. [Google Scholar] [CrossRef]
- Lai, X.L.; Vega-Leon, R.; Hugouvieux, V.; Blanc-Mathieu, R.; Wal, F.; Lucas, J.; Silva, C.S.; Jourdain, A.; Muino, J.M.; Nanao, M.H.; et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021, 12, 4760. [Google Scholar] [CrossRef]
- Davies, B.; Egea-Cortines, M.; Silva, E.A.; Saedler, H.; Sommer, H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 1996, 15, 4330–4343. [Google Scholar] [CrossRef]
- Fan, H.Y.; Hu, Y.; Tudor, M.; Ma, H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 1997, 12, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.H.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Boer, B.G.; Montagu, M.V.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene apetala2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Du, X.L. Leafy cotyledons: Connecting different stages of plant development. Front. Plant Sci. 2022, 13, 916831. [Google Scholar] [CrossRef]
- Krizek, B.A.; Meyerowitz, E.M. The Arabidopsis homeotic genes apetala3 and pistillata are sufficient to provide the B class organ identity function. Development 1996, 122, 11–22. [Google Scholar] [CrossRef]
- Pelayo, M.A.; Yamaguchi, N.; Ito, T. One factor, many systems: The floral homeotic protein agamous and its epigenetic regulatory mechanisms. Curr. Opin. Plant Biol. 2021, 61, 102009. [Google Scholar] [CrossRef]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Zahn, L.M.; Kong, H.Z.; Leebens-Mack, J.H.; Kim, S.; Soltis, P.S.; Landherr, L.L.; Soltis, D.E.; DePamphilis, C.W.; Ma, H. The evolution of the sepallata subfamily of MADS-box genes. Genetics 2005, 169, 2209–2223. [Google Scholar] [CrossRef]
- Malcomber, S.T.; Kellogg, E.A. Sepallata gene diversification: Brave new whorls. Trends Plant Sci. 2005, 10, 435–472. [Google Scholar] [CrossRef]
- Mandel, M.A.; Yanofsky, M.F. The Arabidopsis AGL9 MADS box gene is expressed in young flower primordial. Sex Plant Repord. 1998, 11, 22–28. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require sepallata MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 21, 1935–1940. [Google Scholar] [CrossRef]
- Carpenter, J.L.; Ploense, S.E.; Snustad, D.P.; Silflow, C.D. Preferential expression of an alpha-tubulin gene of Arabidopsis in pollen. Plant Cell 1992, 4, 557–571. [Google Scholar] [CrossRef]
- Oakley, R.V.; Wang, Y.S.; Ramakrishna, W.; Harding, S.A.; Tsai, C.J. Differential expansion and expression of alpha- and beta-tubulin gene families in Populus. Plant Physiol. 2007, 145, 961–973. [Google Scholar] [CrossRef]
- Hu, T.Q.; Li, X.H.; Du, L.R.; Manuela, D.; Xu, M.L. Leafy and apetala1 down-regulated zinc finger protein 1 and 8 to release their repression on class B and C floral homeotic genes. Proc. Natl. Acad. Sci. USA 2023, 120, e2221181120. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, J.; Li, H.; Ding, Y.; Wu, W.; Qin, R.; Ni, J.; Wei, P.; Yang, J. OsGSTU5 and OsGSTU37 encoding glutathione reductases are required for cadmium tolerance in rice. Int. J. Environ. Sci. Technol. 2023, 20, 10253–10260. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, L.; An, H.L.; Wu, C.G.; Guo, X.Q. GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 2011, 12, 22. [Google Scholar] [CrossRef]
- Zhao, M.H.; Li, X.; Zhang, X.X.; Zhang, H.; Zhao, X.Y. Mutation mechanism of leaf color in plants. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Masuda, T.; Ohta, H.; Shioi, Y.; Tsuji, H.; Takamiya, K. Stimulation of glutamy 1-tRNA reductase activity by benzyladenine in greening cucumber cotyledons. Plant Cell Physiol. 1995, 36, 1237–1243. [Google Scholar]
- Nagai, S.; Koide, M.; Takahashi, S.; Kikuta, A.; Aono, M.; Sasaki-Sekimoto, Y.; Ohta, H.; Takamiya, K.; Masuda, T. Induction of isoforma of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol. 2007, 144, 1039–1051. [Google Scholar] [CrossRef]
- Sinha, N.; Eirich, J.; Finkemeier, I.; Grimm, B. Glutamate 1-semialdehyde aminotransferase is connected to GluTR by GluTR-binding protein and contributes to the rate-limiting step of 5-aminolevulinic acid synthesis. Plant Cell 2022, 34, 4623–4640. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Q.; Tang, W.J.; Ma, T.T.; Niu, D.; Jin, J.B.; Wang, H.Y.; Lin, R.C. A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 2016, 58, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Quesada, V.; Sarmiento-Manus, R.; Gonzalez-Bayon, R.; Hricova, A.; Ponce, M.R.; Micol, J.L. Prophobilinogen deaminase deficiency alters vegetative and reproductive development and causes lesions in Arabidopsis. PLoS ONE 2013, 8, e53378. [Google Scholar] [CrossRef]
- Aizencang, G.I.; Bishop, D.F.; Forrest, D.; Astrin, K.H.; Desnick, R.J. Uroporphyrinogen III synthase: An alternative promoter controls erythroid-specific expression in the murine gene. J. Biol. Chem. 2000, 275, 2295–2304. [Google Scholar] [CrossRef]
- Shalygo, N.V.; Mock, H.P.; Averina, N.G.; Grimm, B. Photodynamic action of uroporphyrin and protochlorophyllide in greening barley leaves treated with cesium chloride. J. Photochem. Photobiol. B Biol. 1998, 42, 151–158. [Google Scholar] [CrossRef]
- Kiba, T.; Henriques, R.; Sakakibara, H.; Chua, N.H. Targeted degradation of pseudo-response regulator 5 by an SCFZTL complex regulated clock function and photomorphogenesis in Arabidopsis thalianan. Plant Cell 2007, 19, 2516–2530. [Google Scholar] [CrossRef]
- Narita, S.; Tanaka, R.; Ito, T.; Okada, K.; Taketani, S.; Inokuchi, H. Molecular cloning and characterization of a cDNA that encodes protoporphyrinogen oxidase of Arabidopsis thaliana. Gene 1996, 182, 169–175. [Google Scholar] [CrossRef]
- Molina, A.; Volrath, S.; Guyer, D.; Maleck, K.; Ryals, J.; Ward, E. Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J. 1999, 17, 667–678. [Google Scholar] [CrossRef]
- Kruse, E.; Mock, H.P.; Grimm, B. Isolation and characterization of tobacco (Nicotiana tabacum) cDNA clones encoding proteins involved in magnesium chelation into protoporphyrin IX. Plant Mol. Biol. 1997, 35, 1053–1056. [Google Scholar] [CrossRef] [PubMed]
- Hiriart, J.B.; Lehto, K.; Tyystjarvi, E.; Junttila, T.; Aro, E.M. Suppression of a key gene involved in chlorophyll biosynthesis by means of virus-inducing gene silencing. Plant Mol. Biol. 2002, 50, 213–224. [Google Scholar] [CrossRef]
- Matsumoto, F.; Obayashi, T.; Sasaki, S.Y.; Ohta, H.; Takamiya, K.; Masuda, T. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 2004, 135, 2379–2391. [Google Scholar] [CrossRef] [PubMed]
- Pontier, D.; Albrieux, C.; Joyard, J.; Lagrange, T.; Block, M.A. Knockout of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. J. Biol. Chem. 2007, 282, 2297–2304. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, N.; Brusslan, J.A.; Larkin, R.; Nagatani, A.; Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 2001, 98, 2053–2058. [Google Scholar] [CrossRef]
- Bang, W.Y.; Jeong, I.S.; Kim, D.W.; Im, C.H.; Ji, C.; Hwang, S.M.; Kim, S.W.; Son, Y.S.J.J.; Shiina, T.; Bahk, J.D. Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant Cell Physiol. 2008, 49, 1350–1363. [Google Scholar] [CrossRef]
- Xue, Y.J.; Dong, H.X.; Huang, H.R.; Li, S.P.; Shan, X.H.; Li, H.; Liu, H.K.; Xia, D.; Su, S.Z.; Yuan, Y.P. Mutation in Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase gene ZmCRD1 causes chlorophyll-deficiency in maize. Front. Plant Sci. 2022, 13, 912215. [Google Scholar] [CrossRef]
- Nakanishi, H.; Nozue, H.; Suzuki, K.; Kaneko, Y.; Taguchi, G.; Hayashida, N. Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol. 2005, 46, 467–473. [Google Scholar] [CrossRef]
- Paddock, T.N.; Mason, M.E.; Lima, D.F.; Armstrong, G.A. Arabidopsis protochlorophyllide oxidoreductase A (PORA) restores bulk chlorophyll synthesis and normal development to a porbporc double mutant. Plant Mol. Biol. 2010, 72, 445–457. [Google Scholar] [CrossRef]
- Frick, G.; Su, Q.; Apel, K.; Armstrong, G.A. An Arabidopsis porbporc double mutant lacking light dependent NADPH: Protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J. 2003, 35, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.T.; Gu, D.C.; Lie, Z.Y.; Yang, Y.Y.; Lu, L.X.; Dai, G.Y.; Peng, T.; Deng, L.; Zheng, F.; Liu, X.C. Regulation of chlorphyll biosynthesis by light-dependent acetylation of NADPH: protochlorophyll oxidoreductase A in Arabidopsis. Plant Sci. 2023, 330, 111641. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Koshino, Y.; Sawa, S.; Ishiguro, S.; Okada, K.; Tanaka, A. Overexpression of chlorophyllide a oxygenase (CAO) enlarges the antenna size of photosystem II in Arabidopsis thaliana. Plant J. 2001, 26, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, C.; Bartsch, S.; Eggink, L.L.; Hoober, K.; Brusslan, J.; Andrsde-Paz, R.; Monnet, J.; Reinbothe, S. A role for chlorophyllide a oxygenase in the regulated import and stabilization of light-harvesting chlorophyll a/b proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 4777–4782. [Google Scholar] [CrossRef]
- Lian, X.P.; Zhang, H.C.; Zeng, J.; Wang, Y.K.; Bai, X.J.; Liu, Q.Y.; Zuo, T.H.; Zhang, Y.Z.; Converse, R.; Zhu, L.Q. C2H2-like zinc finger protein 1 cause pollen and pistil malformation through the auxin pathway. Plant Growth Regul. 2020, 90, 505–518. [Google Scholar] [CrossRef]

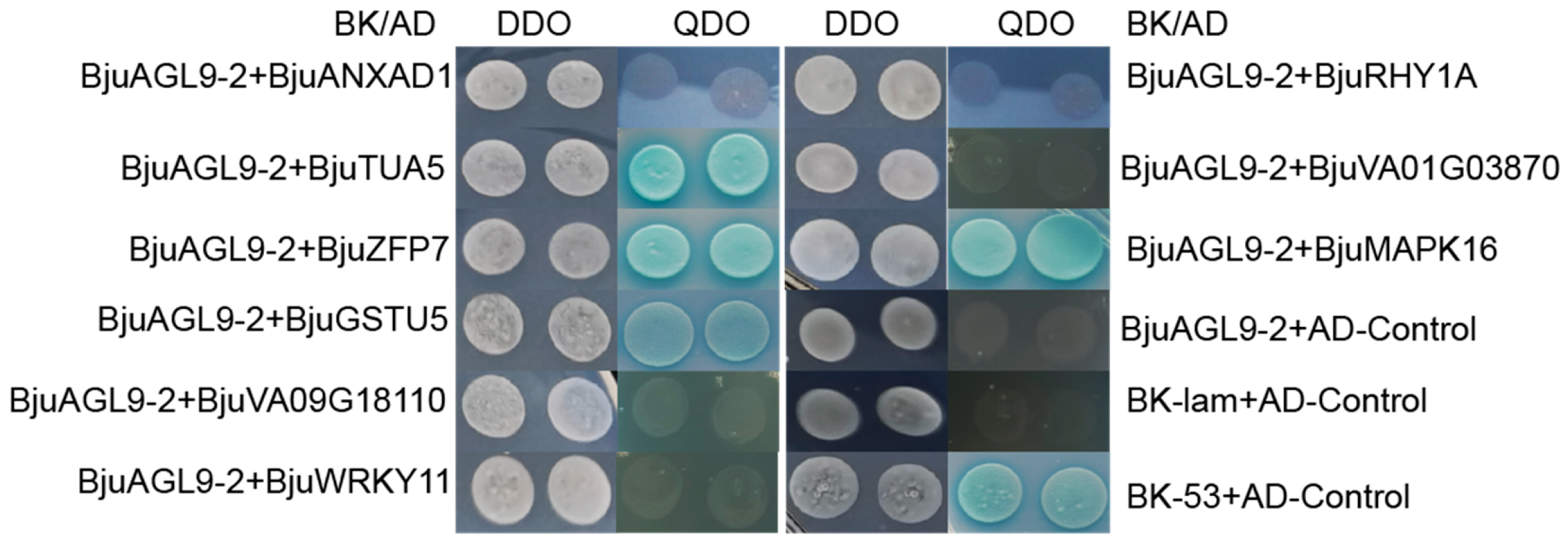

| Gene ID | Gene Names | Similarity | Function |
|---|---|---|---|
| BjuVA05G23270 | Annexin D1 | 100% | Abiotic stress, seedling development |
| BjuVA10G20230 | Tubulin alpha-5 chain (TUA5) | 99.19% | Pollen development |
| BjuVA08G27100 | Zinc finger protein 7 (ZFP7) | 97.96% | Trichome development |
| BjuVA03G25670 | Glutathione S-transferase U5 (GSTU5) | 100% | Defense response |
| BjuVA09G18110 | Uncharacterized protein | 100% | Unknown |
| BjuVA03G59720 | WRKY transcription factor 11 | 98.17% | Abiotic stress responses |
| BjuVA08G03470 | E3 ubiquitin-protein ligase RHY1A | 100% | Protein degradation |
| BjuVA01G03870 | Uncharacterized protein | 93.42% | Unknown |
| BjuVA10G20860 | Mitogen-activated protein kinase 16 (MAPK16) | 100% | Defense response |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, G.; Ren, K.; He, R.; Mo, R.; Zeng, J.; Sui, M. Ectopic Over-Expression of BjuAGL9-2 Promotes Flowering and Pale-Yellow Phenotype in Arabidopsis. Plants 2025, 14, 3502. https://doi.org/10.3390/plants14223502
Han G, Ren K, He R, Mo R, Zeng J, Sui M. Ectopic Over-Expression of BjuAGL9-2 Promotes Flowering and Pale-Yellow Phenotype in Arabidopsis. Plants. 2025; 14(22):3502. https://doi.org/10.3390/plants14223502
Chicago/Turabian StyleHan, Guoqiang, Keran Ren, Rongyan He, Ruirui Mo, Jing Zeng, and Mingming Sui. 2025. "Ectopic Over-Expression of BjuAGL9-2 Promotes Flowering and Pale-Yellow Phenotype in Arabidopsis" Plants 14, no. 22: 3502. https://doi.org/10.3390/plants14223502
APA StyleHan, G., Ren, K., He, R., Mo, R., Zeng, J., & Sui, M. (2025). Ectopic Over-Expression of BjuAGL9-2 Promotes Flowering and Pale-Yellow Phenotype in Arabidopsis. Plants, 14(22), 3502. https://doi.org/10.3390/plants14223502






