Molecular Phylogenetics and Morphological Analyses Support Dolichopoda, a New Neotropical Genus of Marantaceae (Zingiberales)
Abstract
1. Introduction
2. Results
2.1. Phylogenetic Relationships
2.2. Ancestral State Evolution
3. Discussion
Taxonomic Treatment
- New Combination
| 1. Leaves homotropic..............................................................................................................................2 |
| 1’. Leaves antitropic................................................................................................................................3 |
| 2. Rachis slightly flexuous with obvious scars.........................................................................Saranthe |
| 2’. Rachis absent to straight without obvious scars.................................................................Maranta |
| 3. Florescence rachis strongly flexuous................................................................................................4 |
| 3’. Florescence rachis straight................................................................................................................5 |
| 4. Cymule peduncle ≥ 2 cm, unequal outer staminodes...................................................Dolichopoda |
| 4’. Cymule peduncle ≤ 1.8 cm, outer staminodes equal if two..........................................Stromanthe |
| 5. Bracteole present, corolla tube as long as wide, shorter than or equaling the sepals...Ctenanthe |
| 5’. Bracteole absent, corolla tube longer than wide, longer than sepals...............................Maranta |
4. Materials and Methods
4.1. Sampling
4.2. DNA Extraction Amplification and Sequencing
4.3. Alignment and Phylogenetic Analysis
4.4. Ancestral State Reconstruction
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PP | Posterior Probabilities |
References
- Andersson, L. Marantaceae. In The Families and Genera of Vascular Plants, Vol. IV. Flowering Plants, Monocotyledons, Alismatanae and Commelinanae (Except Gramineae); Kubitzki, K., Ed.; Springer: Berlin, Germany, 1998; pp. 278–293. [Google Scholar]
- Tanaka, N.; Ohi-Toma, T.; Suksathan, P.; Aung, M.M.; Poulsen, A.D.; Mohamad, S.; Armstrong, K.E. Myanmaranthus roseiflorus, a new genus and species of Marantaceae from Myanmar. J. Jpn. Bot. 2022, 97, 187–196. [Google Scholar]
- Fernandes, G.C.; Luna, N.K.; Fraga, E.; Barros, M.C.; Chase, M.W.; Pessoa, E.M. Molecular phylogenetics of Maranta (Marantaceae: Zingiberales): Non-monophyly and support for a wider circumscription. Bot. J. Linn. Soc. 2023, 20, 181–194. [Google Scholar] [CrossRef]
- Luna, N.K.M.; Fernandes, G.C.; Christenhusz, M.J.M.; Pessoa, E.M. Taxonomic notes on Maranta (Marantaceae): A new combination and an updated infrageneric classification for the genus. Phytotaxa 2025, 684, 135–138. [Google Scholar] [CrossRef]
- Plants of the World Online (POWO). Facilitated by the Royal Botanic Gardens, Kew. 2025. Available online: https://powo.science.kew.org (accessed on 30 October 2024).
- Kress, W.J. The phylogeny and classification of the Zingiberales. Ann. Mo. Bot. Gard. 1990, 77, 698–721. [Google Scholar] [CrossRef]
- Andersson, L.; Chase, M.W. Phylogeny and classification of Marantaceae. Bot. J. Linn. Soc. 2001, 135, 275–287. [Google Scholar] [CrossRef][Green Version]
- Prince, L.M.; Kress, W.J. Phylogenetic relationships and classification in Marantaceae: Insights from plastid DNA sequence data. Taxon 2006, 55, 281–296. [Google Scholar] [CrossRef]
- Suksathan, P.; Gustafsson, M.H.; Borchsenius, F. Phylogeny and generic delimitation of Asian Marantaceae. Bot. J. Linn. Soc. 2009, 159, 381–395. [Google Scholar] [CrossRef]
- Al-Gharaibeh, H.M.M. Seed Germination and Genetic Structure of Two Salvia Species in Response to Environmental Variables Among Phytogeographic Regions in Jordan (Part I) and Phylogeny of the Pan-Tropical Family Marantaceae (Part II). Ph.D. Dissertation, Martin Luther University, Halle, Germany, 2017. [Google Scholar]
- Yoshida-Arns, K.N.; Mayo, S.J.; Braga, J.M.A. Stromanthe bahiensis sp. nov. (Marantaceae), endemic to southern Bahia State, Brazil. Nord. J. Bot. 2011, 29, 357–360. [Google Scholar] [CrossRef]
- Schumann, K.M. Marantaceae. In Das Pflanzenreich IV; Engler, A., Ed.; Verlag von Wilhem Engelmann: Leipzig, Germany, 1902; pp. 1–184. [Google Scholar]
- Futuyma, D.J. Evolutionary constraint and ecological consequences. Evolution 2010, 64, 1865–1884. [Google Scholar] [CrossRef]
- Fraga, F.R.M. Saranthe. In Flora e Funga Do Brasil; Jardim Botânico do Rio de Janeiro: Rio de Janeiro, Brazil, 2025. Available online: https://floradobrasil.jbrj.gov.br/FB9371 (accessed on 30 October 2024).
- Yoshida-Arns, K.N. Revisão Taxonômica de Ctenanthe Eichler e Stromanthe Sond. (Marantaceae). Ph.D. Dissertation, Universidade Federal de Pernambuco, Recife, Brazil, 2003. [Google Scholar]
- Wiley, E.O.; Lieberman, B.S. Phylogenetics: Theory and Practice of Phylogenetic Systematics, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2011; 406p. [Google Scholar]
- Braga, J.M.A. A new combination in the genus Stromanthe (Marantaceae). Eugeniana 1995, 21, 22–24. [Google Scholar]
- Fraga, F.R.M.; Braga, J.M.A. Two new species of Marantaceae from Reserva Natural Vale, Espírito Santo State, Brazil. Phytotaxa 2025, 682, 112–120. [Google Scholar] [CrossRef]
- Fraga, F.R.M. Ctenanthe. In Flora e Funga Do Brasil; Jardim Botânico do Rio de Janeiro: Rio de Janeiro, Brazil, 2025. Available online: https://floradobrasil.jbrj.gov.br/FB9331 (accessed on 30 October 2024).
- Luna, N.K.M.; Gomes-Silva, F.; Pessoa, E.M.; Felix, L.P. Revised typification of Ctenanthe and Stromanthe (Marantaceae). Phytotaxa 2024, 659, 295–300. [Google Scholar] [CrossRef]
- Chase, M.W.; Hills, H.H. Silica gel: An ideal material for field preservation of leaf samples for DNA studies. Taxon 1991, 40, 215–220. [Google Scholar] [CrossRef]
- Andersson, L. The Neotropical genera of Marantaceae: Circumscription and relationships. Nord. J. Bot. 1981, 1, 218–245. [Google Scholar] [CrossRef]
- Andersson, L. Revision of Maranta subgen. Maranta (Marantaceae). Nord. J. Bot. 1986, 6, 729–756. [Google Scholar] [CrossRef]
- Luna, N.; Pessoa, E.; Alves, M. Flora da Usina São José, Igarassu, Pernambuco: Zingiberales. Rodriguésia 2016, 67, 261–273. [Google Scholar] [CrossRef][Green Version]
- Luna, N.; Pessoa, E.; Alves, M. Sinopse de Marantaceae no estado de Pernambuco, Brasil. Rodriguésia 2020, 70, e02682018. [Google Scholar] [CrossRef]
- Luna, N.; Pessoa, E.; Alves, M. Taxonomic study of the species of Maranta Plum. ex L. (Marantaceae) from northeastern Brazil: A neglected diversity center for the genus with five new species. Syst. Bot. 2021, 46, 582–610. [Google Scholar] [CrossRef]
- Vieira, S. Estudos Filogenéticos e Taxonômicos em Marantaceae, Com Ênfase em Maranta L. Ph.D. Dissertation, Universidade de São Paulo, São Paulo, Brazil, 2005. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation, method for small quantities of fresh tissues. Phytochem. Bull. Bot. Soc. Am. 1987, 19, 11–15. [Google Scholar]
- Borchsenius, F.; Suárez, L.S.S.; Prince, L.M. Molecular phylogeny and redefined generic limits of Calathea (Marantaceae). Syst. Bot. 2012, 37, 620–635. [Google Scholar] [CrossRef]
- Steele, K.P.; Vilgalys, R. Phylogenetic analysis of Polemoniaceae using nucleotide sequences of the plastid gene matK. Syst. Bot. 1994, 19, 126–142. [Google Scholar] [CrossRef]
- Saka, M.N. Revisão Taxonômica e Análise Filogenética Das Espécies Extra-Amazônicas de Goeppertia Nees, Caldo Breviscapus (Marantaceae). Ph.D. Dissertation, Universidade Estadual Paulista “Júlio de Mesquita Filho”, Rio Claro, Brazil, 2016; 347p. [Google Scholar]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Oxelman, B.; Lidén, M.; Berglund, D. Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Syst. Evol. 1996, 206, 393–410. [Google Scholar] [CrossRef]
- Schloötterer, C. Ribosomal DNA probes and primers. In Molecular Tools for Screening Biodiversity: Plants and Animals; Karp, A., Isaac, P.G., Ingram, D.S., Eds.; Chapman & Hall: London, UK, 1998; pp. 267–276. [Google Scholar]
- Poulsen, A.D.; Mathisen, H.B.; Newman, M.F.; Ardiyani, M.; Lofthus, Ø.; Bjorå, C.S. Sulettaria: A new ginger genus disjunct from Elettaria cardamomum. Taxon 2018, 67, 725–738. [Google Scholar] [CrossRef]
- Pessoa, E.M.; Nollet, F.; Magalhães, R.F.; Viruel, J.; Pinheiro, F.; Chase, M.W. Nuclear–plastid discordance indicates past introgression in Epidendrum species (Laeliinae, Orchidaceae) with highly variable chromosome numbers. Bot. J. Linn. Soc. 2022, 199, 357–371. [Google Scholar] [CrossRef]
- Vieira, T.L.; Salazar, G.A.; Van den Berg, C. Phylogeny of Prosthechea (Laeliinae, Orchidaceae) based on nrITS and plastid DNA sequences: Reassessing the lumper–splitter debate and shedding light on the evolution of this Neotropical genus. Taxon 2024, 73, 142–160. [Google Scholar] [CrossRef]
- Gostel, M.R.; Loeuille, B.; Santana, M.H.; Kelloff, C.L.; Chan, R.; Simões, A.R.G.; Larridon, I.; Funk, V.A. Molecular phylogenetics of Distephanus supports the recognition of a new tribe, Distephaneae (Asteraceae). Bot. J. Linn. Soc. 2024, 206, 296–312. [Google Scholar] [CrossRef]
- Preez, B.; Schrire, B.D.; Dreyer, L.L.; Stirton, C.H.; Chimphango, S.B.M.; Muasya, A.M. Phylogeny and new sectional classification for the Cape Clade of the genus Indigofera (Fabaceae: Indigofereae). Taxon 2025, 74, 819–856. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Holder, M.; Vos, R.; Midford, P.E.; Liebowitz, T.; Chan, L.; Hoover, P.; Warnow, T. The CIPRES Portals. 2010. Available online: http://www.phylo.org/sub_sections/portal (accessed on 30 October 2024).
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Constructing a significance test for incongruence. Syst. Biol. 1995, 44, 570–572. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP *: Phylogenetic Analysis Using Parsimony (* and Other Methods), Version 4; Sinauer: Sunderland, MA, USA, 2002. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.4, 2018. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 30 October 2024).
- Cummings, M.P.; Handley, S.A.; Myers, D.S.; Reed, D.L.; Rokas, A.; Winka, K. Comparing bootstrap and posterior probability values in the four-taxon case. Syst. Biol. 2003, 52, 477–487. [Google Scholar] [CrossRef]
- Erixon, P.; Svennblad, B.; Britton, T.; Oxelman, B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst. Biol. 2003, 52, 665–673. [Google Scholar] [CrossRef]
- Simmons, M.P.; Pickett, K.M.; Miya, M. How meaningful are Bayesian support values? Mol. Biol. Evol. 2004, 21, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Costa, F.; Espinelli, F.P.; Figueiredo, F.O.G. Guia de Zingiberales Dos Sítios PPBio na Amazônia Ocidental Brasileira; Áttema Design Editorial: Manaus, Brazil, 2011; 284p. [Google Scholar]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Yu, Y.; Blair, C.; He, X. RASP 4: Ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 2020, 37, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Janssens, S.B.; Vandelook, F.; Langhe, E.D.; Verstraete, B.; Smets, E.; Vandenhouwe, I.; Swennen, R. Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol. 2016, 210, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Iles, W.J.D.; Sass, C.; Lagomarsino, L.; Benson-Martin, G.; Driscoll, H.; Specht, C.D. The phylogeny of Heliconia (Heliconiaceae) and the evolution of floral presentation. Mol. Phylogenetics Evol. 2017, 117, 150–167. [Google Scholar] [CrossRef]
- Ley, A.C.; Röser, M. Phylogeny of the climber genus Haumania (Marantaceae) endemic to the tropical lowland rainforest in Central Africa. Phytotaxa 2018, 379, 143–152. [Google Scholar] [CrossRef]
- Givnish, T.J.; Zuluaga, A.; Spalink, D.; Gomez, M.S.; Lam, V.K.Y.; Saarela, J.M.; Sass, C.; Iles, W.J.D.; de Sousa, D.J.L.; Leebens-Mack, J.; et al. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses and a functional model for the origin of monocots. Am. J. Bot. 2018, 105, 1888–1910. [Google Scholar] [CrossRef] [PubMed]
- Gostel, M.R.; Carlsen, M.M.; Devine, A.; Barker, K.B.; Coddington, J.A.; Steier, J. Data Release: DNA Barcodes of Plant Species Collected for the Global Genome Initiative for Gardens (GGI-Gardens) II. Diversity 2022, 14, 234. [Google Scholar] [CrossRef]
- Li, D.M.; Liu, H.L.; Pan, Y.G.; Yu, B.; Huang, D.; Zhu, G. Comparative chloroplast genomics of 21 species in Zingiberales with implications for their phylogenetic relationships and molecular dating. Int. J. Mol. Sci. 2023, 24, 15031. [Google Scholar] [CrossRef] [PubMed]

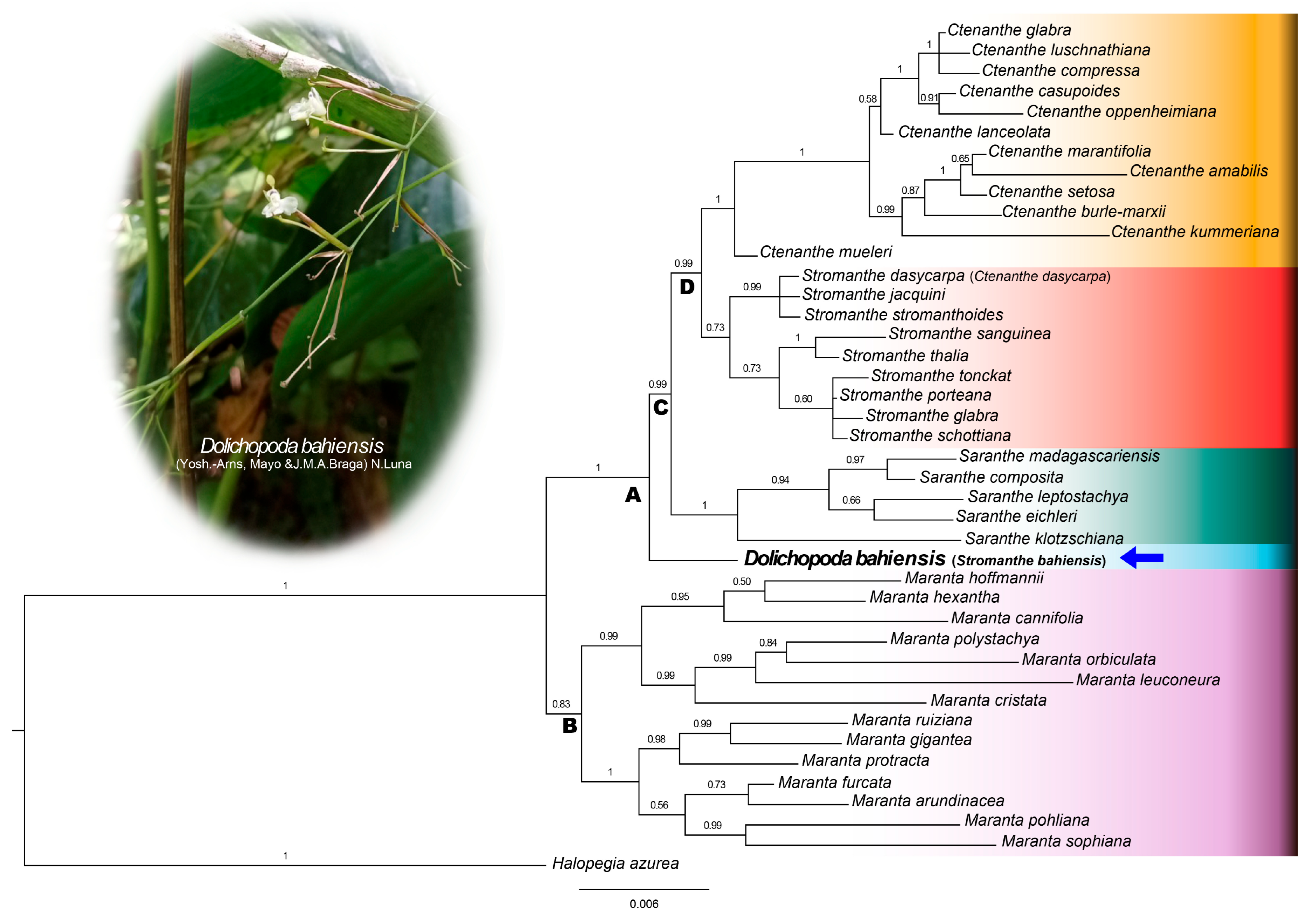
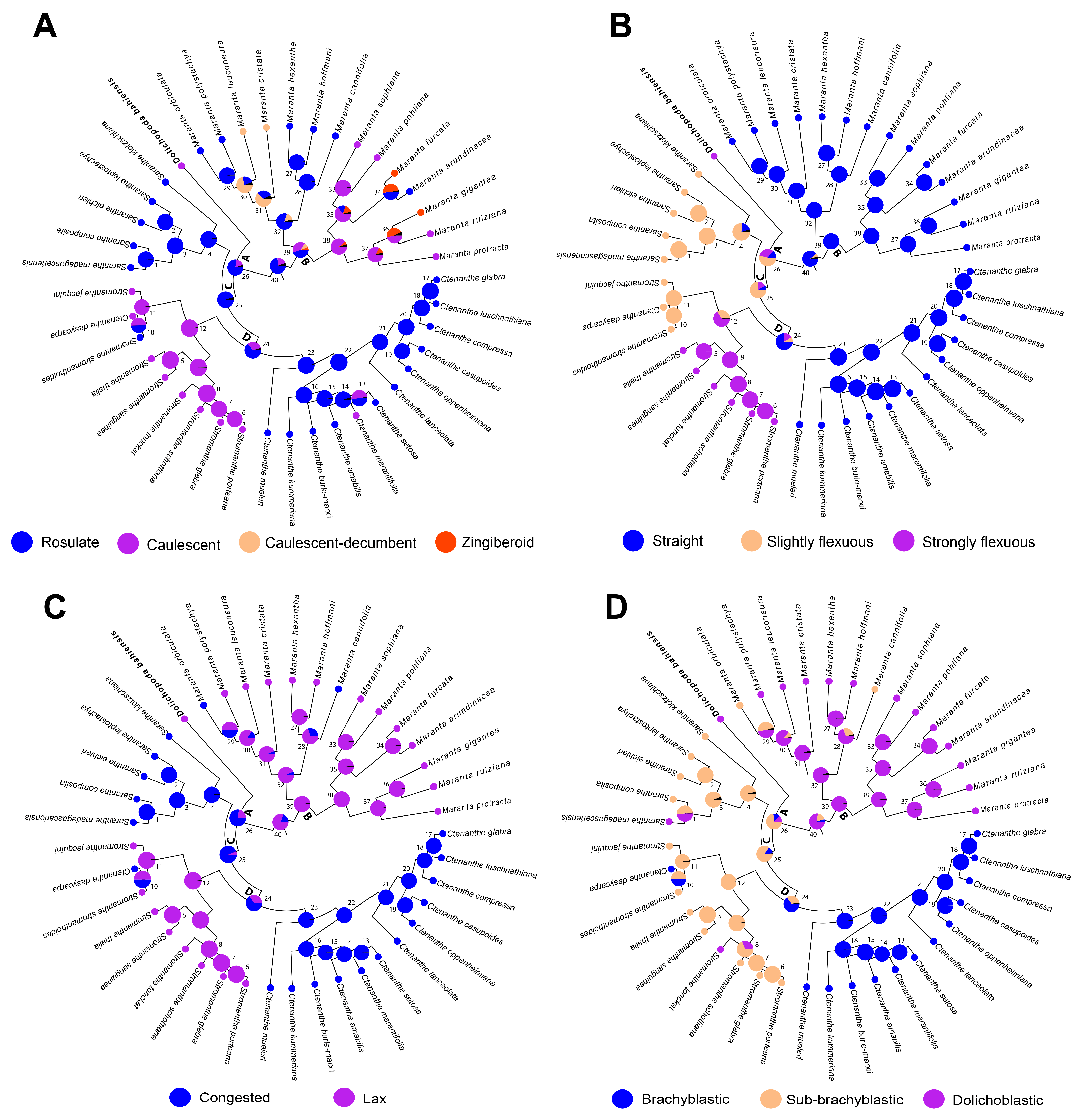

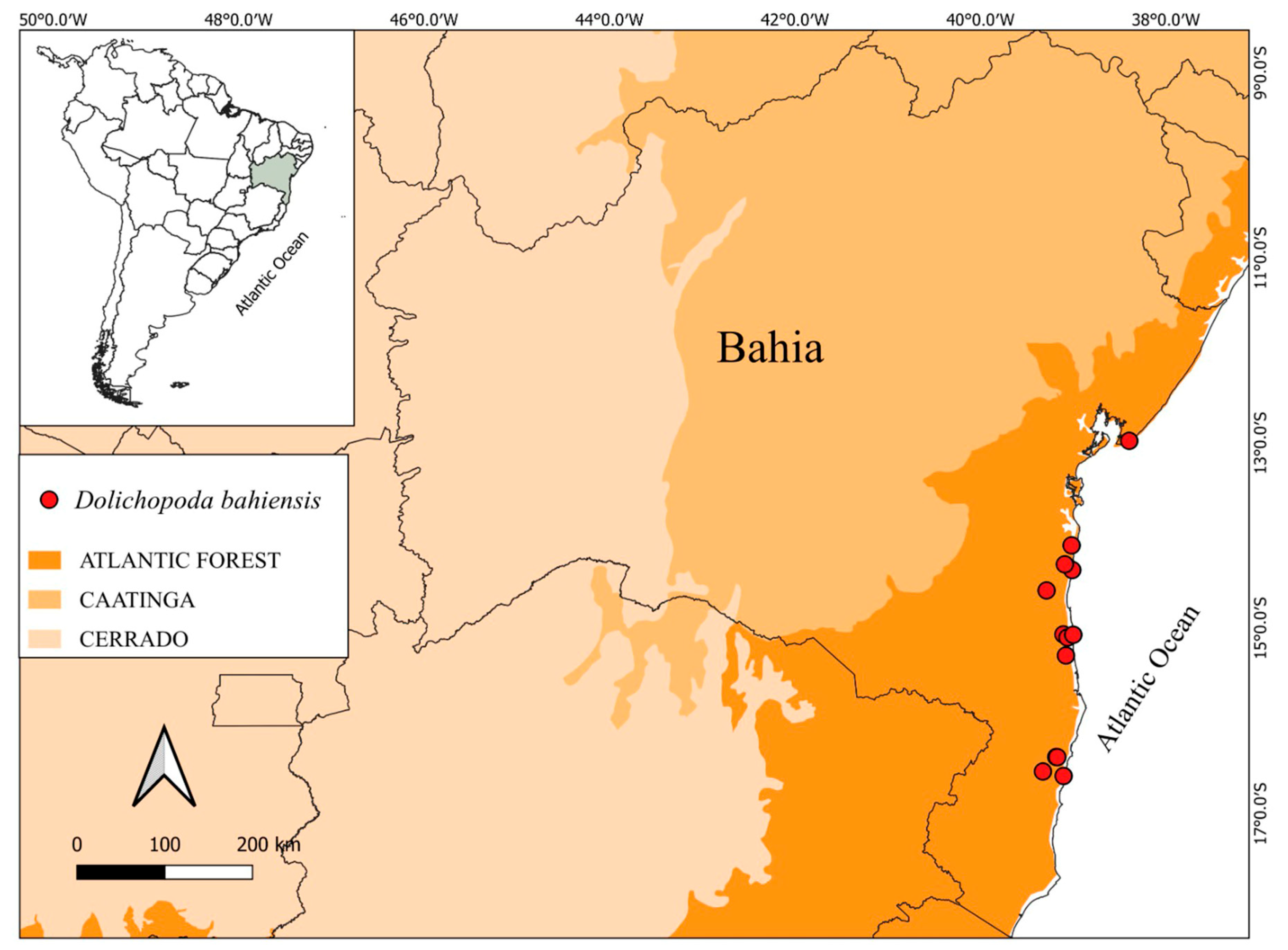
| nrITS | matK | rps16 | trnL-F | Combined Plastid | All Markers Combined | |
|---|---|---|---|---|---|---|
| Taxon number | 29 | 23 | 45 | 40 | 41 | 42 |
| Alignment length | 681 | 1602 | 902 | 948 | 3452 | 4133 |
| Number of variable positions | 226 (33.2%) | 123 (7.5%) | 136 (15%) | 103 (10.8%) | 345 (10%) | 561 (13.6%) |
| Number of informative sites | 109 (16%) | 36 (2.2%) | 49 (5.4%) | 14 (1.5%) | 95 (2.7%) | 196 (4.7%) |
| Substitution model | TIM3+G | GTR+G | TPM2uf+G | TIM1 | - | - |
| Genera/ Characters | Dolichopoda | Ctenanthe | Maranta s.l. | Saranthe | Stromanthe |
|---|---|---|---|---|---|
| Habit | Caulescent | Rosulate or caulescent | Rosulate, caulescent, zingiberoid or scandent | Rosulate | Caulescent |
| Synflorescence | Axillary, branched, lax, 1–4 florescence per node | Axillary or terminal-basal, simple to branched, congested, 1–4 florescences | Axillary, terminal-apical, or arising from rhizome, simple to branched, congested or lax 1–6 florescence per node | Axillary or terminal-basal, branched, congested, 1–3 florescence per node | Axillary or terminal-apical, simple to branched, lax, 1–4 florescences per node |
| Inflorescence penducle | 4–35 cm long., pendulous | Sessile to 22 cm long, erect | Sessile to 18 cm long, erect | Sessile to 15 cm long, erect | 0.7–22.5 cm long, erect |
| Spathe | Deciduous, chartaceous | Persistent, chartaceous or membranous | Persistent, chartaceous or membranous | Persistent or deciduous, membranous | Persistent or deciduous, membranous or papyraceous |
| Rachis | Strongly flexuous, hispid to hispid at nodes, 7–35 cm long | Straight, glabrous or pilose, 2–15 cm long | Absent, if present, straight, glabrous to glabrescent, puberules, hirsute to hirsute only at the base, 3–8 cm long | Slightly flexuous, puberulent to sericeous, 3–10 cm long. | Slightly to strongly f lexuous, glabrous, villous, lanuginous, 0.9–18.0 cm long, |
| Cymules | 2–4 per spathe, dolichoblastic | 2–7 per spathe, brachiblastic | 1–6 per spathe, brachiblastic or dolichoblastic | 1 per spathe, dolichoblastic | 1–7 per spathe, brachiblastic to dolichoblastic |
| Bracteole | Absent | Present | Absent | Absent | Absent, rarely present |
| Outer staminodes | 2 unequal | 2 slightly unequal | 2 equal or unequal | 2 equal or unequal | Absent, 1–2 equal |
| Callose staminode | 1 callus | 1 to 2 calluses | 1 or 3 calluses | 1 or 2 | 1 callus |
| Sepals in fruits | Deciduous | Deciduous or persistent | Deciduous | Deciduous | Deciduous or persistent |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luna, N.K.M.; Coutinho, T.S.; Chase, M.W.; Felix, L.P.; Pessoa, E.M. Molecular Phylogenetics and Morphological Analyses Support Dolichopoda, a New Neotropical Genus of Marantaceae (Zingiberales). Plants 2025, 14, 3486. https://doi.org/10.3390/plants14223486
Luna NKM, Coutinho TS, Chase MW, Felix LP, Pessoa EM. Molecular Phylogenetics and Morphological Analyses Support Dolichopoda, a New Neotropical Genus of Marantaceae (Zingiberales). Plants. 2025; 14(22):3486. https://doi.org/10.3390/plants14223486
Chicago/Turabian StyleLuna, Naédja K. M., Thales S. Coutinho, Mark W. Chase, Leonardo P. Felix, and Edlley M. Pessoa. 2025. "Molecular Phylogenetics and Morphological Analyses Support Dolichopoda, a New Neotropical Genus of Marantaceae (Zingiberales)" Plants 14, no. 22: 3486. https://doi.org/10.3390/plants14223486
APA StyleLuna, N. K. M., Coutinho, T. S., Chase, M. W., Felix, L. P., & Pessoa, E. M. (2025). Molecular Phylogenetics and Morphological Analyses Support Dolichopoda, a New Neotropical Genus of Marantaceae (Zingiberales). Plants, 14(22), 3486. https://doi.org/10.3390/plants14223486







