Functional Analysis of Maize SDG102 Gene in Response to Setosphaeria turcica
Abstract
1. Introduction
2. Results
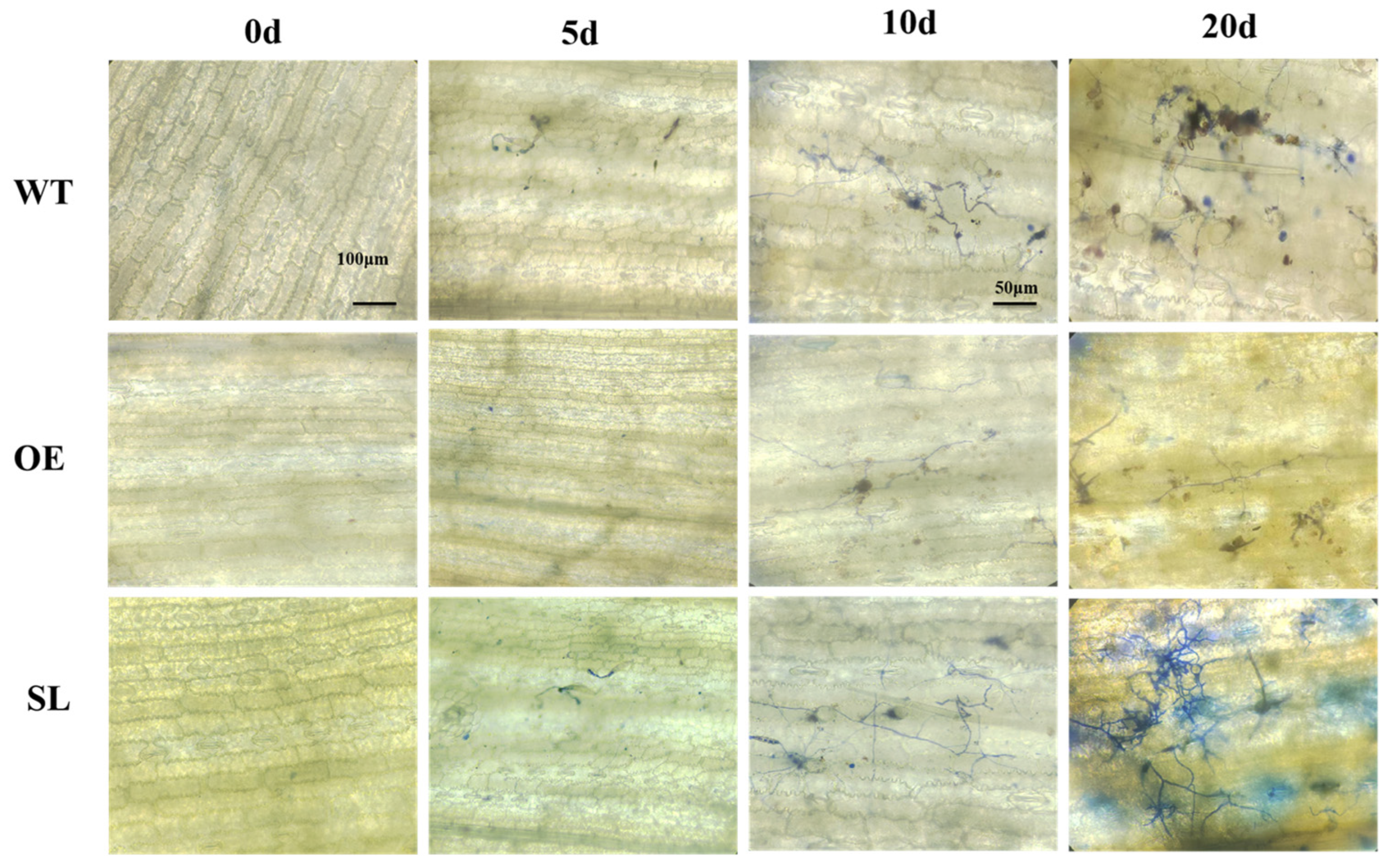
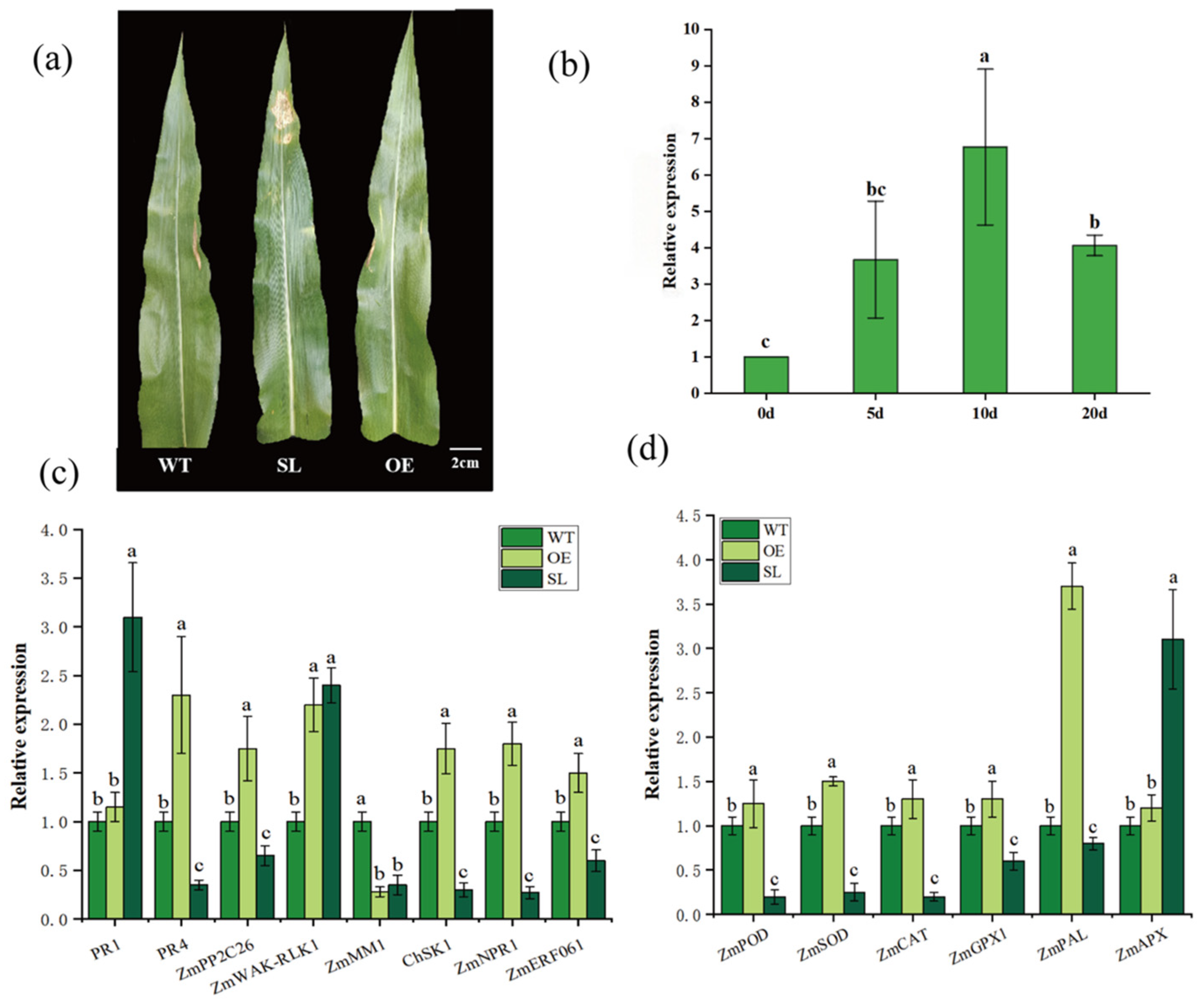
2.1. The Effect of SDG102 Gene to Maize Resistance Against S. turcica
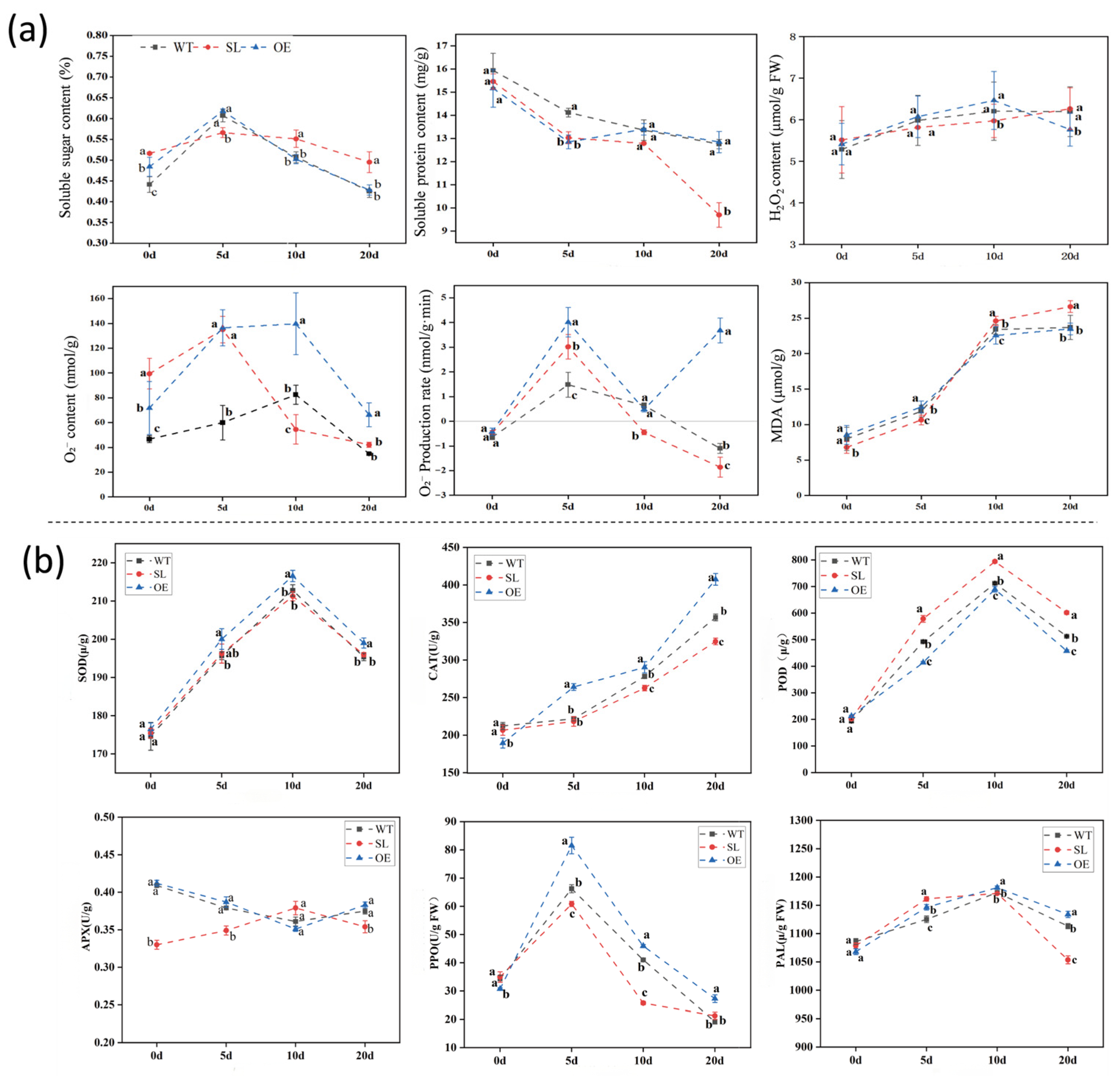
2.2. Effects of S. turcica Infection on Physiological Parameters of Maize
2.3. SDG102’s Resistance to NCLB
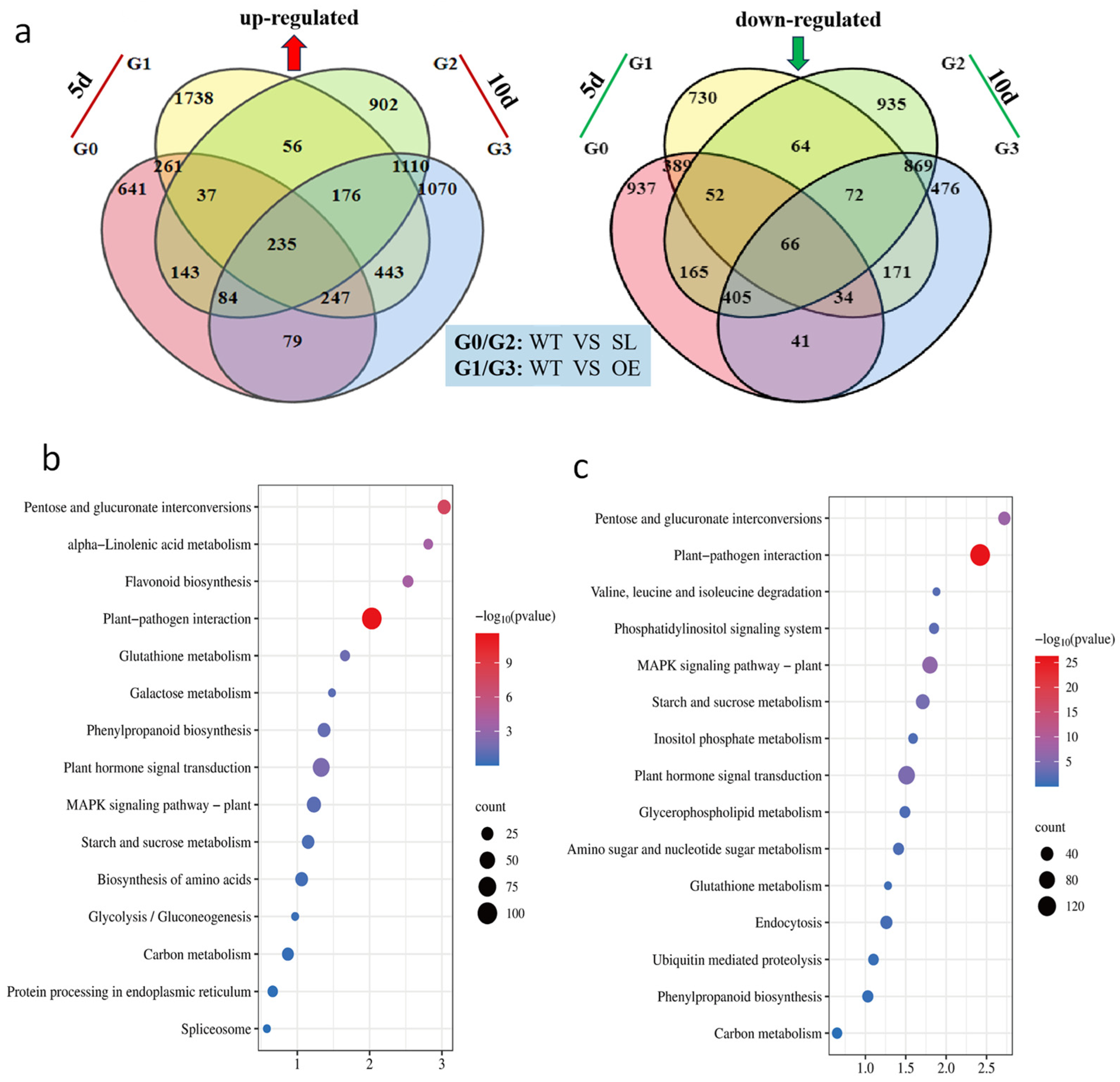
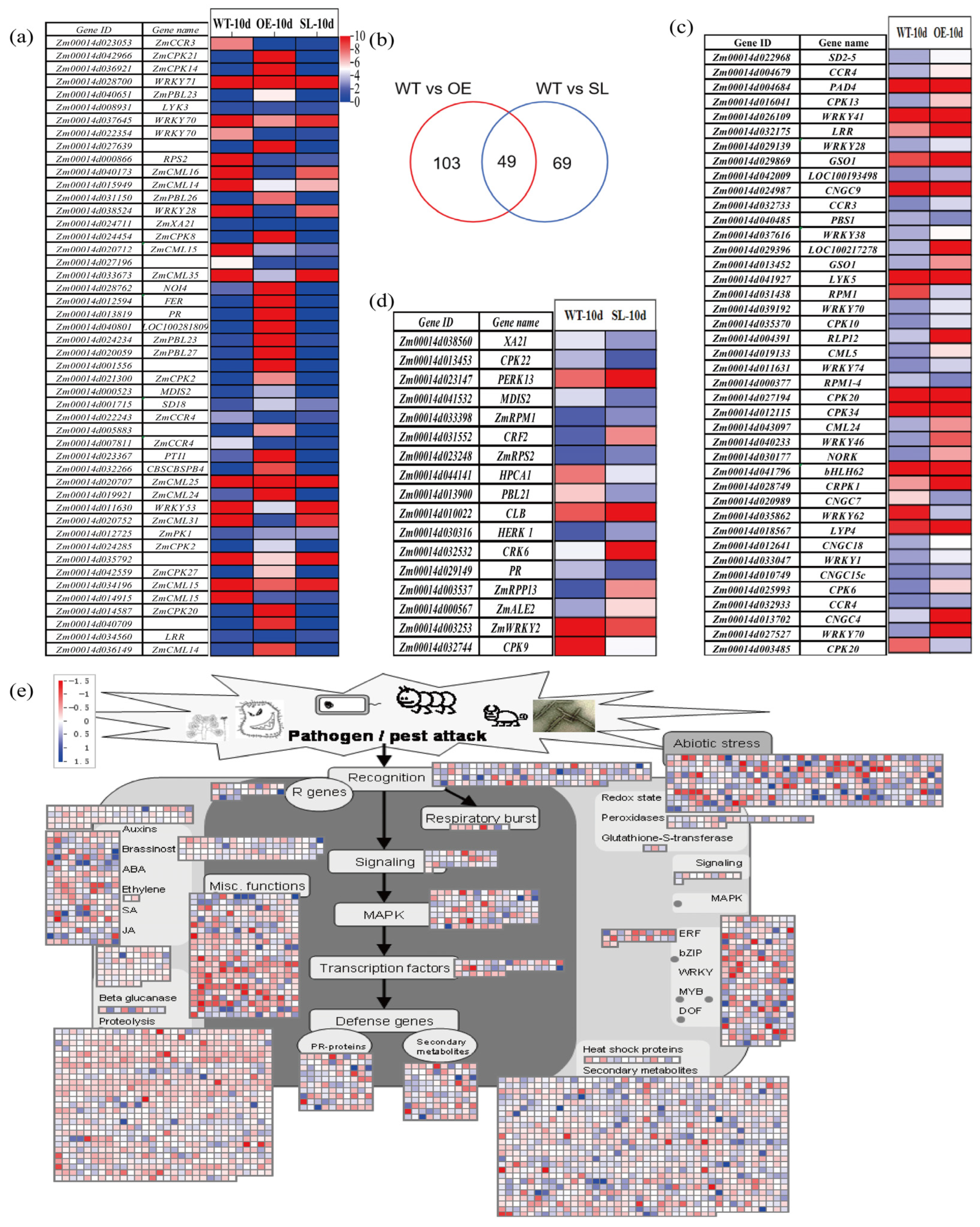
2.4. Analysis of Differentially Expressed Genes in Response to S. turcica Infection and KEGG Classification Annotation
2.5. Analysis of SDG102-Mediated Regulation of Plant–Pathogen Interaction Pathway Genes
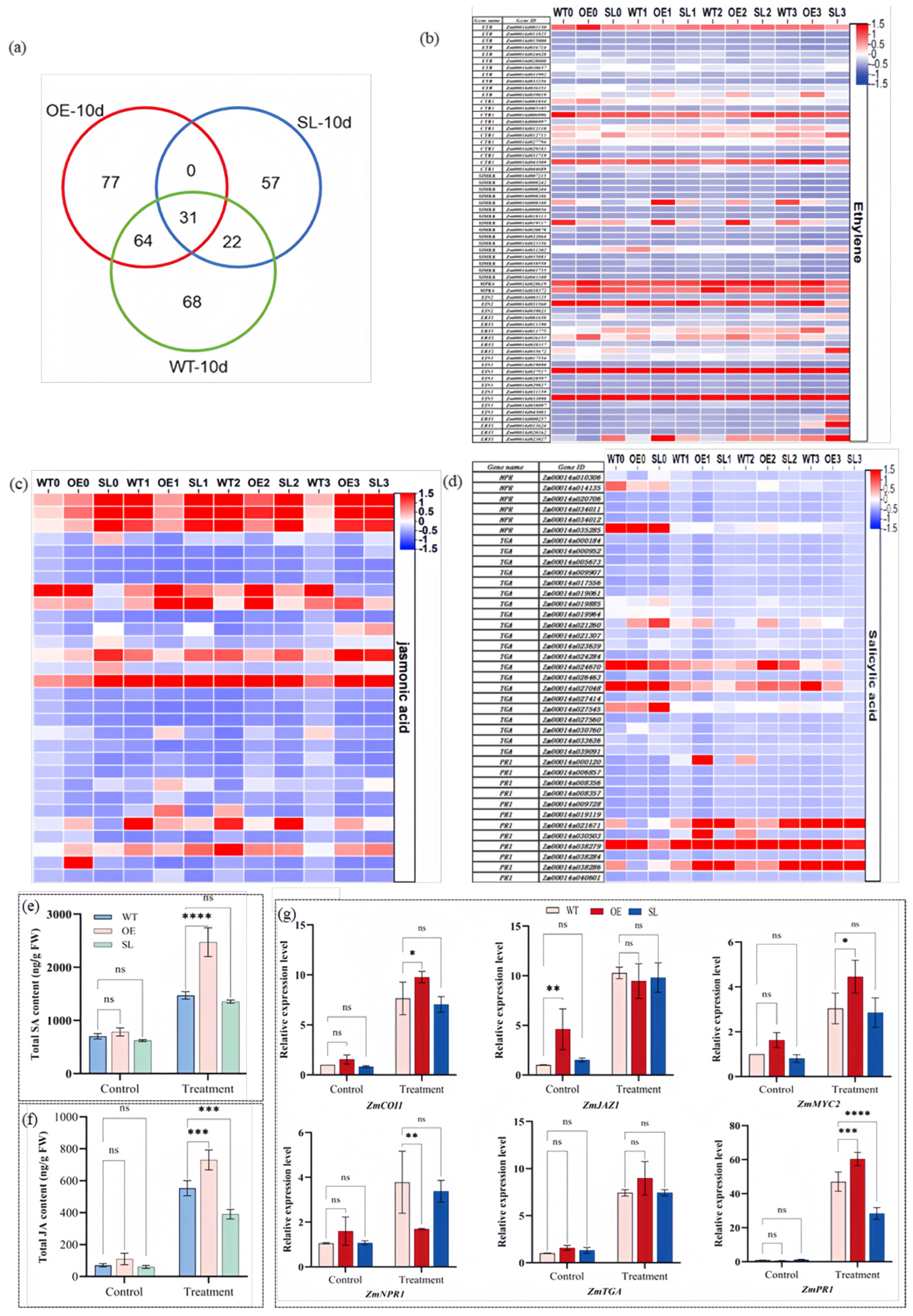
2.6. SDG102 Modulates Hormone-Mediated Defense Responses Against S. turcica Infection
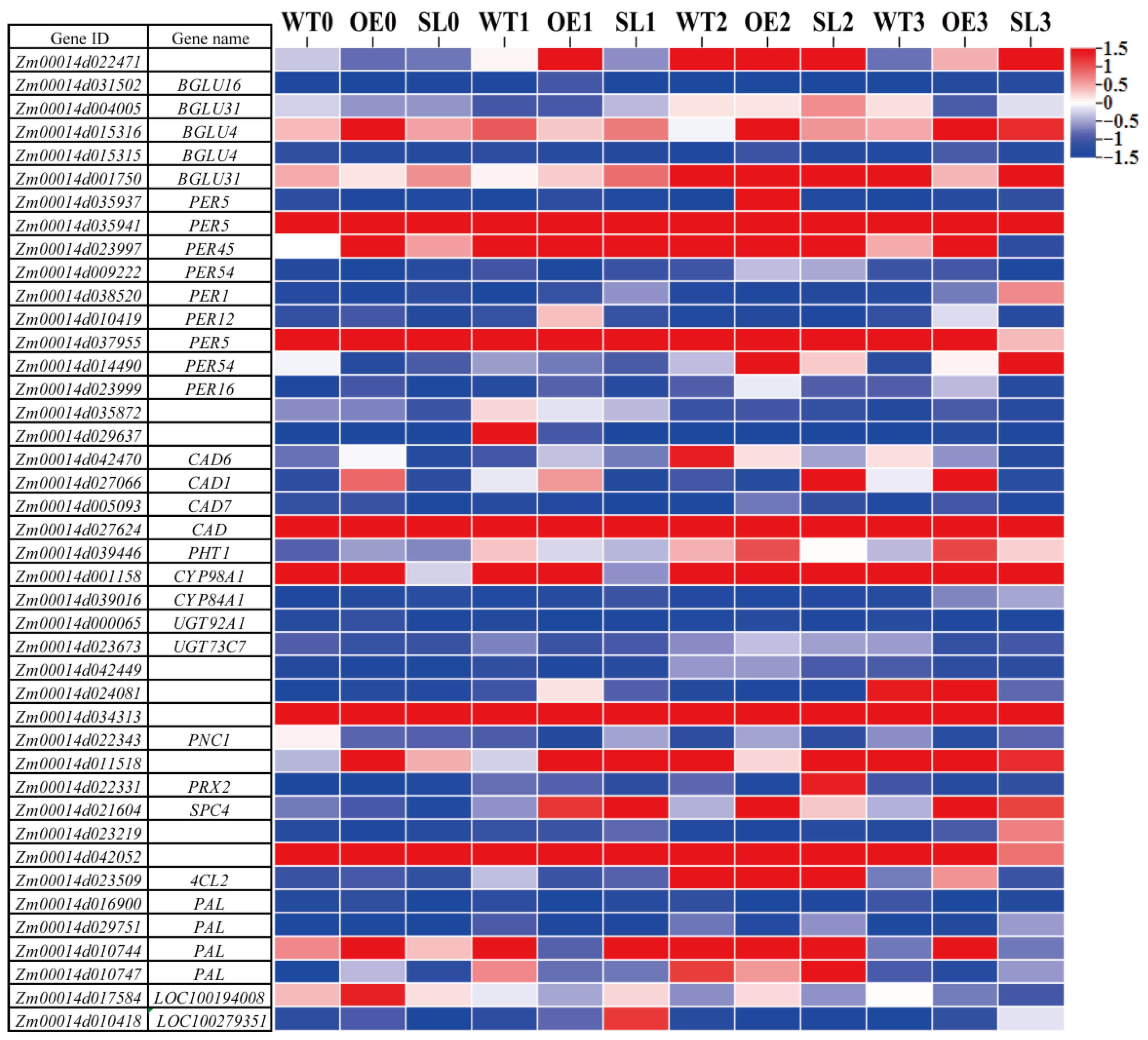
2.7. Effects on Differential Gene Expression in Phenylpropanoid Metabolic Pathways
2.8. Impact of SDG102 on Corn Yield
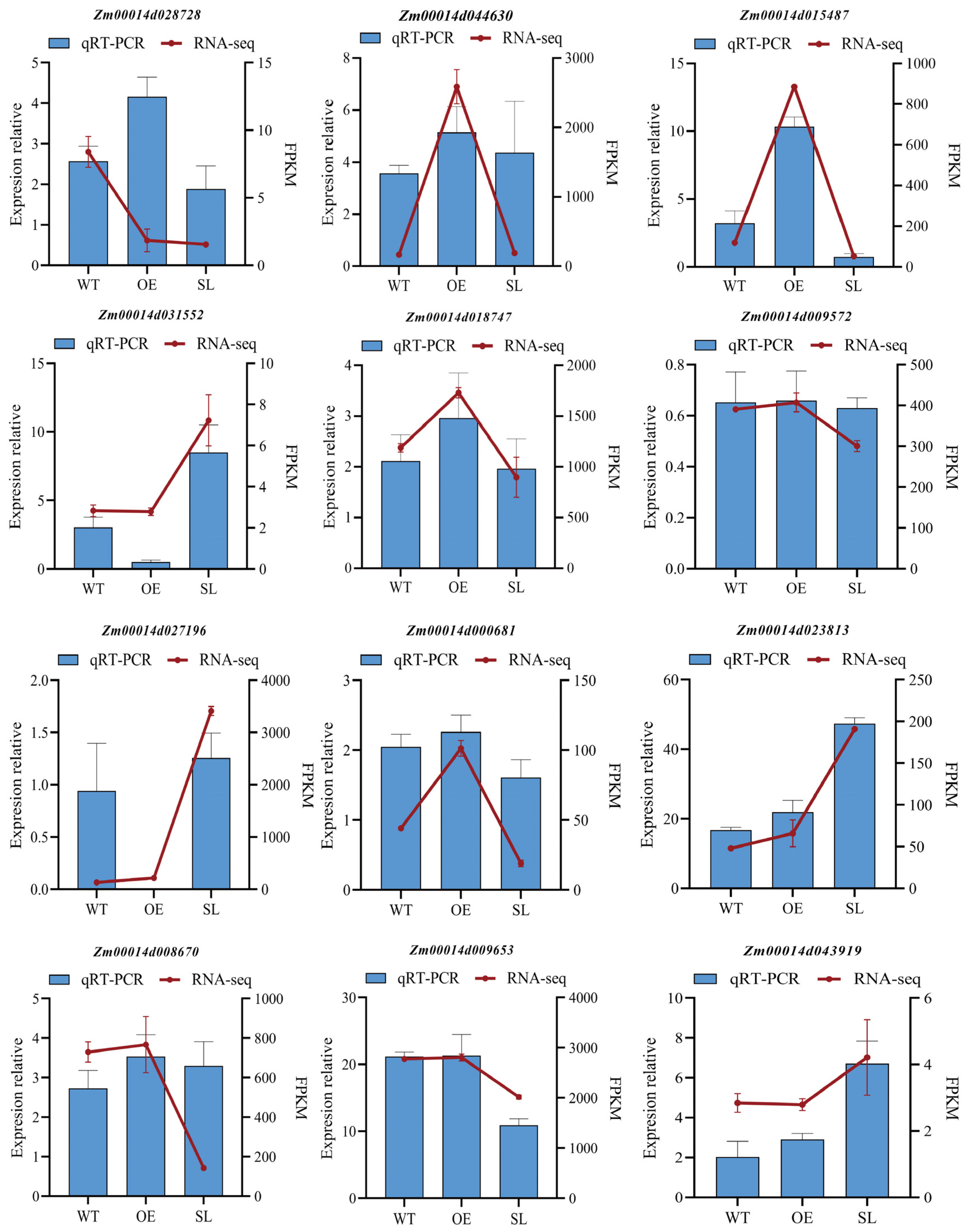
2.9. Validation of RNA-Seq Results by qRT-PCR Analysis
3. Discussion
3.1. Effects of SDG102 Gene on Agronomic Traits and Physiological–Biochemical Indices in Maize Under S. turcica Stress
3.2. Transcriptomic Analysis of SDG102-Mediated Responses to S. turcica Infection
3.3. SDG102 Regulates Expression of Genes Involved in Hormone Signaling and Secondary Metabolite Biosynthesis
4. Materials and Methods
4.1. Plant Materials
4.2. Pathogen Inoculation and Disease Phenotyping
4.3. Histopathological Examination and Symptom Quantification
4.4. Physiological Characteristics and Hormonal Analyses
4.5. Transcriptome Sequencing and Analysis
4.6. Quantitative PCR Validation
4.7. Assessment of Yield-Related Agronomic Traits
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, M.; Tong, L.; Xu, M.; Zhong, T. Genetic dissection of maize disease resistance and its applications in molecular breeding. Mol. Breed. 2021, 41, 32. [Google Scholar] [CrossRef]
- Kotze, R.; Van der Merwe, C.; Crampton, B.; Kritzinger, Q. A histological assessment of the infection strategy of Exserohilum turcicum in maize. Plant Pathol. 2019, 68, 504–512. [Google Scholar] [CrossRef]
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, C.Y.; Khalid, H.; Li, N.; Sun, Q.; Miao, Q.; Wu, S.; Li, F. Pyramiding resistance genes to northern leaf blight and head smut in maize. Int. J. Agric. Biol. 2012, 14, 430–434. [Google Scholar]
- Welz, H.G.; Geiger, H.H. Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breed. 2000, 119, 1–14. [Google Scholar] [CrossRef]
- Ogliari, J.B.; Guimarães, M.; Geraldi, I.; Camargo, L. New resistance genes in the Zea mays: Exserohilum turcicum pathosystem. Genet. Mol. Biol. 2005, 28, 435–439. [Google Scholar] [CrossRef]
- Xie, S.S.; Duan, C.G. Epigenetic regulation of plant immunity: From chromatin codes to plant disease resistance. aBIOTECH 2023, 4, 124–139. [Google Scholar] [CrossRef]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Du, J. Structure and mechanism of histone methylation dynamics in Arabidopsis. Curr. Opin. Plant Biol. 2022, 67, 102211. [Google Scholar] [CrossRef]
- Zheng, S.Y.; Guan, B.B.; Yuan, D.Y.; Zhao, Q.Q.; Ge, W.; Tan, L.M.; Chen, S.S.; Li, L.; Chen, S.; Xu, R.M.; et al. Dual roles of the Arabidopsis PEAT complex in histone H2A deubiquitination and H4K5 acetylation. Mol. Plant 2023, 16, 1847–1865. [Google Scholar] [CrossRef]
- Xia, H.; Gao, W.; Qu, J.; Dai, L.; Gao, Y.; Lu, S.; Zhang, M.; Wang, P.; Wang, T. Genetic mapping of northern corn leaf blight-resistant quantitative trait loci in maize. Medicine 2020, 99, e21326. [Google Scholar] [CrossRef]
- Cheng, L.; Shafiq, S.; Xu, W.; Sun, Q. Early Flowering in Short Days (EFS) regulates the seed size in Arabidopsis. Sci. China Life Sci. 2018, 61, 214–224. [Google Scholar] [CrossRef]
- Chen, K.; Du, K.; Shi, Y.; Yin, L.; Shen, W.H.; Yu, Y.; Liu, B.; Dong, A. H3K36 methyltransferase SDG708 enhances drought tolerance by promoting abscisic acid biosynthesis in rice. New Phytol. 2021, 230, 1967–1984. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, Z.; Dong, A.; Soubigou-Taconnat, L.; Renou, J.P.; Steinmetz, A.; Shen, W.H. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell Biol. 2008, 28, 1348–1360. [Google Scholar] [CrossRef]
- Kramer, H.M.; Cook, D.E.; Seidl, M.F.; Thomma, B.P.H.J. Epigenetic regulation of nuclear processes in fungal plant pathogens. PLoS Pathog. 2023, 19, e1011525. [Google Scholar] [CrossRef]
- Berr, A.; McCallum, E.J.; Alioua, A.; Heintz, D.; Heitz, T.; Shen, W.H. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 2010, 154, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ménard, R.; Li, Y.; Coruzzi, G.M.; Heitz, T.; Shen, W.H.; Berr, A. Arabidopsis SDG8 Potentiates the Sustainable Transcriptional Induction of the Pathogenesis-Related Genes PR1 and PR2 During Plant Defense Response. Front. Plant Sci. 2020, 11, 277. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Y.; Wang, B.; Luo, Q.; Shi, J.; Gan, J.; Shen, W.H.; Yu, Y.; Dong, A. The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment. Nat. Commun. 2019, 10, 2999. [Google Scholar] [CrossRef] [PubMed]
- Sui, P.; Jin, J.; Ye, S.; Mu, C.; Gao, J.; Feng, H.; Shen, W.H.; Yu, Y.; Dong, A. H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 2012, 70, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Liu, K.; Shen, T.; Shi, J.; Liu, B.; Han, M.; Peng, M.; Fu, H.; Song, Y.; Zhu, J.; et al. Position-specific intron retention is mediated by the histone methyltransferase SDG725. BMC Biol. 2018, 16, 44. [Google Scholar] [CrossRef]
- Li, T.; Chen, X.; Zhong, X.; Zhao, Y.; Liu, X.; Zhou, S.; Cheng, S.; Zhou, D.X. Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 2013, 25, 4725–4736. [Google Scholar] [CrossRef]
- Mierziak, J.; Wojtasik, W. Epigenetic weapons of plants against fungal pathogens. BMC Plant Biol. 2024, 24, 175. [Google Scholar] [CrossRef]
- Lee, S.; Fu, F.; Xu, S.; Lee, S.Y.; Yun, D.J.; Mengiste, T. Global regulation of plant immunity by Histone Lysine Methyl Transferases. Plant Cell 2016, 28, 1640–1661. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E.; et al. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Perrella, G.; Bäurle, I.; van Zanten, M. Epigenetic regulation of thermomorphogenesis and heat stress tolerance. New Phytol. 2022, 234, 1144–1160. [Google Scholar] [CrossRef]
- Banerjee, A.; Wani, S.H.; Roychoudhury, A. Epigenetic Control of Plant Cold Responses. Front. Plant Sci. 2017, 8, 1643. [Google Scholar] [CrossRef]
- Qi, X.; Wan, C.; Zhang, X.; Sun, W.; Liu, R.; Wang, Z.; Wang, Z.; Ling, F. Effects of histone methylation modification on low temperature seed germination and growth of maize. Sci. Rep. 2023, 13, 5196. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Xue, S.; Quan, T.; Cui, D.; Han, L.; Cong, W.; Li, M.; Yun, D.J.; Liu, B.; et al. SET DOMAIN GROUP 721 protein functions in saline-alkaline stress tolerance in the model rice variety Kitaake. Plant Biotechnol. J. 2021, 19, 2576–2588. [Google Scholar] [CrossRef]
- Kasote, D.M.; Lee, J.; Jayaprakasha, G.K. Seed Priming with Iron Oxide Nanoparticles Modulate Antioxidant Potential and Defense-Linked Hormones in Watermelon Seedlings. ACS Sustain. Chem. Eng. 2019, 7, 5142–5151. [Google Scholar] [CrossRef]
- Arbona, V.; Flors, V.; Jacas, J.; García-Agustín, P.; Gómez-Cadenas, A. Enzymatic and non-enzymatic antioxidant responses of Carrizo citrange, a salt-sensitive citrus rootstock, to different levels of salinity. Plant Cell Physiol. 2003, 44, 388–394. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, F.; Wang, Z.; Cao, H.; Li, X.; Deng, X.; Soppe, W.J.J.; Li, Y.; Liu, Y. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012, 193, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, P.; Zhang, F.; Luo, X.; Xie, J. Histone deacetylase HDA19 interacts with histone methyltransferase SUVH5 to regulate seed dormancy in Arabidopsis. Plant Biol. 2020, 22, 1062–1071. [Google Scholar] [CrossRef]
- Bouyer, D.; Roudier, F.; Heese, M.; Andersen, E.D.; Gey, D.; Nowack, M.K.; Goodrich, J.; Renou, J.P.; Grini, P.E.; Colot, V.; et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011, 7, e1002014. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Ma, D.P.; Li, J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 2008, 373, 659–664. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Ankala, A.; Luthe, D.S.; Williams, W.P.; Wilkinson, J.R. Integration of ethylene and jasmonic acid signaling pathways in the expression of maize defense protein Mir1-CP. Mol. Plant Microbe Interact. 2009, 22, 1555–1564. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Wang, J.; Eulgem, T. The Arabidopsis RRM domain proteins EDM3 and IBM2 coordinate the floral transition and basal immune responses. Plant J. 2023, 116, 128–143. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, L.; Duan, J.; Miki, B.; Wu, K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 2005, 17, 1196–1204. [Google Scholar] [CrossRef]
- Fromm, M.; Avramova, Z. ATX1/AtCOMPASS and the H3K4me3 marks: How do they activate Arabidopsis genes? Curr. Opin. Plant Biol. 2014, 21, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mukherjee, I.; Thum, K.E.; Tanurdzic, M.; Katari, M.S.; Obertello, M.; Edwards, M.B.; McCombie, W.R.; Martienssen, R.A.; Coruzzi, G.M. The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol. 2015, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Cazzonelli, C.I.; Roberts, A.C.; Carmody, M.E.; Pogson, B.J. Transcriptional control of SET DOMAIN GROUP 8 and CAROTENOID ISOMERASE during Arabidopsis development. Mol. Plant 2010, 3, 174–191. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, W.; Wang, Z.; Wan, C.; Zhang, J.; Qi, X.; Zhang, J. SDG102, a H3K36-Methyltransferase-Encoding Gene, Plays Pleiotropic Roles in Growth and Development of Maize (Zea mays L.). Int. J. Mol. Sci. 2022, 23, 7458. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Soft Cover; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Durgbanshi, A.; Arbona, V.; Pozo, O.; Miersch, O.; Sancho, J.V.; Gómez-Cadenas, A. Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. J. Agric. Food Chem. 2005, 53, 8437–8442. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Levin, J.Z.; Ureta-Vidal, A. RNA-Seq sample preparation, sequencing, and data analysis. Cold Spring Harb. Protoc. 2015, pdb.top082234. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Name | Length of Spike (cm) | Diameter of Spike (cm) | Weight of Spike (g) | Length of Grain (cm) | Width of Grain (cm) | Number of Panicle Rows | Number of Grains in Row | Diameter of Cob (cm) | Yield of Seed (%) | 100-Kernel Weight (g) | Plot Yield (kg) | Yield/667 m2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | 15.76 ± 0.43 b | 3.93 ± 0.09 c | 100.79 ± 1.67 c | 1.00 ± 0.06 b | 0.76 ± 0.05 a | 15.30 ± 0.94 b | 24.5 ± 0.50 a | 2.75 ± 0.27 a | 75.70 ± 1.44 b | 25.60 ± 0.10 b | 12.21 ± 1.14 b | 298.31 ± 27.85 b |
| OE | 17.22 ± 0.69 a | 4.30 ± 0.08 a | 118.98 ± 2.80 b | 1.12 ± 0.06 b | 0.82 ± 0.07 a | 16.00 a | 23.75 ± 0.83 a | 2.72 ± 0.15 a | 79.09 ± 0.20 a | 27.00 ± 0.71 a | 13.61 ± 1.98 a | 332.52 ± 48.37 a |
| SL | 15.08 ± 0.94 b | 4.20 ± 0.05 b | 90.28 ± 2.60 a | 1.17 ± 0.07 a | 0.60 ± 0.10 b | 15.00 ± 0.92 b | 23.00 ± 1.58 b | 2.97 ± 0.41 a | 79.01 ± 0.42 a | 25.16 ± 0.47 b | 11.52 ± 2.86 b | 281.45 ± 69.87 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, X.; Zhang, X.; Ma, X.; Zhao, X.; Liu, X.; Wei, X.; Tian, H.; Liu, Y.; Zhang, J.; Wang, Z. Functional Analysis of Maize SDG102 Gene in Response to Setosphaeria turcica. Plants 2025, 14, 3463. https://doi.org/10.3390/plants14223463
Qi X, Zhang X, Ma X, Zhao X, Liu X, Wei X, Tian H, Liu Y, Zhang J, Wang Z. Functional Analysis of Maize SDG102 Gene in Response to Setosphaeria turcica. Plants. 2025; 14(22):3463. https://doi.org/10.3390/plants14223463
Chicago/Turabian StyleQi, Xin, Xing Zhang, Xiaoxiao Ma, Xinyi Zhao, Xinyang Liu, Xiaoshuang Wei, Huai Tian, Yang Liu, Jianhua Zhang, and Zhenhui Wang. 2025. "Functional Analysis of Maize SDG102 Gene in Response to Setosphaeria turcica" Plants 14, no. 22: 3463. https://doi.org/10.3390/plants14223463
APA StyleQi, X., Zhang, X., Ma, X., Zhao, X., Liu, X., Wei, X., Tian, H., Liu, Y., Zhang, J., & Wang, Z. (2025). Functional Analysis of Maize SDG102 Gene in Response to Setosphaeria turcica. Plants, 14(22), 3463. https://doi.org/10.3390/plants14223463







