Conservation and Divergence of E(z) Genes in Green Plants
Abstract
1. Introduction
2. Results
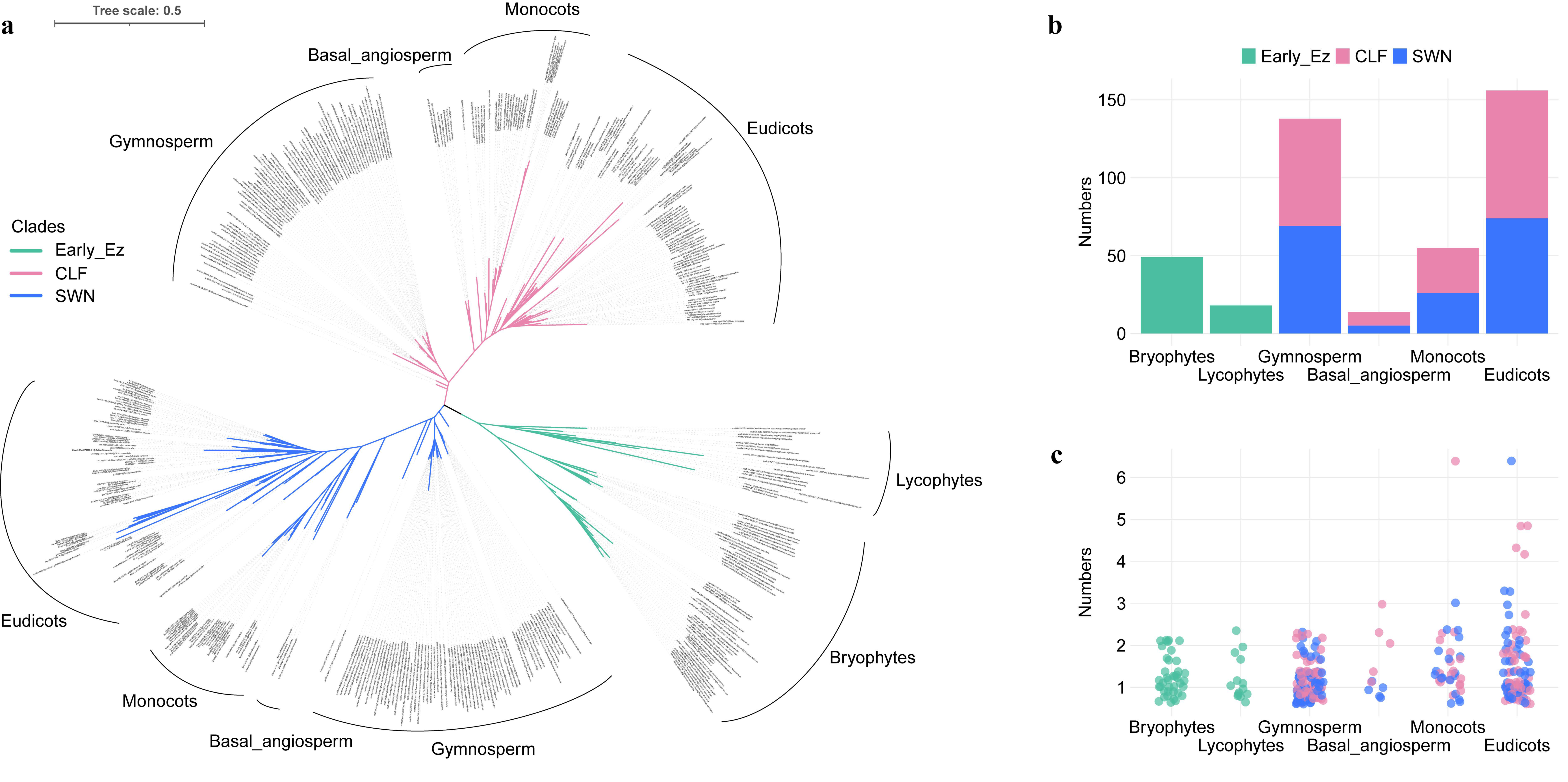
2.1. Identification of E(z) Genes in Green Plants
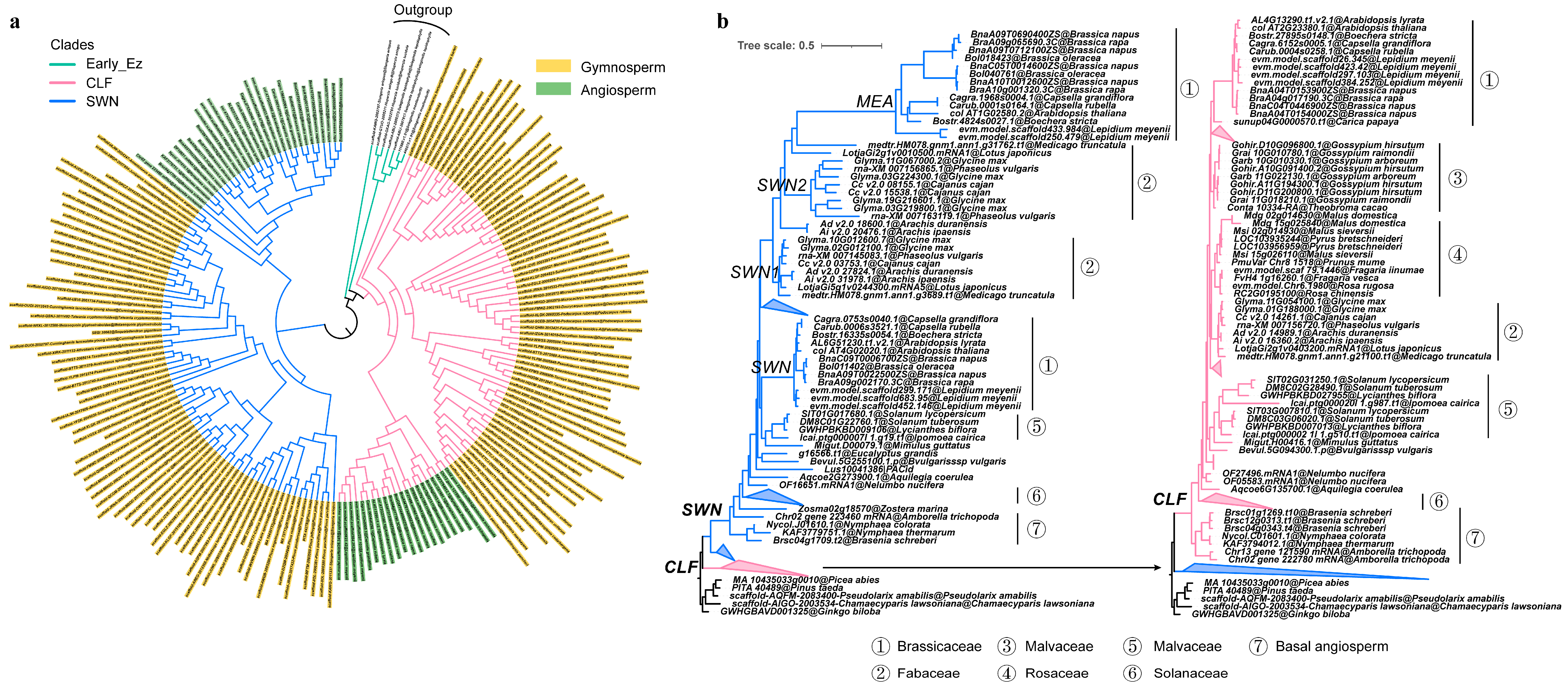
2.2. The Duplication and Divergence of E(z) Genes in Seed Plants
2.3. The Divergence of E(z) Genes in Angiosperms
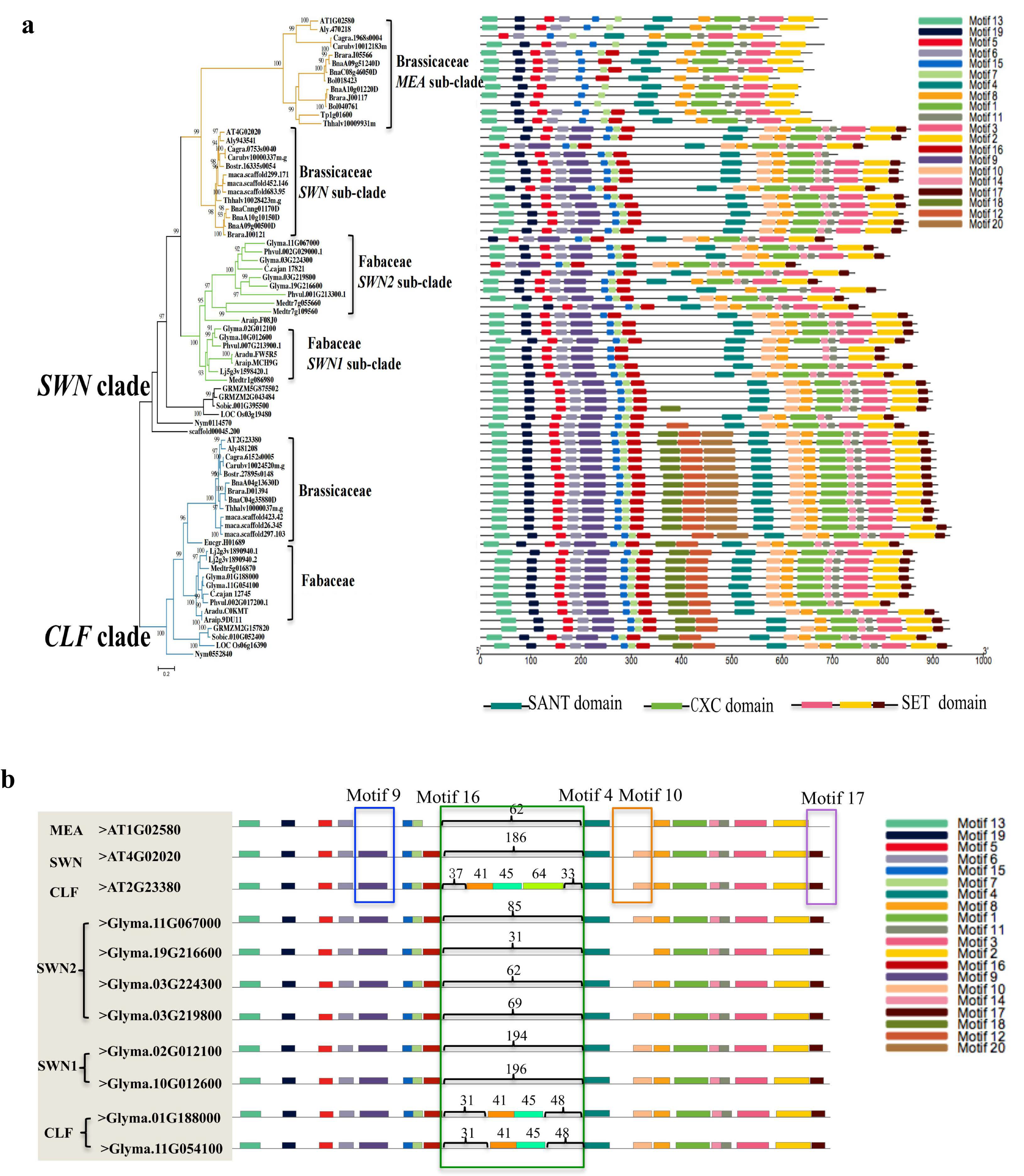
2.4. Motif Patterns and Gene Structure of E(z) Genes in Fabaceae and Brassicaceae
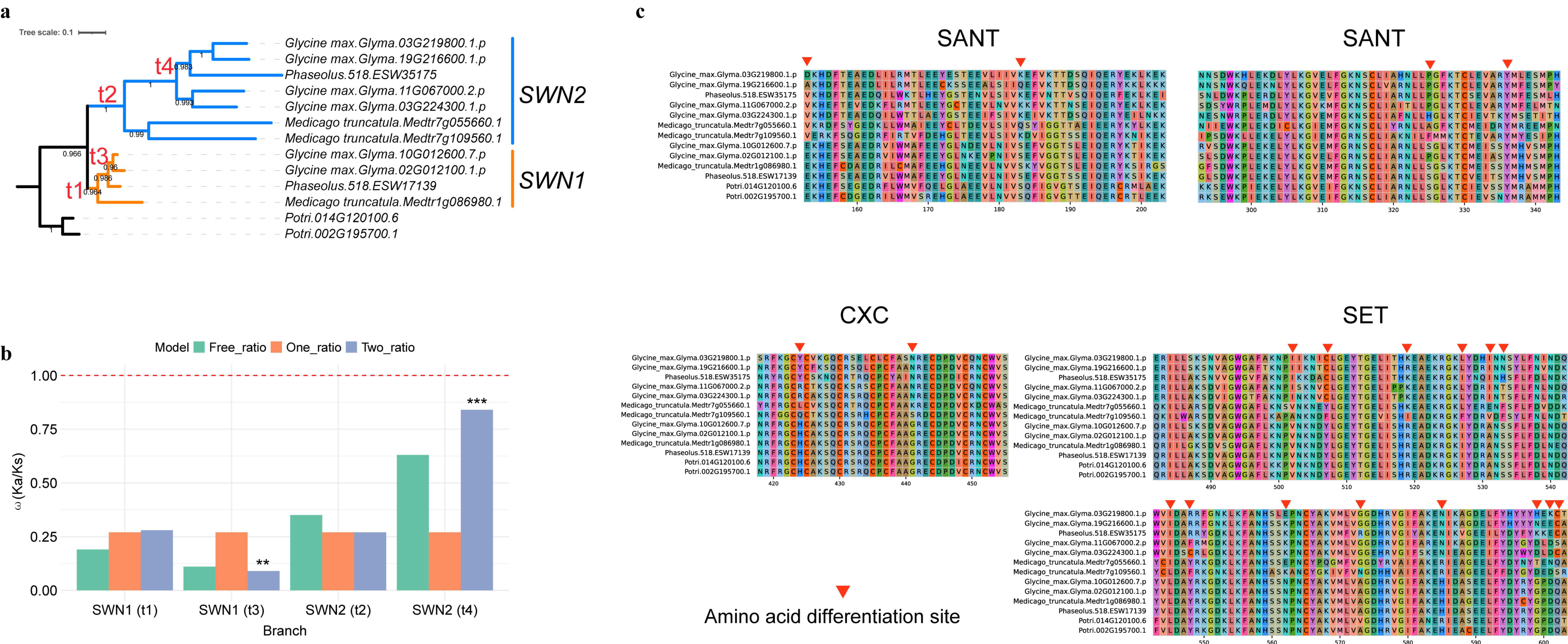
2.5. SWN2 Show Accelerated Amino Acid Substitution Rates and Evidence for Positive Selection in Fabaceae
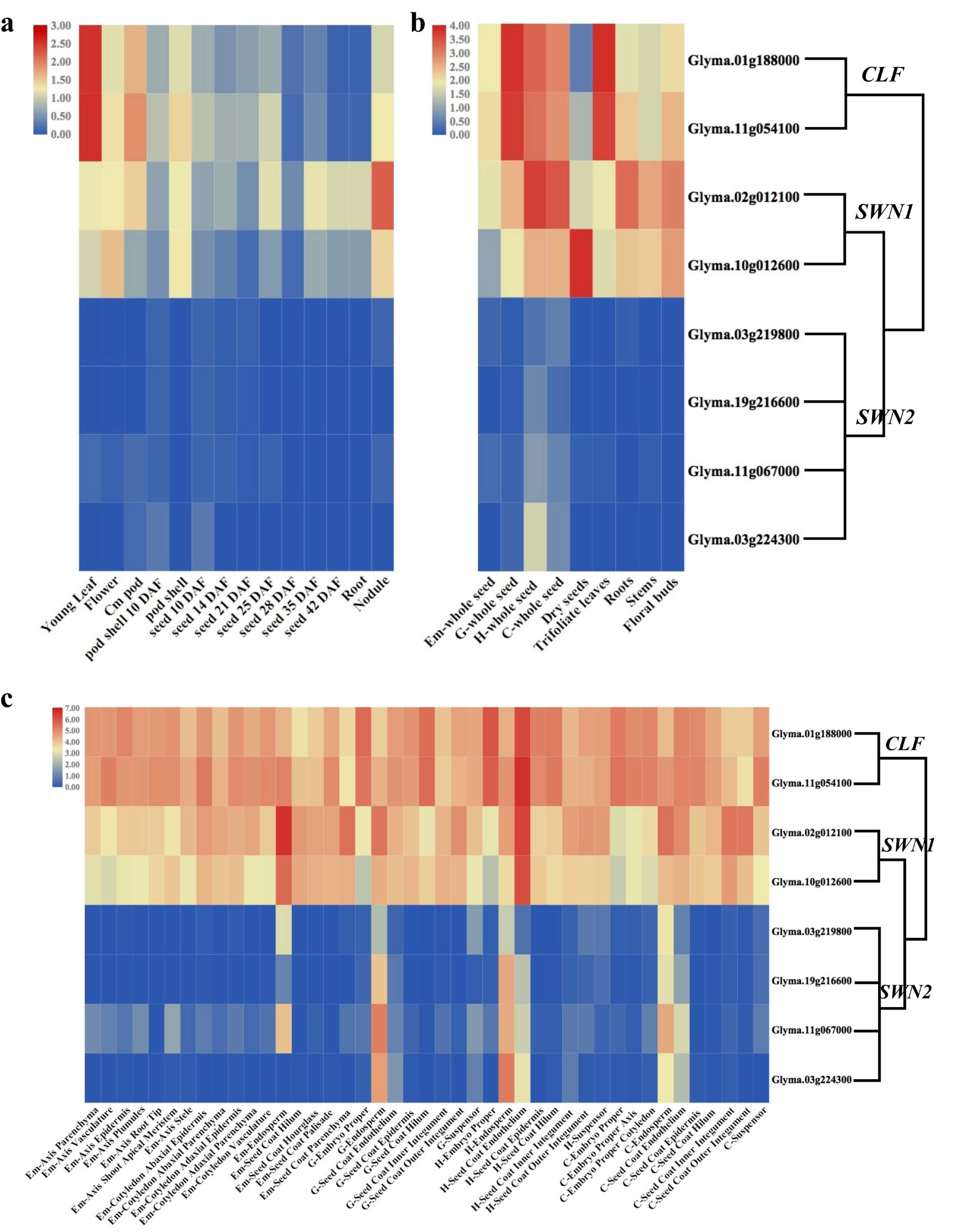
2.6. Expression Profiling of E(z) Genes in G. max
3. Discussion
3.1. Conservation and Divergence of E(z) Genes in Green Plants
3.2. Neo-Functionalization of E(z) in Fabaceae
4. Materials and Methods
4.1. Data Collection
4.2. Identification of E(z) Homolog Genes
4.3. Phylogenetic Analysis
4.4. Motif Patterns and Gene Structure
4.5. Synteny and Selection Analysis
4.6. Expression Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuroda, M.I.; Kang, H.; De, S.; Kassis, J.A. Dynamic Competition of Polycomb and Trithorax in Transcriptional Programming. Annu. Rev. Biochem. 2020, 89, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015, 66, 269. [Google Scholar] [CrossRef]
- Xiao, J.; Wagner, D. Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 2015, 23, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, Y. The functions of E (Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004, 14, 155–164. [Google Scholar] [CrossRef]
- Cheng, X.; Pan, M.; Zhou, Y.; Niu, B.; Chen, C. The maternally expressed polycomb group gene OsEMF2a is essential for endosperm cellularization and imprinting in rice. Plant Commun. 2021, 2, 100092. [Google Scholar] [CrossRef]
- Delaval, K.; Feil, R. Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 2004, 14, 188–195. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007, 8, 9–22. [Google Scholar] [CrossRef]
- Mozgova, I.; Köhler, C.; Hennig, L. Keeping the gate closed: Functions of the polycomb repressive complex PRC 2 in development. Plant J. 2015, 83, 121–132. [Google Scholar] [CrossRef]
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.; Villar, C.B. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008, 18, 236–243. [Google Scholar] [CrossRef]
- Kassis, J.A.; Kennison, J.A.; Tamkun, J.W. Polycomb and Trithorax Group Genes in Drosophila. Genetics 2017, 206, 1699–1725. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, D.H.; Liu, B.Y.; Shen, W.H.; Ruan, Y. Conservation and diversification of polycomb repressive complex 2 (PRC2) proteins in the green lineage. Brief Funct. Genom. 2017, 16, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Bemer, M.; Grossniklaus, U. Dynamic regulation of Polycomb group activity during plant development. Curr. Opin. Plant Biol. 2012, 15, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Aichinger, E.; Villar, C.B.; Farrona, S.; Reyes, J.C.; Hennig, L.; Köhler, C. CHD3 Proteins and Polycomb Group Proteins Antagonistically Determine Cell Identity in Arabidopsis. PLoS Genet. 2009, 5, e1000605. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Dong, L.; Wang, C.; Hao, K.; Wang, J.; Zhao, L.; Xu, L.; Xia, Y.; Jiang, Q.; Qin, J. Functional redundancy among Polycomb complexes in maintaining the pluripotent state of embryonic stem cells. Stem Cell Rep. 2022, 17, 1198–1214. [Google Scholar] [CrossRef]
- Scott, S.; CasasMollano, J.A.; Ronald, L.C.; Cerutti, H. Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics 2010, 5, 301–312. [Google Scholar] [CrossRef]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Shen, X.; Liu, Y.; Hsu, Y.J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.C.; Orkin, S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef]
- Chen, Y.H.; Hung, M.C.; Li, L.Y. EZH2: A pivotal regulator in controlling cell differentiation. Am. J. Transl. Res. 2012, 4, 364–375. [Google Scholar]
- Goodrich, J.; Puangsomlee, P.; Martin, M.; Long, D.; Meyerowitz, E.M.; Coupland, G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 1997, 386, 44–51. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Viellecalzada, J.P.; Hoeppner, M.A.; Gagliano, W.B. Maternal Control of Embryogenesis by MEDEA, a Polycomb Group Gene in Arabidopsis. Science 1998, 280, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Bilodeau, P.; Koltunow, A.; Dennis, E.S.; Peacock, W.J.; Chaudhury, A.M. Genes Controlling Fertilization-Independent Seed Development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 296–301. [Google Scholar] [CrossRef]
- Wang, X.-X.; Ma, L.-G. Polycomb-group (Pc-G) Proteins Control Seed Development in Arabidopsis thaliana L. J. Integr. Plant Biol. 2007, 49, 52–59. [Google Scholar] [CrossRef]
- Okano, Y.; Aono, N.; Hiwatashi, Y.; Murata, T.; Nishiyama, T.; Ishikawa, T.; Kubo, M.; Hasebe, M. A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 16321–16326. [Google Scholar] [CrossRef]
- Hemenway, E.A.; Gehring, M. Epigenetic Regulation During Plant Development and the Capacity for Epigenetic Memory. Annu. Rev. Plant. Biol. 2023, 74, 87–109. [Google Scholar] [CrossRef]
- Spillane, C.; Schmid, K.J.; Laoueillé-Duprat, S.; Pien, S.; Escobar-Restrepo, J.M.; Baroux, C.; Gagliardini, V.; Page, D.R.; Wolfe, K.H.; Grossniklaus, U.J.N. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 2007, 448, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Berr, A.; Xu, L.; Gao, J.; Cognat, V.; Steinmetz, A.; Dong, A.; Shen, W.H. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009, 151, 1476–1485. [Google Scholar] [CrossRef]
- Zhang, D.; Martyniuk, C.J.; Trudeau, V.L. SANTA domain: A novel conserved protein module in Eukaryota with potential involvement in chromatin regulation. Bioinformatics 2006, 22, 2459–2462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Y.; Liang, Y.; Zhou, D.; Li, S.; Lin, S.; Dong, H.; Huang, L. The function of histone lysine methylation related SET domain group proteins in plants. Protein Sci. 2020, 29, 1120–1137. [Google Scholar] [CrossRef]
- Severin, A.J.; Woody, J.L.; Bolon, Y.T.; Joseph, B.; Shoemaker, R.C. RNA-seq atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef]
- Danzer, J.; Mellott, E.; Bui, A.Q.; Le, B.H.; Martin, P.; Hashimoto, M.; Perez-Lesher, J.; Chen, M.; Pelletier, J.M.; Somers, D.A.; et al. Down-Regulating the Expression of 53 Soybean Transcription Factor Genes Uncovers a Role for SPEECHLESS in Initiating Stomatal Cell Lineages during Embryo Development. Plant Physiol. 2015, 168, 1025–1035. [Google Scholar] [CrossRef]
- Simonini, S.; Bemer, M.; Bencivenga, S.; Gagliardini, V.; Pires, N.D.; Desvoyes, B.; Graaff, E.V.D.; Gutierrez, C.; Grossniklaus, U. The Polycomb group protein MEDEA controls cell proliferation and embryonic patterning in Arabidopsis. Dev. Cell. 2021, 56, 945–1960. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.J.; Ploetz, L. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2007, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Lyons, E.; Freeling, M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008, 53, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Leebens-Mack, J.; Wickett, N.; Deyholos, M.K.; Degironimo, L.; Pires, J.C.J.N. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Sneddon, T.P.; Si Zhe, X.; Edmunds, S.C.; Li, P.; Goodman, L.; Hunter, C.I.J.D. GigaDB: Promoting data dissemination and reproducibility. Database J. Biol. Databases Curation 2014, 2014, bau018. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.; Chenhao, Z.; Jinghua, H.; Chunjin, L.; Yanhong, F.; Yongfeng, Z.; Rui, C.; Haibin, L.; Xiaoming, S.J.H.R. plantGIR: A genomic database of plants. Hortic. Res. 2024, 12, uhae342. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Birney, E.; Durbin, R.; Eddy, S.R.; Howe, K.L.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2000, 28, 263–266. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2022, 51, D384–D388. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020, 49, D458–D460. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. Bmc Bioinform. 2004, 5, 113. [Google Scholar]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R.J.M.p. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, 1152–1158. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete Transcriptome of the Soybean Root Hair Cell, a Single-Cell Model, and Its Alteration in Response to Bradyrhizobium japonicum Infection. Plant Physiol. 2009, 152, 541–552. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, X.; Chen, Z.; Zhang, L.; Chang, X. Conservation and Divergence of E(z) Genes in Green Plants. Plants 2025, 14, 3444. https://doi.org/10.3390/plants14223444
Gan X, Chen Z, Zhang L, Chang X. Conservation and Divergence of E(z) Genes in Green Plants. Plants. 2025; 14(22):3444. https://doi.org/10.3390/plants14223444
Chicago/Turabian StyleGan, Xiaolong, Zihua Chen, Liangsheng Zhang, and Xiaojun Chang. 2025. "Conservation and Divergence of E(z) Genes in Green Plants" Plants 14, no. 22: 3444. https://doi.org/10.3390/plants14223444
APA StyleGan, X., Chen, Z., Zhang, L., & Chang, X. (2025). Conservation and Divergence of E(z) Genes in Green Plants. Plants, 14(22), 3444. https://doi.org/10.3390/plants14223444





