Genome-Wide Identification of DnaJ Gene Family and VIGS Analysis Reveal the Function of GhDnaJ316 in Floral Development for Upland Cotton
Abstract
1. Introduction
2. Results
2.1. Identification and Physiochemical Properties of GhDnaJs
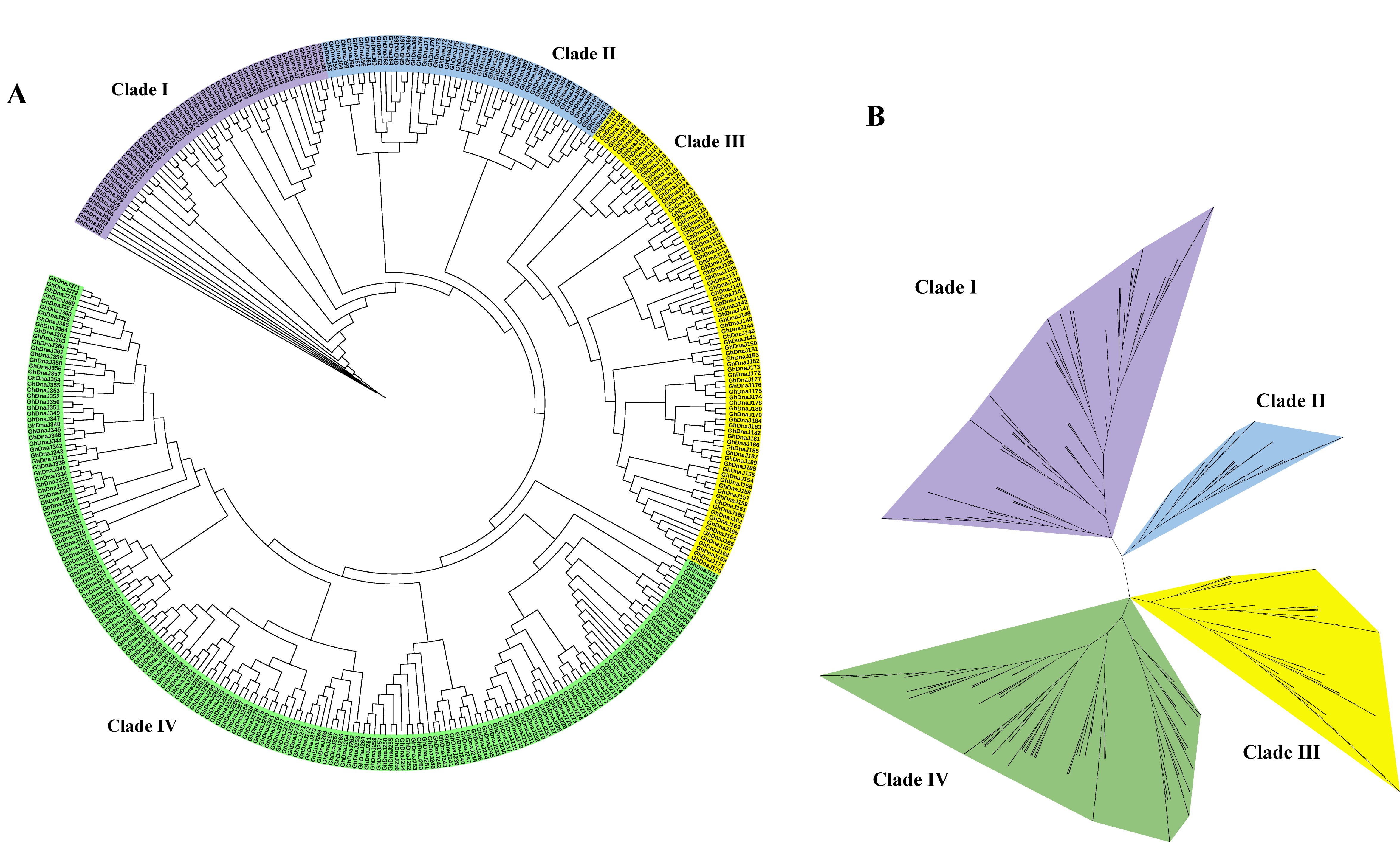
2.2. Phylogenetic Tree Analysis
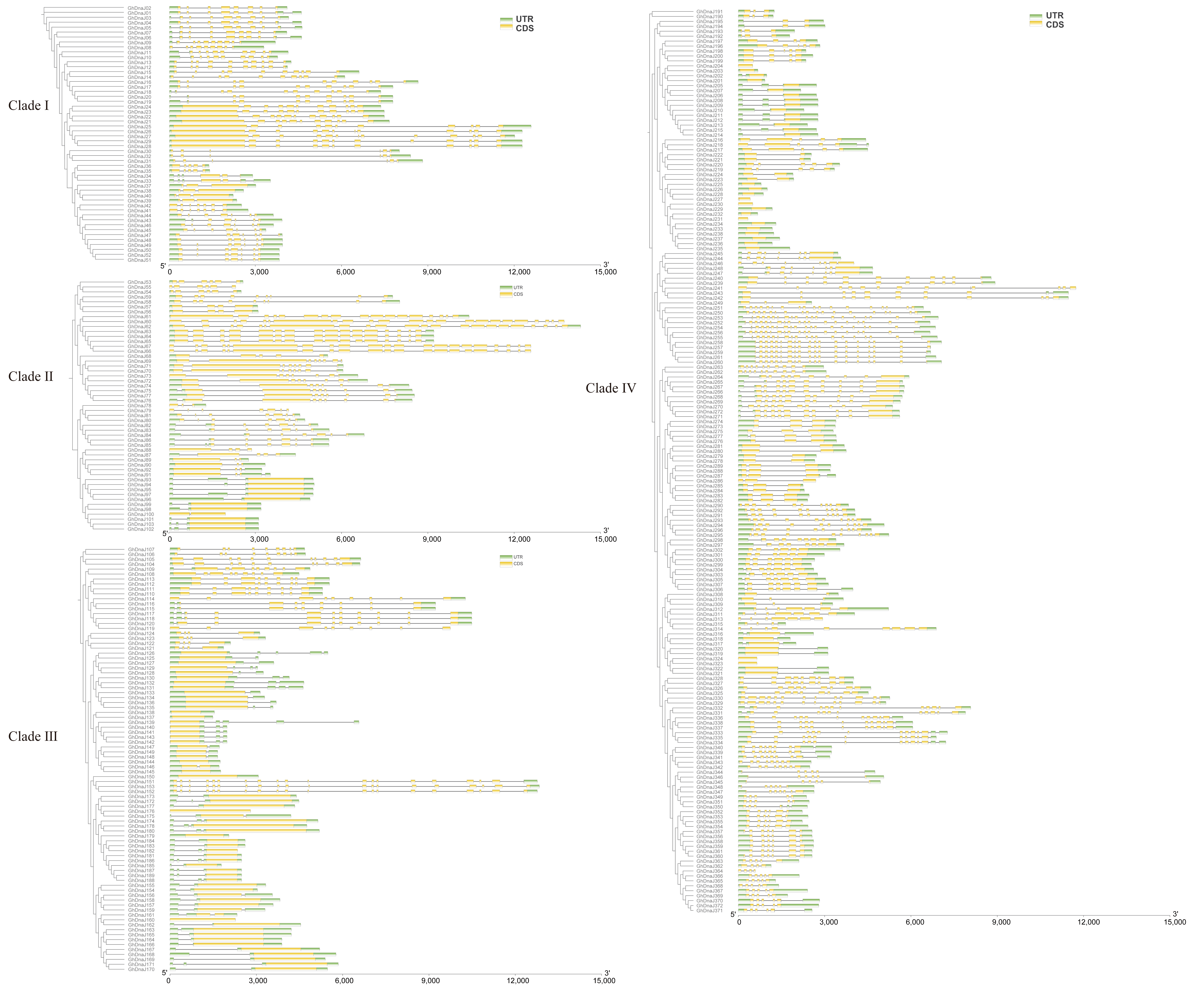
2.3. Gene Structure and Conserved Domain
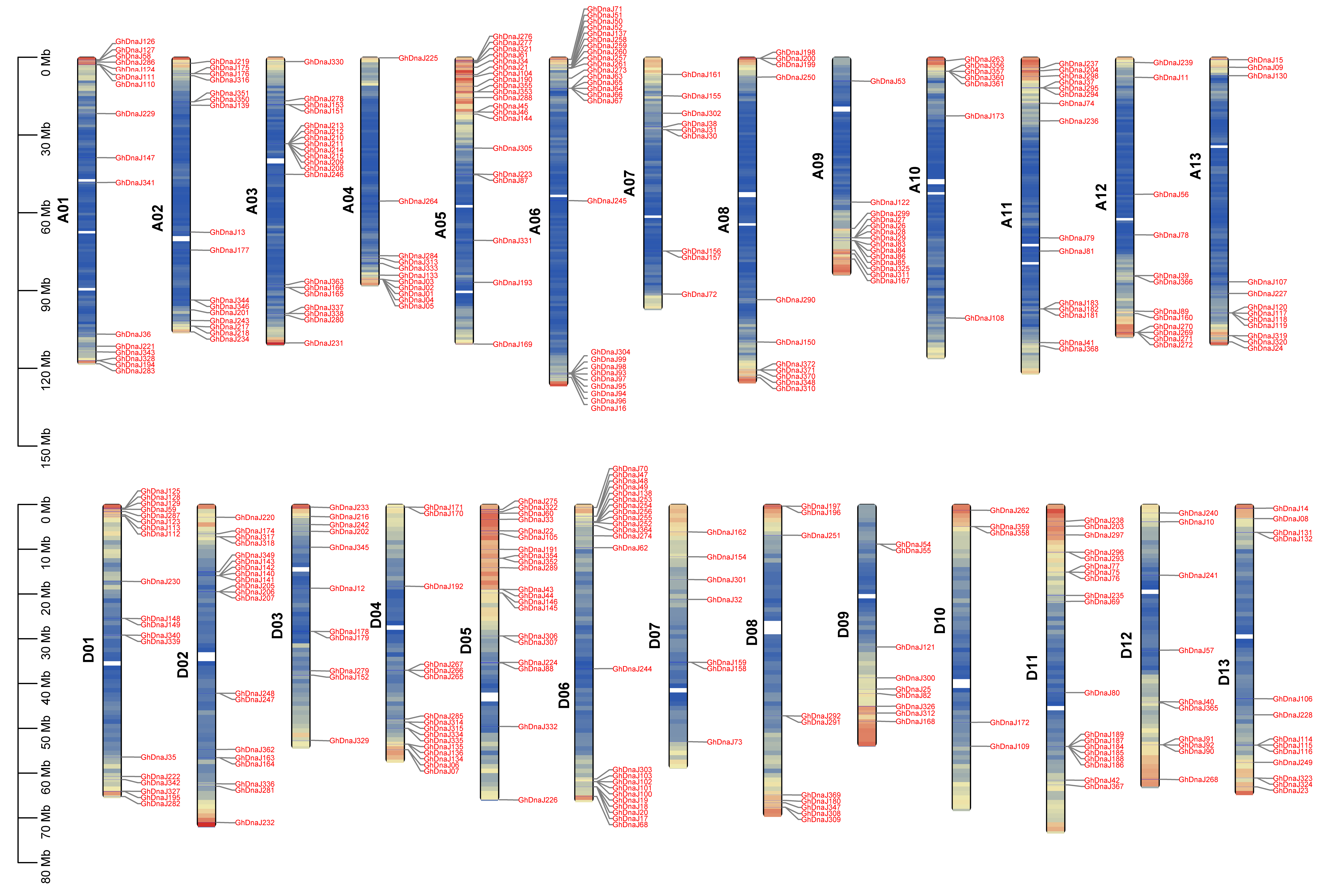
2.4. Chromosome Location, Gene Structures, and Cis-Acting Elements
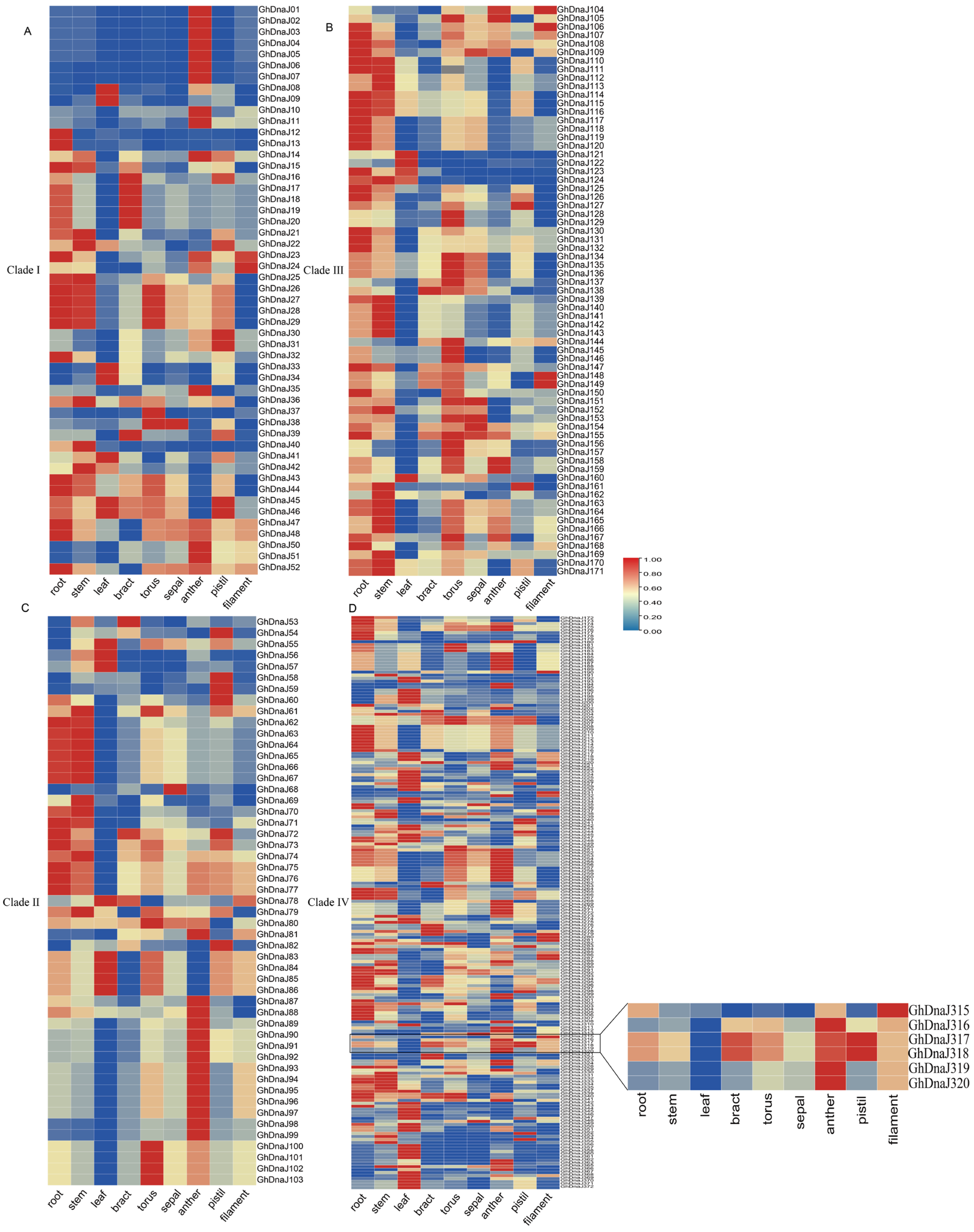
2.5. Analysis of the DnaJs Expression Pattern
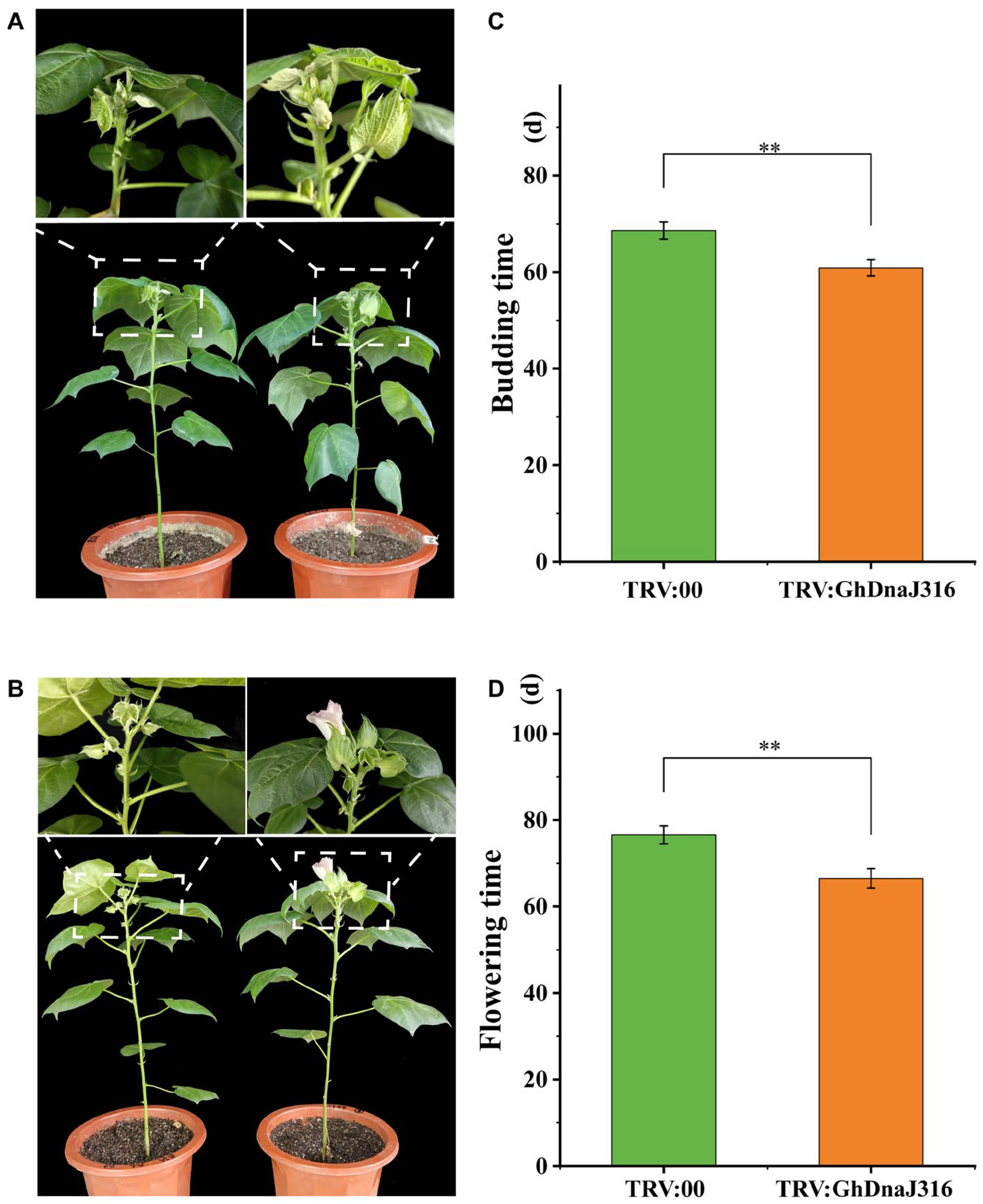
2.6. Functional Validation of GhDnaJ316 in the Floral Development of Upland Cotton
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of DnaJ Gene Family
4.2. Physicochemical Properties and Subcellular Localization
4.3. Chromosomal Localization and Structural Prediction
4.4. Analysis of Conserved Motifs, Gene Structure, and Cis-Acting Elements
4.5. Phylogenetic Analysis of the DnaJ Gene Family
4.6. Tissue-Specific Expression Analysis
4.7. Silencing of GhDnaJ316 by VIGS in Upland Cotton
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Georgopoulos, C.P.; Lundquist-Heil, A.; Yochem, J.; Feiss, M. Identification of the E. coli DnaJ gene product. Mol. Genet. Genom. 1980, 178, 583–588. [Google Scholar] [CrossRef]
- Liberek, K.; Marszalek, J.; Ang, D.; Georgopoulos, C.; Zylicz, M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA 1991, 88, 2874–2878. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, F.; Nicoll, W.S.; Zimmermann, R.; Cheetham, M.E.; Blatch, G.L. Not all J domains are created equal: Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005, 14, 1697–1709. [Google Scholar] [CrossRef]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Rajan, V.B.V.; D’Silva, P. Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genom. 2009, 9, 433–446. [Google Scholar] [CrossRef]
- Verma, A.K.; Tamadaddi, C.; Tak, Y.; Lal, S.S.; Cole, S.J.; Hines, J.K.; Sahi, C. The expanding world of plant J-domain proteins. Crit. Rev. Plant Sci. 2019, 38, 382–400. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A. Role of DnaK-DnaJ proteins in PSII repair. Plant Physiol. 2020, 182, 1804–1805. [Google Scholar] [CrossRef]
- Chen, K.M.; Holmström, M.; Raksajit, W.; Suorsa, M.; Piippo, M.; Aro, E.-M. Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 43. [Google Scholar] [CrossRef]
- Zhang, J.; Bai, Z.; Ouyang, M.; Xu, X.; Xiong, H.; Wang, Q.; Grimm, B.; Rochaix, J.D.; Zhang, L. The DnaJ proteins DJA6 and DJA5 are essential for chloroplast iron-sulfur cluster biogenesis. EMBO J. 2021, 40, e106742. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Jia, T.; Jiao, Q.; Hu, X. Research Progress in J-Proteins in the chloroplast. Genes 2022, 13, 1469. [Google Scholar] [CrossRef]
- Vitha, S.; Froehlich, J.E.; Koksharova, O.; Pyke, K.A.; Erp, H.; Osteryoung, K.W. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 2003, 15, 1918–1933. [Google Scholar] [CrossRef]
- Takano, A.; Suetsugu, N.; Wada, M.; Kohda, D. Crystallographic and functional analyses of J-domain of JAC1 essential for chloroplast photorelocation movement in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1372–1376. [Google Scholar] [CrossRef]
- Pulido, P.; Toledo-Ortiz, G.; Phillips, M.A.; Wright, L.P.; Rodríguez-Concepción, M. Arabidopsis J-Protein J20 Delivers the First Enzyme of the Plastidial Isoprenoid Pathway to Protein Quality Control. Plant Cell 2013, 25, 4183–4194. [Google Scholar] [CrossRef]
- Kong, F.; Deng, Y.; Zhou, B.; Wang, G.; Wang, Y.; Meng, Q. A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 2014, 65, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Pulido, P.; Llamas, E.; Llorente, B.; Ventura, S.; Wright, L.P.; Rodríguez-Concepción, M. Specific Hsp100 chaperones determine the fate of the first enzyme of the plastidial isoprenoid pathway for either refolding or degradation by the stromal Clp protease in Arabidopsis. PLoS Genet. 2016, 27, e1005824. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ren, Y.; Luo, H.; Liu, H.; Wu, J.; Hu, S.; Jiang, Y.; Wang, G. Identification and functional analysis of J-domain proteins involved in determining flowering time in Phalaenopsis orchids. Plant Physiol. Biochem. 2025, 226, 110081. [Google Scholar] [CrossRef]
- Zhi, L.; Zhang, H.; Mao, Y.; Liang, L.; Ni, H.; Li, P.; Huang, J.; Zhu, Y.; Hu, X. A J-domain protein J3 antagonizes ABI5-binding protein2 to Regulate constans stability and flowering time. Plant Cell Environ. 2025, 48, 4743–4755. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Uji, S.; Sugiyama, T.; Sakamoto, T.; Kimura, S.; Endo, T.; Nishikawa, S. ERdj3B-mediated quality control maintains anther development at high temperatures. Plant Physiol. 2020, 182, 1979–1990. [Google Scholar] [CrossRef]
- Park, M.Y.; Kim, S.Y. The Arabidopsis J protein AtJ1 is essential for seedling growth, flowering time control and ABA response. Plant Cell Physiol. 2014, 55, 2152–2163. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, L.; Wang, J.; Lu, N.; Ma, W.; Ma, J.; Zhang, Y.; Fu, P.; Yao, C.; Hu, J.; et al. Genome-wide identification of DnaJ gene family in Catalpa bungei and functional analysis of CbuDnaJ49 in leaf color formation. Front. Plant Sci. 2023, 14, 1116063. [Google Scholar] [CrossRef]
- Lee, K.-W.; Rahman, M.A.; Kimi, K.-Y.; Choi, G.J.; Cha, J.-Y.; Cheong, M.S.; Shohael, A.M.; Jones, C.; Lee, S.-H. Overexpression of the alfalfa DnaJ-like protein (MsDJLP) gene enhances tolerance to chilling and heat stresses in transgenic tobacco plants. Turk. J. Biol. 2018, 42, 12–22. [Google Scholar] [CrossRef]
- Wang, G.; Cai, G.; Xu, N.; Zhang, L.; Sun, X.; Guan, J.; Meng, Q. Novel DnaJ protein facilitates thermotolerance of transgenic tomatoes. Int. J. Mol. Sci. 2019, 20, 367. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.K.; Dubeaux, G.; Takahashi, Y.; Schroeder, J.I. Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 2021, 105, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, M.; Yu, J.; Ma, A.; Wang, J.; Yun, D.-J.; Xu, Z.-Y. Plasma membrane-localized Hsp40/DNAJ chaperone protein facilitates OsSUVH7-OsBAG4-OsMYB106 transcriptional complex formation for OsHKT1;5 activation. J. Integr. Plant Biol. 2023, 65, 265–279. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Jans, Y.; Bloh, W.V.; Schaphoff, S.; Müller, C. Global cotton production under climate change-Implications for yield and water consumption. Hydrol. Earth Syst. Sci. 2021, 25, 2027–2044. [Google Scholar] [CrossRef]
- Feng, S.; Xu, M.; Liu, F.; Cui, C.; Zhou, B. Reconstruction of the full-length transcriptome atlas using PacBio Iso-Seq provides insight into the alternative splicing in Gossypium australe. BMC Plant Biol. 2019, 19, 365. [Google Scholar] [CrossRef]
- Pan, Y.; Meng, F.; Wang, X. Sequencing multiple cotton genomes reveals complex structures and lays foundation for breeding. Front. Plant Sci. 2020, 11, 560096. [Google Scholar] [CrossRef]
- Farooq, M.; Naqvi, R.Z.; Amin, I.; Rehman, A.U.; Asif, M.; Mansoor, S. Transcriptome diversity assessment of Gossypium arboreum (FDH228) leaves under control, drought and whitefly infestation using PacBio long reads. Gene 2023, 852, 147065. [Google Scholar] [CrossRef]
- Su, J.; Li, D.; Yuan, W.; Li, Y.; Ju, J.; Wang, N.; Ling, P.; Feng, K.; Wang, C. Integrating RTM-GWAS and meta-QTL data revealed genomic regions and candidate genes associated with the first fruit branch node and its height in upland cotton. Theor. Appl. Genet. 2024, 137, 207. [Google Scholar] [CrossRef]
- Wang, C.; Liu, J.; Xie, X.; Wang, J.; Ma, Q.; Chen, P.; Yang, D.; Ma, X.; Hao, F.; Su, J. GhAP1-D3 positively regulates flowering time and early maturity with no yield and fiber quality penalties in upland cotton. J. Integr. Plant Biol. 2023, 65, 985–1002. [Google Scholar] [CrossRef]
- Zhang, X.; Li, D.; Guo, X.; Yang, Q.; Xu, W.; Yu, X.; Yang, J.; Wang, F.; Su, J.; Wang, C. Comprehensive evolutionary, differential expression and VIGS analyses reveal the function of GhNST1 in regulating drought tolerance and early maturity in upland cotton. Funct. Integr. Genom. 2025, 25, 195. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G.; et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Reddy, K.R.; Brand, D.; Wijewardana, C.; Gao, W. Temperature effects on cotton seedling emergence, growth, and development. Agron. J. 2017, 109, 1379–1387. [Google Scholar] [CrossRef]
- Zafar, M.M.; Chattha, W.S.; Khan, A.I.; Zafar, S.; Subhan, M.; Saleem, H.; Ali, A.; Ijaz, A.; Zunaira, A.; Qiao, F.; et al. Drought and heat stress on cotton genotypes suggested agro-physiological and biochemical features for climate resilience. Front. Plant Sci. 2023, 14, 1265700. [Google Scholar] [CrossRef]
- Gapare, W.; Conaty, W.; Zhu, Q.; Liu, S.; Stiller, W.; Llewellyn, D.; Wilson, I. Genome-wide association study of yield components and fibre quality traits in a cotton germplasm diversity panel. Euphytica 2017, 213, 66. [Google Scholar] [CrossRef]
- Wang, X.; Kong, F.; Gao, L.; Shen, G.; Duan, B.; Wang, Z.; Xu, D.; Fan, D.; Deng, Y.; Han, Z. Identification of QTLs for early maturity-related traits based on RIL population of two elite cotton cultivars. BMC Plant Biol. 2024, 24, 1243. [Google Scholar] [CrossRef]
- Wang, P.; Wan, J.; Guo, L.; Xiong, X.; Zhao, C.; Liu, Q.; Yu, J.; Xiang, L.; Liu, J.; Li, W.; et al. A MADS-box protein GhAGL8 promotes early flowering and increases yield without compromising fiber quality in cotton. Ind. Crops Prod. 2025, 225, 120545. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Gierasch, L.M. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2018, 294, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Shahrajabian, M.H.; Sun, W.; Cheng, Q. Molecular breeding and the impacts of some important genes families on agronomic traits, a review. Genet. Resour. Crop Evol. 2021, 68, 1709–1730. [Google Scholar] [CrossRef]
- Nestor, B.J.; Bayer, P.E.; Fernandez, C.G.T.; Edwards, D.; Finnegan, P.M. Approaches to increase the validity of gene family identification using manual homology search tools. Genetica 2023, 151, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chen, Z.; Guo, X.; Tian, D.; Li, C.; Lin, M.; Hu, C.; Yan, J. Genome-wide identification and analysis of maize DnaJ family genes in response to salt, heat, and cold at the seedling stage. Plants 2024, 13, 2488. [Google Scholar] [CrossRef]
- Fan, F.F.; Liu, F.; Yang, X.; Wan, H.; Kang, Y. Global analysis of expression profile of members of DnaJ gene families involved in capsaicinoids synthesis in pepper (Capsicum annuum L). BMC Plant Biol. 2020, 20, 326. [Google Scholar] [CrossRef]
- Liu, T.T.; Xu, M.; Gao, S.; Zhang, Y.; Hu, Y.; Jin, P.; Cai, L.; Cheng, Y.; Chen, J.; Yang, J.; et al. Genome-wide identification and analysis of the regulation wheat DnaJ family genes following wheat yellow mosaic virus infection. J. Integr. Agric. 2022, 21, 153–169. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, Z.; Zhou, R.; Cheng, P.; Li, H.; Wang, Z.; Liu, Y.; Li, M.; Zhao, Z.; Hu, Z.; et al. Genome-wide analysis of soybean DnaJA-family genes and functional characterization of GmDnaJA6 responses to saline and alkaline stress. Crop J. 2023, 11, 1230–1241. [Google Scholar] [CrossRef]
- Chang, X.; He, X.; Li, J.; Liu, Z.; Pi, R.; Luo, X.; Wang, R.; Hu, X.; Lu, S.; Zhang, X.; et al. High-quality Gossypium hirsutum and Gossypium barbadense genome assemblies reveal the landscape and evolution of centromeres. Plant Commun. 2024, 5, 100722. [Google Scholar] [CrossRef]
- Tamadaddi, C.; Verma, A.K.; Zambare, V.; Vairagkar, A.; Diwan, D.; Sahi, C. J-like protein family of Arabidopsis thaliana: The enigmatic cousins of J-domain proteins. Plant Cell Rep. 2022, 41, 1343–1355. [Google Scholar] [CrossRef]
- Chen, T.; Xu, T.; Zhang, T.; Liu, T.; Shen, L.; Chen, Z.; Wu, Y.; Yang, J. Genome-Wide Identification and Characterization of DnaJ Gene Family in Grape (Vitis vinifera L.). Horticulturae 2021, 7, 589. [Google Scholar] [CrossRef]
- Chen, P.; Xiao, X.; Li, Y.; Gu, L.; Zhang, Y.; Peng, Y.; Wei, F. Genome-wide identification and expression analysis of the DnaJ genes in Gossypium hirsutum. Ind. Crops Prod. 2025, 228, 120904. [Google Scholar] [CrossRef]
- Zhang, X.X.; Li, X.X.; Zhao, R.; Zhou, Y.; Jiao, Y.N. Evolutionary strategies drive a balance of the interacting gene products for the CBL and CIPK gene families. New Phytol. 2020, 226, 1506–1516. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Cheng, S.; Chen, P.; Su, Z.; Ma, L.; Hao, P.; Zhang, J.; Ma, Q.; Liu, G.; Liu, J.; Wang, H.; et al. High-resolution temporal dynamic transcriptome landscape reveals a GhCAL-mediated flowering regulatory pathway in cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2021, 19, 153–166. [Google Scholar] [CrossRef]
- Li, L.; Zhang, C.; Huang, J.; Liu, Q.; Wei, H.; Wang, H.; Liu, G.; Gu, L.; Yu, S. Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2021, 19, 109–123. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.-T.; Guo, X.-F.; Li, D.-D.; Jia, Y.; Wang, C.-H.; Fan, Y.-N.; Wang, C.-X.; Su, J.-J. Genome-Wide Identification of DnaJ Gene Family and VIGS Analysis Reveal the Function of GhDnaJ316 in Floral Development for Upland Cotton. Plants 2025, 14, 3380. https://doi.org/10.3390/plants14213380
Zhang T-T, Guo X-F, Li D-D, Jia Y, Wang C-H, Fan Y-N, Wang C-X, Su J-J. Genome-Wide Identification of DnaJ Gene Family and VIGS Analysis Reveal the Function of GhDnaJ316 in Floral Development for Upland Cotton. Plants. 2025; 14(21):3380. https://doi.org/10.3390/plants14213380
Chicago/Turabian StyleZhang, Ting-Ting, Xue-Feng Guo, Dan-Dan Li, Yun Jia, Chen-Hui Wang, Yu-Nuo Fan, Cai-Xiang Wang, and Jun-Ji Su. 2025. "Genome-Wide Identification of DnaJ Gene Family and VIGS Analysis Reveal the Function of GhDnaJ316 in Floral Development for Upland Cotton" Plants 14, no. 21: 3380. https://doi.org/10.3390/plants14213380
APA StyleZhang, T.-T., Guo, X.-F., Li, D.-D., Jia, Y., Wang, C.-H., Fan, Y.-N., Wang, C.-X., & Su, J.-J. (2025). Genome-Wide Identification of DnaJ Gene Family and VIGS Analysis Reveal the Function of GhDnaJ316 in Floral Development for Upland Cotton. Plants, 14(21), 3380. https://doi.org/10.3390/plants14213380




