Genome-Wide SNP Analysis Reveals Population Structure and Genetic Diversity in Lycium ruthenicum Murr
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genomic DNA Extraction
2.3. SLAF-Seq Library Construction and Sequencing
2.3.1. Reference Genome and Restriction Enzyme Selection
2.3.2. SLAF-Seq Sequencing
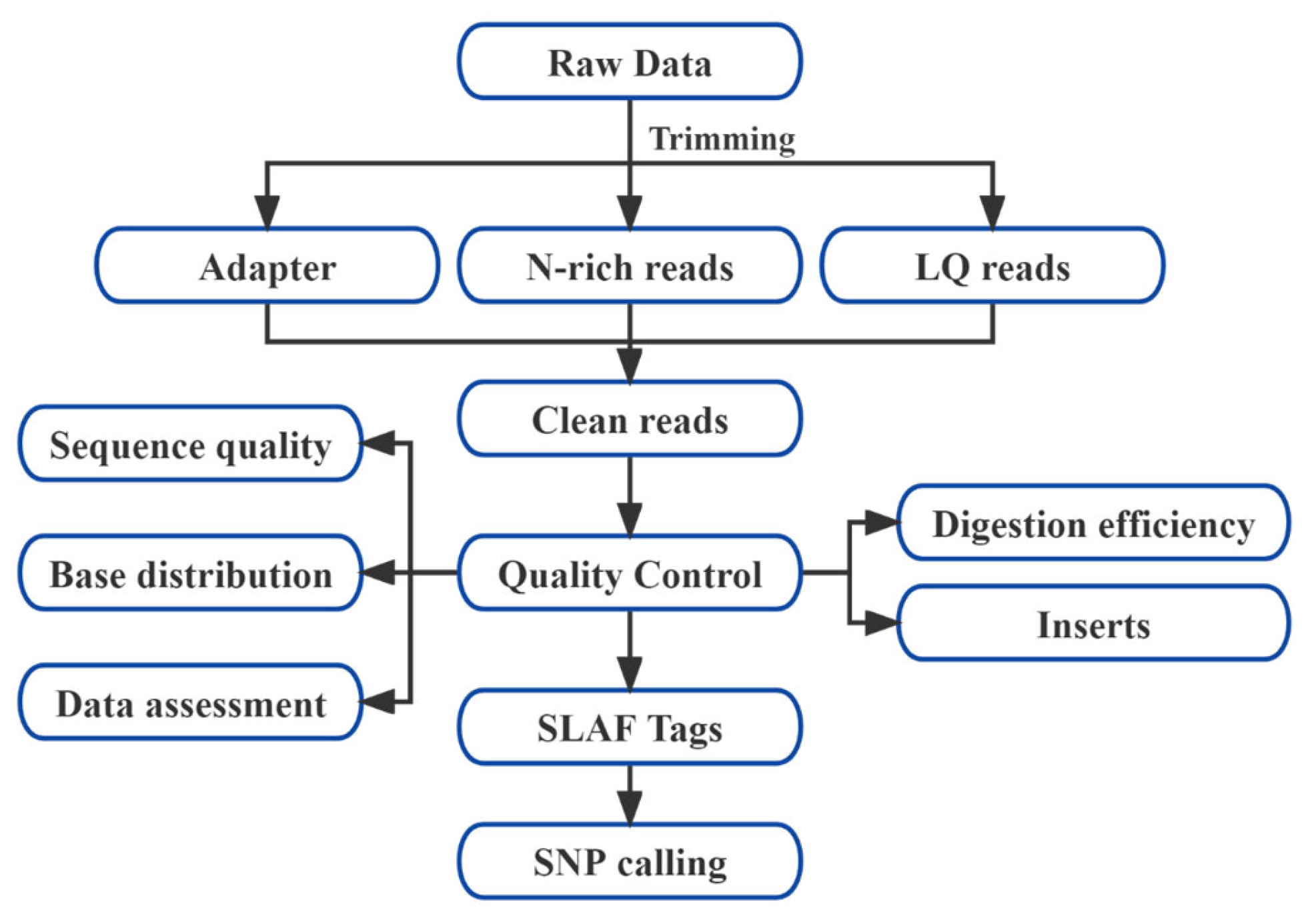
2.4. Data Processing and SNP Discovery
2.5. Population Genetic Structure Analysis
2.6. Genetic Diversity Analysis
2.7. Construction of L. ruthenicum DNA Fingerprints
3. Results and Analysis
3.1. Restriction Enzyme Selection and Library Construction Evaluation
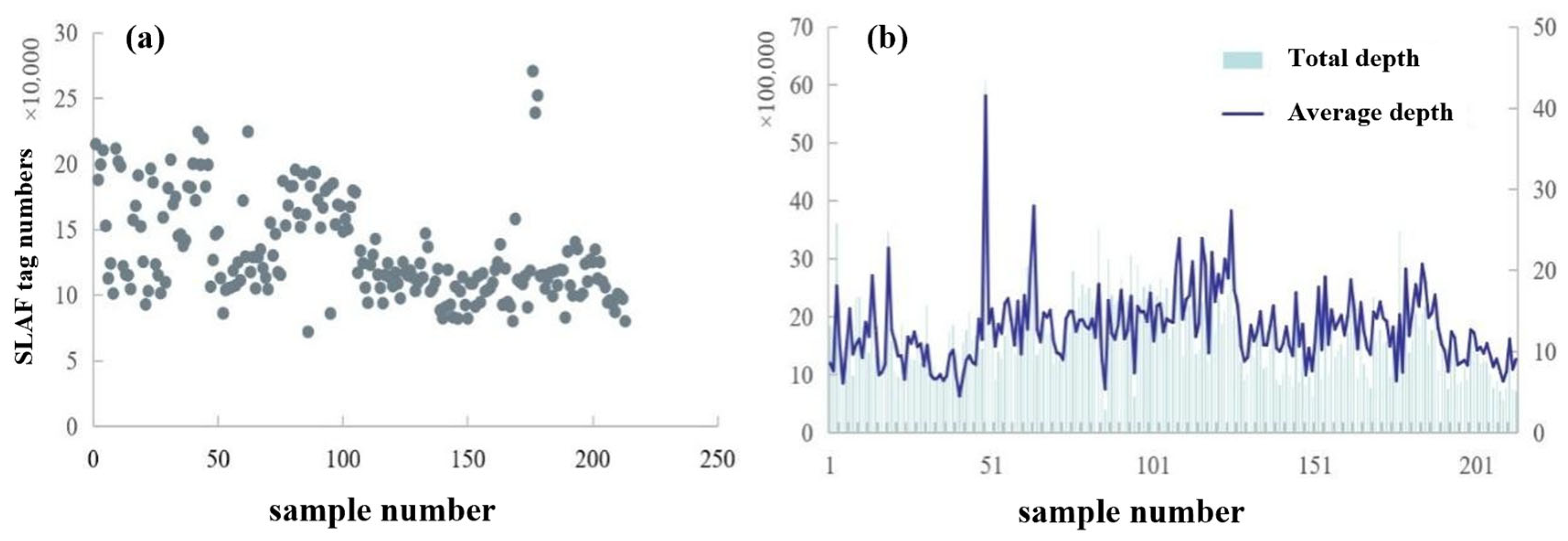
3.2. Statistical Analysis and Quality Assessment of Reduced-Representation Genome Sequencing
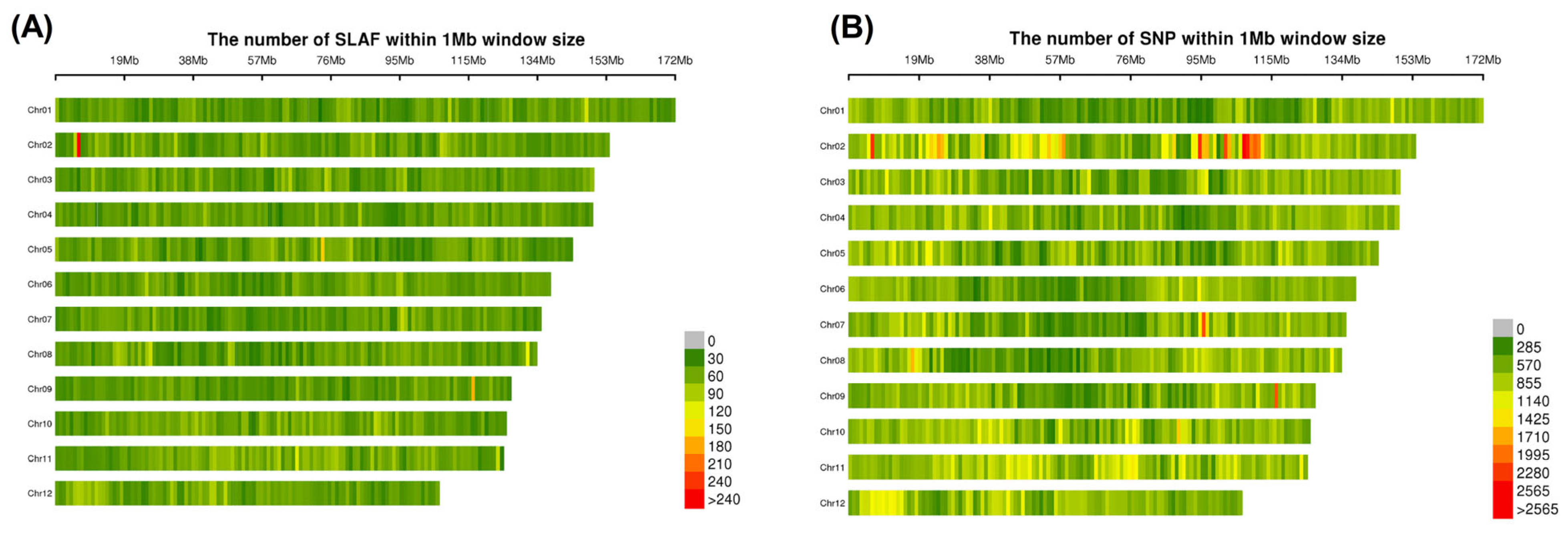
3.3. Development of SLAF Tags and SNP Markers
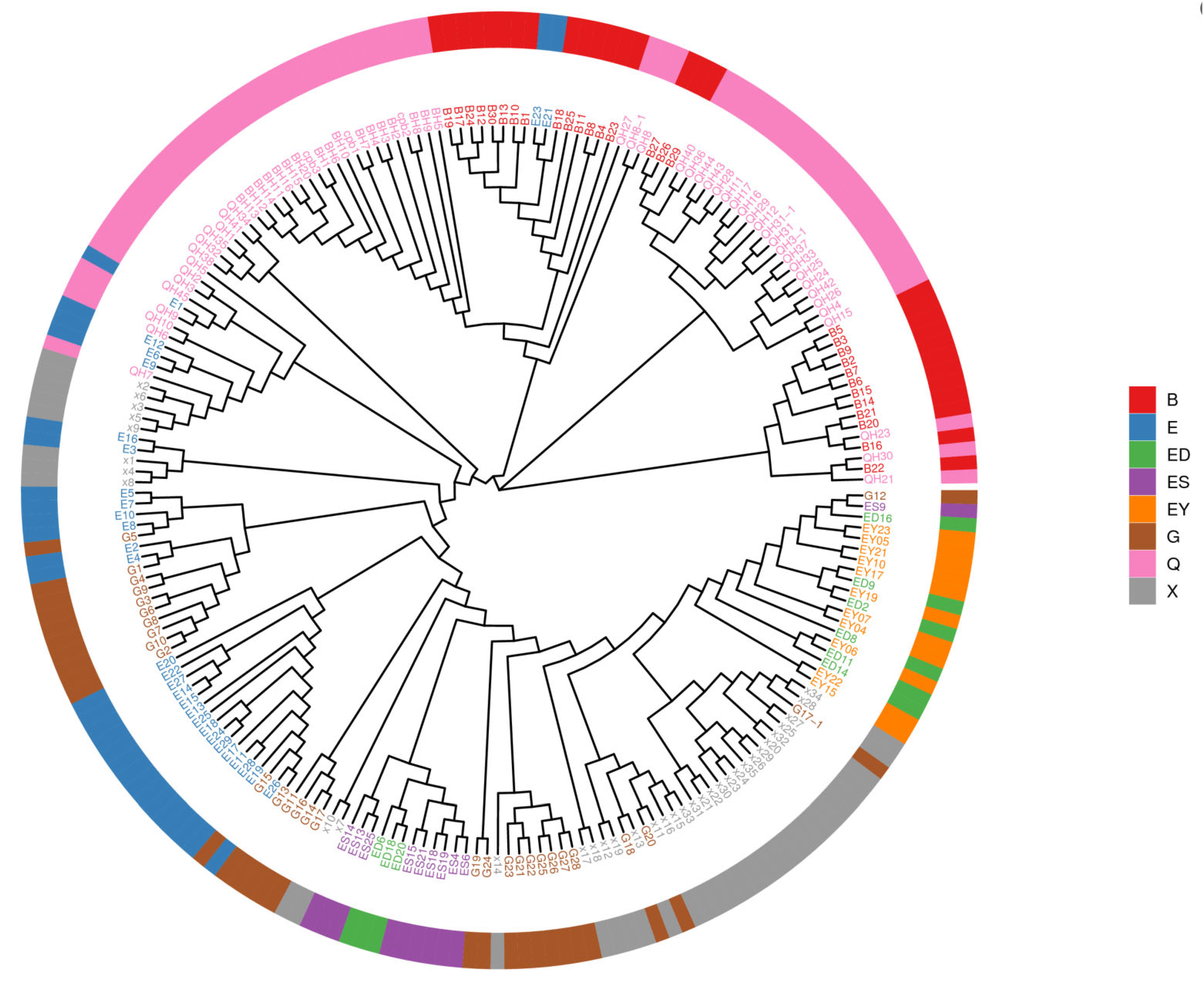
3.4. Phylogenetic Analysis of L. ruthenicum
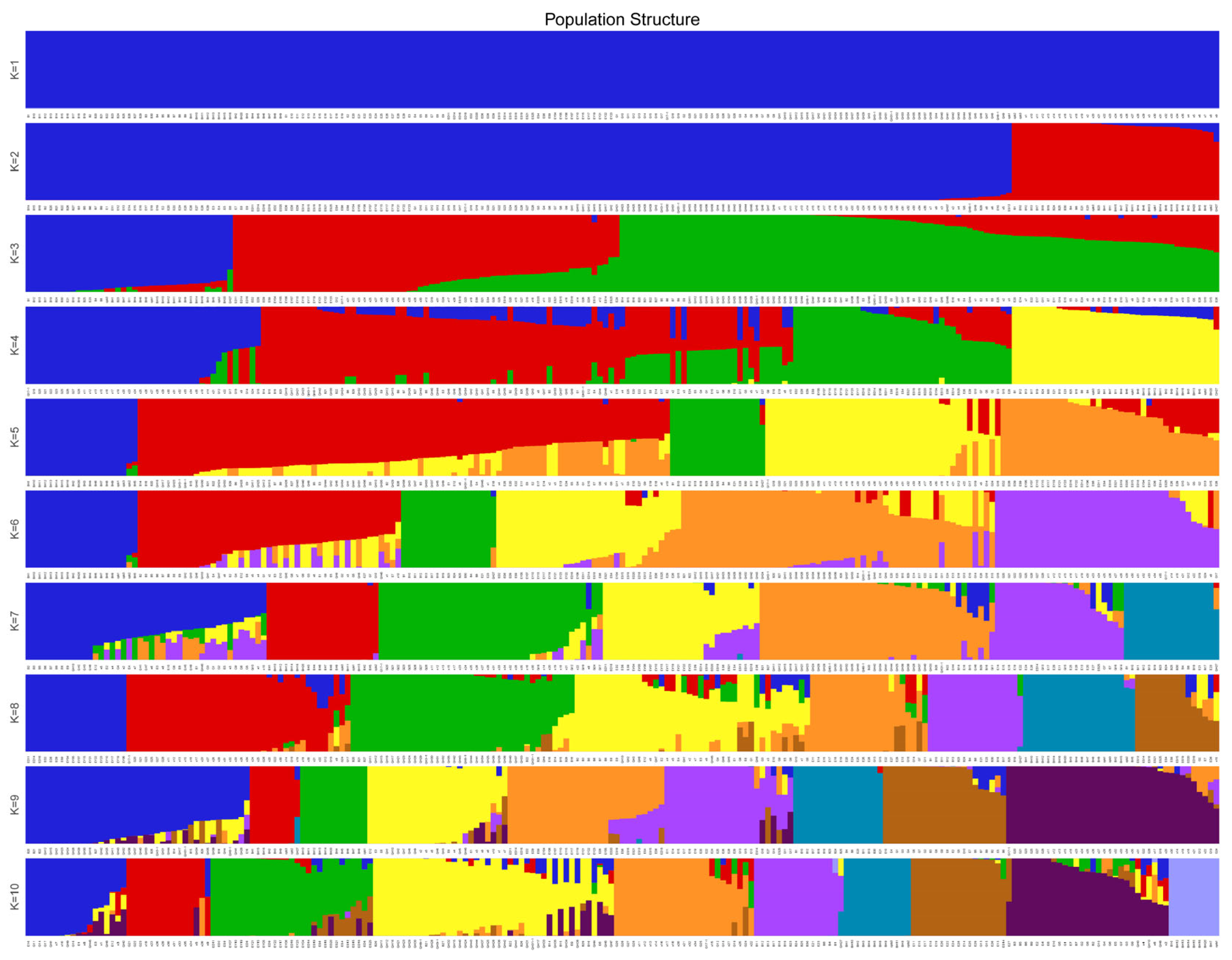
3.5. Population Structure Analysis
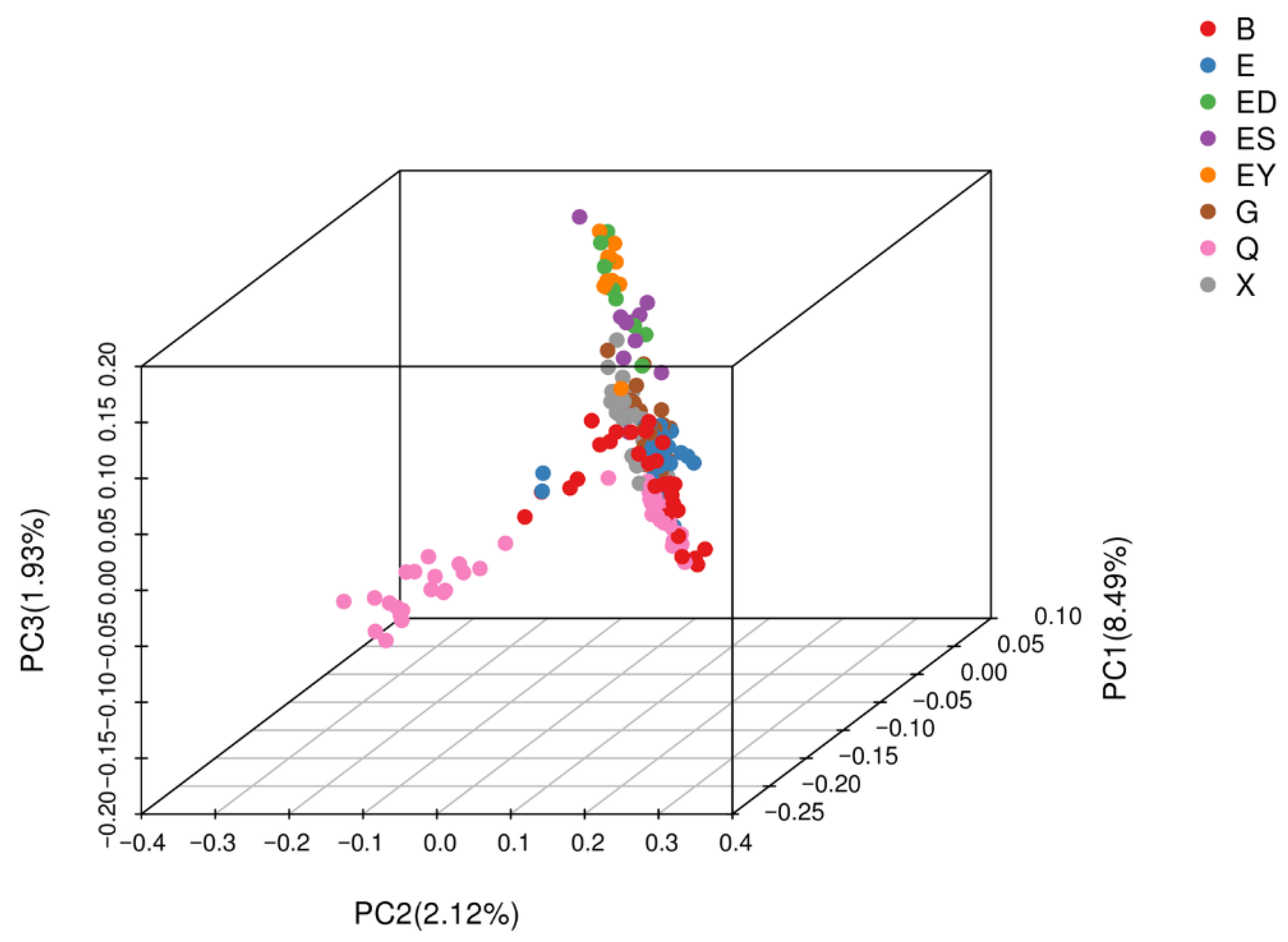
3.6. PCA of Population Structure
3.7. Analysis of Genetic Diversity
3.8. Construction of DNA Fingerprints
4. Discussion
4.1. SNP-Based Identification of L. ruthenicum Germplasm
4.2. Population Structure of L. Germplasm
4.3. Genetic Diversity of L. ruthenicum Germplasm
4.4. Comparative Analysis of SNP-Based and Phenotypic Clustering in L. ruthenicum Germplasm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yao, R.; Heinrich, M.; Weckerle, C.S. The Genus Lycium as Food and Medicine: A Botanical, Ethnobotanical and Historical Review. J. Ethnopharmacol. 2018, 212, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; An, L.; Yu, H.; Yang, M. Endogenous Hormones and Biochemical Changes during Flower Development and Florescence in the Buds and Leaves of Lycium ruthenicum Murr. Forests 2022, 13, 763. [Google Scholar] [CrossRef]
- Tiika, R.J.; Wei, J.; Ma, R.; Yang, H.; Cui, G.; Duan, H.; Ma, Y. Identification and Expression Analysis of the WRKY Gene Family during Different Developmental Stages in Lycium ruthenicum Murr. Fruit. PeerJ 2020, 8, e10207. [Google Scholar] [CrossRef]
- Manju Devi, S.; Joel, A.J.; Raveendran, M.; Pushpam, R.; Muthuramu, S.; Pushpa, R.; Sritharan, N.; Prasanna, P.; Suresh, R. Unravelling Population Structure and Marker Trait Association Using SSR Markers among the Identified Drought Tolerant Rice Landraces (Oryza sativa L.). Czech J. Genet. Plant Breed. 2025, 61, 1–22. [Google Scholar] [CrossRef]
- Tahir, N.; Lateef, D.; Rasul, K.; Rahim, D.; Mustafa, K.; Sleman, S.; Mirza, A.; Aziz, R. Assessment of Genetic Variation and Population Structure in Iraqi Barley Accessions Using ISSR, CDDP, and SCoT Markers. Czech J. Genet. Plant Breed. 2023, 59, 148–159. [Google Scholar] [CrossRef]
- Zhou, Y.; Pan, H. Specific-Locus Amplified Fragment Sequencing (SLAF-Seq). In Plant Genotyping: Methods and Protocols; Shavrukov, Y., Ed.; Springer US: New York, NY, USA, 2023; pp. 165–171. ISBN 978-1-0716-3024-2. [Google Scholar]
- Zhu, Z.; Sun, B.; Lei, J. Specific-Locus Amplified Fragment Sequencing (SLAF-Seq) as High-Throughput SNP Genotyping Methods. In Crop Breeding: Genetic Improvement Methods; Tripodi, P., Ed.; Springer US: New York, NY, USA, 2021; pp. 75–87. ISBN 978-1-0716-1201-9. [Google Scholar]
- Wu, X.; Feng, F.; Zhu, Y.; Xie, F.; Yang, J.; Gong, J.; Liu, Y.; Zhu, W.; Gao, T.; Chen, D.; et al. Construction of High-Density Genetic Map and Identification of QTLs Associated with Seed Vigor after Exposure to Artificial Aging Conditions in Sweet Corn Using SLAF-Seq. Genes 2020, 11, 37. [Google Scholar] [CrossRef]
- Liao, Z.; Xia, X.; Zhang, Z.; Nong, B.; Guo, H.; Feng, R.; Chen, C.; Xiong, F.; Qiu, Y.; Li, D.; et al. Genome-Wide Association Study Using Specific-Locus Amplified Fragment Sequencing Identifies New Genes Influencing Nitrogen Use Efficiency in Rice Landraces. Front. Plant Sci. 2023, 14, 1126254. [Google Scholar] [CrossRef]
- Mandozai, A.; Moussa, A.A.; Zhang, Q.; Qu, J.; Du, Y.; Anwari, G.; Al Amin, N.; Wang, P. Genome-Wide Association Study of Root and Shoot Related Traits in Spring Soybean (Glycine max L.) at Seedling Stages Using SLAF-Seq. Front. Plant Sci. 2021, 12, 568995. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, J.; Cao, J.; Ma, X.; Cao, S.; Li, W.; Yang, G.; Li, S. QTL Analysis of Low-Temperature Tolerance in Maize Germination by SLAF-Seq and BSA Technique. Electron. J. Biotechnol. 2024, 70, 14–22. [Google Scholar] [CrossRef]
- Ren, H.; Han, J.; Wang, X.; Zhang, B.; Yu, L.; Gao, H.; Hong, H.; Sun, R.; Tian, Y.; Qi, X.; et al. QTL Mapping of Drought Tolerance Traits in Soybean with SLAF Sequencing. Crop J. 2020, 8, 977–989. [Google Scholar] [CrossRef]
- Mi, X.; Qiao, D.; An, Y.; Yang, C.; Lin, K.; Peng, C.; Chen, J. Genome-Wide Association Study of Tea Plant Based on SLAF-Seq Revealed SNP Variations Regulating Timing of Bud Flush. Plant Gene 2025, 42, 100511. [Google Scholar] [CrossRef]
- Rehman, F.; Gong, H.; Li, Z.; Zeng, S.; Yang, T.; Ai, P.; Pan, L.; Huang, H.; Wang, Y. Identification of Fruit Size Associated Quantitative Trait Loci Featuring SLAF Based High-Density Linkage Map of Goji Berry (Lycium Spp.). BMC Plant Biol. 2020, 20, 474. [Google Scholar] [CrossRef]
- Wang, C.; Qin, K.; Shang, X.; Gao, Y.; Wu, J.; Ma, H.; Wei, Z.; Dai, G. Mapping Quantitative Trait Loci Associated with Self-(in)Compatibility in Goji Berries (Lycium barbarum). BMC Plant Biol. 2024, 24, 441. [Google Scholar] [CrossRef]
- Rehman, F.; Gong, H.; Ma, Y.; Zeng, S.; Ke, D.; Yang, C.; Zhao, Y.; Wang, Y. An Ultra-Dense Linkage Map Identified Quantitative Trait Loci Corresponding to Fruit Quality- and Size-Related Traits in Red Goji Berry. Front. Plant Sci. 2024, 15, 1390936. [Google Scholar] [CrossRef] [PubMed]
- Rehman, F.; Zeng, S.; Zhao, Y.; Zhao, J.; Qin, K.; Yang, C.; Huang, H.; Wang, Y. Preservation and Innovation of Goji Berry Germplasm Resources. Med. Plant Biol. 2024, 3, e022. [Google Scholar] [CrossRef]
- Ciceoi, R.; Asanica, A.; Luchian, V.; Iordachescu, M. Genomic Analysis of Romanian Lycium Genotypes: Exploring BODYGUARD Genes for Stress Resistance Breeding. Int. J. Mol. Sci. 2024, 25, 2130. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, X.; Zhao, J.; An, W.; Li, Y.; Fan, Y.; Wang, Y.; Cao, Y. Genome-Wide Identification of Microsatellite Markers and Their Application in Genetic Studies of Wolfberry (Lycium barbarum). bioRxiv 2022, 2022-05. [Google Scholar] [CrossRef]
- Aboul-Maaty, N.A.F.; Oraby, H.A.S. Extraction of High-Quality Genomic DNA from Different Plant Orders Applying a Modified CTAB-Based Method. Bull. Natl. Res. Cent. 2019, 43, 25. [Google Scholar] [CrossRef]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-Seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Cao, Y.; Li, Y.; Fan, Y.; Li, Z.; Yoshida, K.; Wang, J.; Ma, X.; Wang, N.; Mitsuda, N.; Kotake, T.; et al. Wolfberry Genomes and the Evolution of Lycium (Solanaceae). Commun. Biol. 2021, 4, 671. [Google Scholar] [CrossRef]
- Davey, J.W.; Cezard, T.; Fuentes-Utrilla, P.; Eland, C.; Gharbi, K.; Blaxter, M.L. Special Features of RAD Sequencing Data: Implications for Genotyping. Mol. Ecol. 2013, 22, 3151–3164. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Trees, R.P. The Neighbor-Joining Method: A New Method For. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal Components Analysis Corrects for Stratification in Genome-Wide Association Studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-Wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, H.; Xiang, X.; Yang, A.; Feng, Q.; Dai, P.; Li, Y.; Jiang, X.; Liu, G.; Zhang, X. Construction of a SNP Fingerprinting Database and Population Genetic Analysis of Cigar Tobacco Germplasm Resources in China. Front. Plant Sci. 2021, 12, 618133. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Zhou, G.; Li, J.; Su, B.; Guo, H. Ursolic Acid Activates the Apoptosis of Prostate Cancer via ROCK/PTEN Mediated Mitochondrial Translocation of Cofilin-1. Oncol. Lett. 2018, 15, 3202–3206. [Google Scholar] [CrossRef] [PubMed]
- Abdul Aziz, M.; Masmoudi, K. Molecular Breakthroughs in Modern Plant Breeding Techniques. Hortic. Plant J. 2025, 11, 15–41. [Google Scholar] [CrossRef]
- Shahnazari, N.; Noormohammadi, Z.; Sheidai, M.; Koohdar, F. A New Insight on Genetic Diversity of Sweet Oranges: CAPs-SSR and SSR Markers. J. Genet. Eng. Biotechnol. 2022, 20, 105. [Google Scholar] [CrossRef]
- Zhao, W.-G.; Chung, J.-W.; Cho, Y.-I.; Rha, W.-H.; Lee, G.-A.; Ma, K.-H.; Han, S.-H.; Bang, K.-H.; Park, C.-B.; Kim, S.-M. Molecular Genetic Diversity and Population Structure in Lycium Accessions Using SSR Markers. Comptes rendus. Biol. 2010, 333, 793–800. [Google Scholar] [CrossRef]
- Chen, Z.; He, Y.; Iqbal, Y.; Shi, Y.; Huang, H.; Yi, Z. Investigation of Genetic Relationships within Three Miscanthus Species Using SNP Markers Identified with SLAF-Seq. BMC Genom. 2022, 23, 43. [Google Scholar] [CrossRef]
- AbuHammad, W.A.; Mamidi, S.; Kumar, A.; Pirseyedi, S.; Manthey, F.A.; Kianian, S.F.; Alamri, M.S.; Mergoum, M.; Elias, E.M. Identification and Validation of a Major Cadmium Accumulation Locus and Closely Associated SNP Markers in North Dakota Durum Wheat Cultivars. Mol. Breed. 2016, 36, 112. [Google Scholar] [CrossRef]
- Taranto, F.; D’Agostino, N.; Greco, B.; Cardi, T.; Tripodi, P. Genome-Wide SNP Discovery and Population Structure Analysis in Pepper (Capsicum Annuum) Using Genotyping by Sequencing. BMC Genom. 2016, 17, 943. [Google Scholar] [CrossRef]
- Khan, H.; Krishnappa, G.; Kumar, S.; Mishra, C.N.; Krishna, H.; Devate, N.B.; Rathan, N.D.; Parkash, O.; Yadav, S.S.; Srivastava, P. Genome-Wide Association Study for Grain Yield and Component Traits in Bread Wheat (Triticum aestivum L.). Front. Genet. 2022, 13, 982589. [Google Scholar] [CrossRef]
- Kassa, M.T.; You, F.M.; Hiebert, C.W.; Pozniak, C.J.; Fobert, P.R.; Sharpe, A.G.; Menzies, J.G.; Humphreys, D.G.; Rezac Harrison, N.; Fellers, J.P. Highly Predictive SNP Markers for Efficient Selection of the Wheat Leaf Rust Resistance Gene Lr16. BMC Plant Biol. 2017, 17, 45. [Google Scholar] [CrossRef]
- Liu, Y.; Teng, Y.; Zheng, J.; Khan, A.; Li, X.; Tian, Y.; Cui, J.; Guo, Q. Analysis of Genetic Diversity in Tea Plant Population and Construction of DNA Fingerprint Profile Using SNP Markers Identified by SLAF-Seq. Horticulturae 2025, 11, 529. [Google Scholar] [CrossRef]
- Fang, H.; Liu, H.; Ma, R.; Liu, Y.; Li, J.; Yu, X.; Zhang, H.; Yang, Y.; Zhang, G. Genome-Wide Assessment of Population Structure and Genetic Diversity of Chinese Lou Onion Using Specific Length Amplified Fragment (SLAF) Sequencing. PLoS ONE 2020, 15, e0231753. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Rehman, F.; Ma, Y.; Zeng, S.; Yang, T.; Huang, J.; Li, Z.; Wu, D.; Wang, Y. Germplasm Resources and Strategy for Genetic Breeding of Lycium Species: A Review. Front. Plant Sci. 2022, 13, 802936. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Du, J.; Khan, M.A.; Cheng, J.; Wei, C.; Mei, Z.; Chen, H.; He, T.; Fu, J. Analysis of Genetic Diversity and Similarities between Different Lycium Varieties Based on ISSR Analysis and RAMP-PCR Markers. World Acad. Sci. J. 2020, 2, 83–90. [Google Scholar] [CrossRef][Green Version]
- Guan, J.; Lv, X.; Cui, G.; Fan, M.; Zhao, X. SCoT analysis on germplasm genetic diversity of 13 Lycium barbarum resources in Zhongning County. Lishizhen Med. Mater. Medica Res. 2022, 33, 2268–2271. [Google Scholar][Green Version]
- Gao, X.; Li, J.; Song, J.; Guo, Q. The SSR Genetic Diversity of Wild Red Fruit Lycium (Lycium barbarum) in Northwest China. Forests 2023, 14, 1598. [Google Scholar] [CrossRef]
- Li, Y.; Fan, Y.; Dai, G.; An, W.; Cao, Y. Analysis of Genetic Diversity for Wolfberry Germplasms by AFLP Technology. Chin. Tradit. Herb. Drugs 2011, 42, 770–773. [Google Scholar]






| No. | Seed Source Location | Province | Code | Coordinates | Elevation (m) |
|---|---|---|---|---|---|
| 1 | Urad Rear Banner, Bayannur City | Inner Mongolia | B (B1-29) | E:106.56° N:40.80° | 996.0 |
| 2 | Dalaihubu Town, Ejin Banner | Inner Mongolia | E (E1-29) | E:100.97° N:41.858° | 899.6 |
| 3 | Subonaoer Sumu, Ejin Banner | Inner Mongolia | ES (ES1-10) | E:101.04° N:42.12° | 869.2 |
| 4 | Dongfeng Town, Ejin Banner | Inner Mongolia | ED (ED1-9) | E:101.07° N:42.01° | 873.0 |
| 5 | Saihantala Sumu, Ejin Banner | Inner Mongolia | EY (EY1-11) | E:100.59° N:41.88° | 911.0 |
| 6 | Guazhou County, Gansu | Gansu | G (G1-29) | E:94.99° N:40.26° | 1274.0 |
| 7 | Wushi County, Aksu, Xinjiang | Xinjiang | X (X1-35) | E:78.53° N:41.30° | 1379.0 |
| 8 | Nuomuhong County, Qinghai | Qinghai | Q (Q1-61) | E:96.19° N:36.25° | 2712.0 |
| Chromosome ID | SLAF Tag Count | Polymorphic SLAF Tags |
|---|---|---|
| Chr01 | 8537 | 5074 |
| Chr02 | 8393 | 4288 |
| Chr03 | 7888 | 4901 |
| Chr04 | 7542 | 4625 |
| Chr05 | 7651 | 4780 |
| Chr06 | 7187 | 4489 |
| Chr07 | 6799 | 4212 |
| Chr08 | 6986 | 4336 |
| Chr09 | 6284 | 3910 |
| Chr10 | 7150 | 4515 |
| Chr11 | 7498 | 4024 |
| Chr12 | 6012 | 4062 |
| Phylogenetic Analysis | B | E | ES | ED | EY | Q | G | X | All | Ratio(%) |
|---|---|---|---|---|---|---|---|---|---|---|
| G1 | 14 | 2 | 0 | 0 | 0 | 23 | 0 | 0 | 39 | 18.31 |
| G2 | 15 | 0 | 0 | 0 | 0 | 24 | 0 | 0 | 39 | 18.31 |
| G3 | 0 | 29 | 9 | 8 | 11 | 14 | 29 | 35 | 135 | 63.38 |
| All | 29 | 31 | 9 | 8 | 11 | 61 | 29 | 35 | 213 | 100% |
| Population Structure | B | E | ES | ED | EY | Q | G | X | All |
|---|---|---|---|---|---|---|---|---|---|
| Q1 | 14 | 1 | 1 | 0 | 0 | 21 | 0 | 0 | 37 |
| Q2 | 0 | 2 | 9 | 9 | 11 | 0 | 13 | 25 | 69 |
| Q3 | 14 | 27 | 0 | 0 | 0 | 40 | 16 | 25 | 107 |
| All | 28 | 30 | 10 | 9 | 11 | 61 | 29 | 50 | 213 |
| PCA Groups | B | E | ES | ED | EY | Q | G | X | All |
|---|---|---|---|---|---|---|---|---|---|
| S1 | 14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 16 |
| S2 | 0 | 0 | 0 | 0 | 0 | 21 | 0 | 0 | 21 |
| S3 | 15 | 26 | 10 | 9 | 11 | 40 | 29 | 50 | 176 |
| All | 20 | 28 | 10 | 9 | 11 | 61 | 29 | 50 | 213 |
| Germplasm Resources | MAF | Ne | He | H | A | Na | Ho | PIC | I |
|---|---|---|---|---|---|---|---|---|---|
| B | 0.275 | 1.000–2.000 (1.506) | 0.041–0.500 (0.293) | 0.042–0.667 (0.312) | 26,302 | 1.000–2.000 (1.809) | 0.038–0.684 (0.075) | 0.040–0.375 (0.233) | 0.101–0.693 (0.436) |
| E | 0.235 | 1.000–2.000 (1.432) | 0.038–0.500 (0.260) | 0.038–0.667 (0.275) | 26,622 | 1.000–2.000 (1.805) | 0.038–0.750 (0.070) | 0.037–0.375 (0.211) | 0.095–0.693 (0.397) |
| ED | 0.296 | 1.000–2.000 (1.332) | 0.105–0.500 (0.191) | 0.111–0.667 (0.217) | 14,115 | 1.000–2.000 (1.499) | 0.111–1.000 (0.092) | 0.099–0.375 (0.151) | 0.215–0.693 (0.281) |
| ES | 0.294 | 1.000–2.000 (1.338) | 0.095–0.500 (0.194) | 0.100–0.667 (0.220) | 14,981 | 1.000–2.000 (1.509) | 0.100–1.000 (0.088) | 0.090–0.375 (0.154) | 0.199–0.693 (0.286) |
| EY | 0.29 | 1.000–2.000 (1.310) | 0.087–0.500 (0.178) | 0.091–0.667 (0.200) | 12,732 | 1.000–2.000 (1.473) | 0.091–1.000 (0.078) | 0.083–0.375 (0.142) | 0.185–0.693 (0.264) |
| G | 0.252 | 1.000–2.000 (1.387) | 0.036–0.500 (0.229) | 0.037–0.667 (0.241) | 22,279 | 1.000–2.000 (1.673) | 0.037–0.731 (0.069) | 0.036–0.375 (0.184) | 0.092–0.693 (0.345) |
| Q | 0.256 | 1.000–2.000 (1.541) | 0.024–0.500 (0.318) | 0.024–0.556 (0.327) | 30,640 | 1.000–2.000 (1.925) | 0.023–0.595 (0.070) | 0.024–0.375 (0.255) | 0.066–0.693 (0.477) |
| X | 0.255 | 1.000–2.000 (1.371) | 0.033–0.500 (0.219) | 0.033–0.667 (0.232) | 21,040 | 1.000–2.000 (1.638) | 0.033–0.667 (0.054) | 0.032–0.375 (0.176) | 0.085–0.693 (0.329) |
| Mean | 0.269 | 1.402 | 0.235 | 0.253 | 21,089 | 1.667 | 0.075 | 0.188 | 0.352 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Wu, X.; Bai, Y.; He, Y.; Chang, S.; Hai, L. Genome-Wide SNP Analysis Reveals Population Structure and Genetic Diversity in Lycium ruthenicum Murr. Plants 2025, 14, 3374. https://doi.org/10.3390/plants14213374
Yang R, Wu X, Bai Y, He Y, Chang S, Hai L. Genome-Wide SNP Analysis Reveals Population Structure and Genetic Diversity in Lycium ruthenicum Murr. Plants. 2025; 14(21):3374. https://doi.org/10.3390/plants14213374
Chicago/Turabian StyleYang, Rong, Xiuhua Wu, Yu’e Bai, Yujiao He, Sujuan Chang, and Long Hai. 2025. "Genome-Wide SNP Analysis Reveals Population Structure and Genetic Diversity in Lycium ruthenicum Murr" Plants 14, no. 21: 3374. https://doi.org/10.3390/plants14213374
APA StyleYang, R., Wu, X., Bai, Y., He, Y., Chang, S., & Hai, L. (2025). Genome-Wide SNP Analysis Reveals Population Structure and Genetic Diversity in Lycium ruthenicum Murr. Plants, 14(21), 3374. https://doi.org/10.3390/plants14213374





