Molecular Characterization of Tissue-Specific Anthocyanin Biosynthesis in Potato Stamens
Abstract
1. Introduction
2. Methods
2.1. Plant Materials
2.2. Anthocyanin Composition and Quantitative Analysis
2.3. RNA Isolation and Purification
2.4. cDNA Library Preparation and RNA-Sequencing Analysis
2.5. Quantitative Real-Time PCR Analysis(qRT-PCR)
2.6. Statistical Analysis and Data Visualization
3. Results
3.1. Anthocyanin Composition and Quantitative Analysis in Potato Reproductive and Storage Tissues
3.2. Transcriptomic Analysis of Anthocyanin Biosynthetic Gene Expression
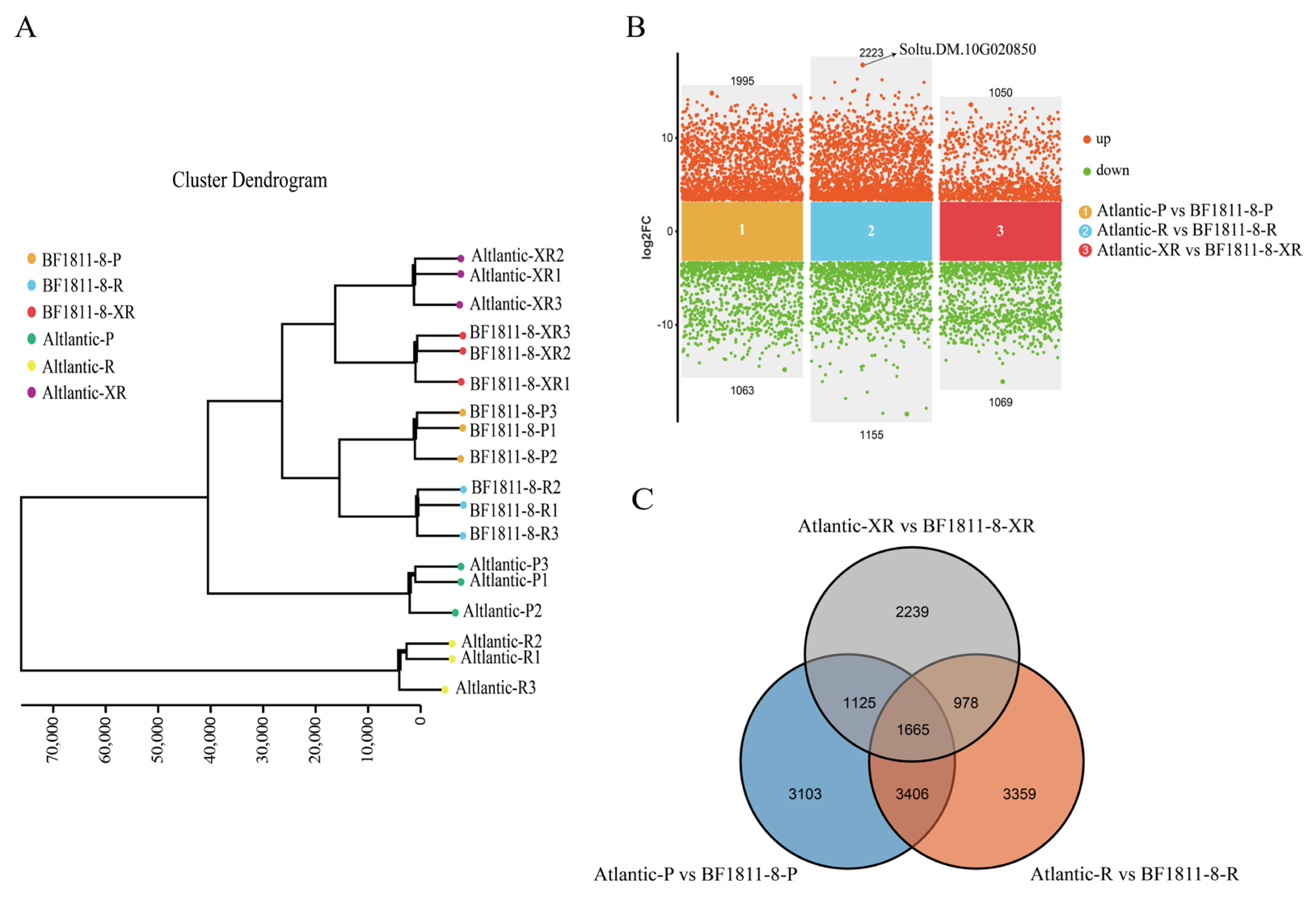
3.2.1. Differential Gene Expression Profiling
3.2.2. Functional Enrichment Analysis of Differentially Expressed Genes
3.2.3. Transcriptional Analysis of Anthocyanin Biosynthetic Genes
3.2.4. Transcriptional Regulation of Anthocyanin Biosynthesis
3.2.5. Regulatory Network Analysis and Gene Correlation Patterns
3.3. Quantitative Validation of Anthocyanin Biosynthetic Gene Expression
4. Discussion
4.1. Tissue-Specific Anthocyanin Accumulation Patterns in Potato
4.2. Transcriptional Level Differences and Expression Analysis of Anthocyanin Synthesis-Related Genes in Potato Stamens and Tuber Tissues
4.2.1. GO and KEGG Enrichment Analysis of Anthocyanin Synthesis-Related Genes
4.2.2. Transcriptional Level Differences and Expression Analysis of Anthocyanin Synthesis-Related Structural Genes
4.2.3. Transcriptional Level Differences and Expression Analysis of Anthocyanin Synthesis-Related Transcription Factors
4.3. Correlation Analysis of Tissue-Specific Expression of Anthocyanin Synthesis-Related Genes in Potato Stamen and Tuber Tissues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Jong, H. Impact of the potato on society. Am. J. Potato Res. 2016, 93, 415–429. [Google Scholar] [CrossRef]
- Seibert, T.; Abel, C.; Wahl, V. Flowering time and the identification of floral marker genes in Solanum tuberosum ssp. andigena. J. Exp. Bot. 2020, 71, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.E.; Hannah, L.; Shrestha, M.; Burd, M.; Dyer, A.G. Fly pollination drives convergence of flower coloration. New Phytol. 2022, 233, 52–61. [Google Scholar] [CrossRef]
- Lawrence, J.; Sauer, J. Metabolic profile of fragrant and non-fragrant ornamental potato flowers (617.5). FASEB J. 2014, 28, 617.5. [Google Scholar] [CrossRef]
- Merecz-Sadowska, A.; Sitarek, P.; Kowalczyk, T.; Zajdel, K.; Jęcek, M.; Nowak, P.; Zajdel, R. Food anthocyanins: Malvidin and its glycosides as promising antioxidant and anti-inflammatory agents with potential health benefits. Nutrients 2023, 15, 3016. [Google Scholar] [CrossRef]
- Tkaczyńska, A.; Sendra, E.; Jiménez-Redondo, N.; Rytel, E. Studying the stability of anthocyanin pigments isolated from juices of colored-fleshed potatoes. Int. J. Mol. Sci. 2024, 25, 11116. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, D.; Li, S.; Pan, J.; Liang, J.; Wu, X.; Wu, X.N.; Krall, L.; Zhu, G. Multiomics analysis reveals the chemical and genetic bases of pigmented potato tuber. J. Agric. Food Chem. 2023, 71, 16402–16416. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Korobczak, A.; Aksamit, A.; Łukaszewicz, M.; Lorenc, K.; Rorat, T.; Szopa, J. The potato glucosyltransferase gene promoter is environmentally regulated. Plant Sci. 2005, 168, 339–348. [Google Scholar] [CrossRef]
- Yang, Y.-n.; Yao, G.-f.; Zheng, D.; Zhang, S.-l.; Wang, C.; Zhang, M.-y.; Wu, J. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2015, 34, 189–198. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liang, J.; Zeng, X.; Guo, H.; Luo, Y.; Kear, P.; Zhang, S.; Zhu, G. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Hortic. Plant J. 2021, 7, 129–141. [Google Scholar] [CrossRef]
- Qi, Y.; Zhou, L.; Han, L.; Zou, H.; Miao, K.; Wang, Y. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony (Paeonia suffruticosa). Plant Physiol. Biochem. 2020, 154, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tang, Y.; Li, Y.; Hu, C.; Li, J.; Lyu, A. Genome-wide identification and bioinformatics analysis of the WD40 transcription factor family and candidate gene screening for anthocyanin biosynthesis in Rhododendron simsii. BMC Genom. 2023, 24, 488. [Google Scholar] [CrossRef]
- He, M.; Ma, X.; Zhou, Y.; Wang, F.; Fang, G.; Wang, J. Combined metabolome and transcriptome analyses reveals anthocyanin biosynthesis profiles between purple and white potatoes. Int. J. Mol. Sci. 2024, 25, 12884. [Google Scholar] [CrossRef]
- Zhu, F. Anthocyanins in cereals: Composition and health effects. Food Res. Int. 2018, 109, 232–249. [Google Scholar] [CrossRef] [PubMed]
- Riaz, B.; Chen, H.; Wang, J.; Du, L.; Wang, K.; Ye, X. Overexpression of maize ZmC1 and ZmR transcription factors in wheat regulates anthocyanin biosynthesis in a tissue-specific manner. Int. J. Mol. Sci. 2019, 20, 5806. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. 2021, 105, 1010–1025. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef]
- Xi, H.; He, Y.; Chen, H. Functional characterization of SmbHLH13 in anthocyanin biosynthesis and flowering in eggplant. Hortic. Plant J. 2021, 7, 73–80. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Liu, J.; Guo, D.; Hou, J.; Chen, S.; Song, B.; Xie, C. Analysis of structural genes and key transcription factors related to anthocyanin biosynthesis in potato tubers. Sci. Hortic. 2017, 225, 310–316. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, J.; Zhao, Y.; Zhao, X.; Liu, J.; Liu, J.; Zhang, J.; Song, B.; Zhang, H. StMYB113 promotes anthocyanin biosynthesis in potato (Solanum tuberosum L.) désirée tubers. Potato Res. 2024, 67, 307–324. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, H.; Liu, T.; Zhao, Y.; Hu, X.; Liu, S.; Lin, Y.; Song, B.; He, C. Transcriptome analysis provides StMYBA1 gene that regulates potato anthocyanin biosynthesis by activating structural genes. Front. Plant Sci. 2023, 14, 1087121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Z.; Zhao, Y.; Guo, D.; Zhao, X.; Gao, W.; Zhang, J.; Song, B.; Cahill, D. StWRKY13 promotes anthocyanin biosynthesis in potato (Solanum tuberosum) tubers. Funct. Plant Biol. 2021, 49, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Moratalla-López, N.; Bagur, M.J.; Lorenzo, C.; Martínez-Navarro, M.E.; Salinas, M.R.; Alonso, G.L. Bioactivity and bioavailability of the major metabolites of Crocus sativus L. flower. Molecules 2019, 24, 2827. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Wei, Z.; Liu, H.; Dong, L. Integrated metabolome and transcriptome analyses reveal that the flavonoid metabolic pathway is associated with pigment differential accumulation in two colors of petaloid staminodes in Canna glauca. Horticulturae 2024, 10, 372. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, L.; Zhang, C.; Kong, X.; Zheng, Y.; Dong, L. Glucose supply induces PsMYB2-mediated anthocyanin accumulation in Paeonia suffruticosa ‘Tai Yang’cut flower. Front. Plant Sci. 2022, 13, 874526. [Google Scholar]
- Cui, L.; Li, M.; Zhang, X.; Guo, Z.; Li, K.; Shi, Y.; Wang, Q.; Guo, H. Enhanced UV-B radiation in potato stems and leaves promotes the accumulation of anthocyanins in tubers. Curr. Issues Mol. Biol. 2023, 45, 9943–9960. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Han, L.; Li, R.; Jin, X.; Li, Y.; Chen, Q.; He, C.; Wang, M. Metabolomic analysis, extraction, purification and stability of the anthocyanins from colored potatoes. Food Chem. X 2024, 22, 101423. [Google Scholar] [CrossRef]
- Mori, M.; Hayashi, K.; Ohara-Takada, A.; Watanuki, H.; Katahira, R.; Ono, H.; Terahara, N. Anthocyanins from skins and fleshes of potato varieties. Food Sci. Technol. Res. 2010, 16, 115–122. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, D.; Wang, Y.; Zhang, L.; Chen, X.; Yang, X.; Xiong, H.; Bhattarai, G.; Ravelombola, W.; Olaoye, D.; et al. Transcript profiling for regulation of sweet potato skin color in Sushu8 and its mutant Zhengshu20. Plant Physiol. Biochem. 2020, 148, 1–9. [Google Scholar] [CrossRef]
- Deguchi, A.; Tatsuzawa, F.; Miyoshi, K. A blackish-flowered cultivar of Catharanthus roseus accumulates high concentrations of a novel anthocyanin with a unique feature of aggregation in weak acid solutions. Dye. Pigment. 2020, 173, 108001. [Google Scholar] [CrossRef]
- Nakayama, M.; Yamaguchi, M.-A.; Urashima, O.; Kan, Y.; Fukui, Y.; Yamaguchi, Y.; Koshioka, M. Anthocyanins in the dark purple anthers of Tulipa gesneriana: Identification of two novel delphinidin 3-O-(6-O-(acetyl-α-rhamnopyranosyl)-β-glucopyranosides). Biosci. Biotechnol. Biochem. 1999, 63, 1509–1511. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tengkun, N.; Dongdong, W.; Xiaohui, M.; Yue, C.; Qin, C. Analysis of key genes involved in potato anthocyanin biosynthesis based on genomics and transcriptomics data. Front. Plant Sci. 2019, 10, 603. [Google Scholar] [CrossRef]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef]
- Jiang, F.; Wang, J.-Y.; Jia, H.-F.; Jia, W.-S.; Wang, H.-Q.; Xiao, M. RNAi-mediated silencing of the flavanone 3-hydroxylase gene and its effect on flavonoid biosynthesis in strawberry fruit. J. Plant Growth Regul. 2013, 32, 182–190. [Google Scholar] [CrossRef]
- Shao, D.; Li, Y.; Zhu, Q.; Zhang, X.; Liu, F.; Xue, F.; Sun, J. GhGSTF12, a glutathione S-transferase gene, is essential for anthocyanin accumulation in cotton (Gossypium hirsutum L.). Plant Sci. 2021, 305, 110827. [Google Scholar] [CrossRef] [PubMed]
- Diretto, G.; Jin, X.; Capell, T.; Zhu, C.; Gomez-Gomez, L. Differential accumulation of pelargonidin glycosides in petals at three different developmental stages of the orange-flowered gentian (Gentiana lutea L. var. aurantiaca). PLoS ONE 2019, 14, e0212062. [Google Scholar] [CrossRef]
- Sun, W.; Sun, S.; Xu, H.; Wang, Y.; Chen, Y.; Xu, X.; Yi, Y.; Ju, Z. Characterization of two key flavonoid 3-O-glycosyltransferases involved in the formation of flower color in Rhododendron delavayi. Front. Plant Sci. 2022, 13, 863482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, H.; Zhang, L.; Duan, S.; Li, G.; Duan, Y. Fine Mapping and Candidate Genes Analysis for Regulatory Gene of Anthocyanin Synthesis in the Corolla, Shedding Light on Wild Potato Evolution. Int. J. Mol. Sci. 2025, 26, 1966. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Wang, S.; Ruan, M.; Li, Y.; Zhu, L.; Dong, Z.; Long, Y. Genome-wide identification and functional roles relating to anthocyanin biosynthesis analysis in maize. BMC Plant Biol. 2025, 25, 57. [Google Scholar] [CrossRef]
- Xu, R.; Pan, R.; Zhang, Y.; Feng, Y.; Nath, U.K.; Gan, Y.; Shi, C.; Akhter, D. RNA-seq-based profiling of pl mutant reveals transcriptional regulation of anthocyanin biosynthesis in rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 9787. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, H.; Zhao, M.; Yang, Z.; Zhou, Z.; Guo, Y.; Lin, Y.; Chen, H. OsMYB3 is a R2R3-MYB gene responsible for anthocyanin biosynthesis in black rice. Mol. Breed. 2021, 41, 51. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021, 107, 198–214. [Google Scholar] [CrossRef]
- Du, H.; Zhai, Z.; Pu, J.; Liang, J.; Wang, R.; Zhang, Z.; Wang, P.; Zhu, Y.; Huang, L.; Li, D.; et al. Two tandem R2R3 MYB transcription factor genes cooperatively regulate anthocyanin accumulation in potato tuber flesh. Plant Biotechnol. J. 2025, 23, 1521–1534. [Google Scholar] [CrossRef]
- Zhang, Y.; Pu, Y.; Zhang, Y.; Li, K.; Bai, S.; Wang, J.; Xu, M.; Liu, S.; Zhou, Z.; Wu, Y.; et al. Tuber transcriptome analysis reveals a novel WRKY transcription factor StWRKY70 potentially involved in potato pigmentation. Plant Physiol. Biochem. 2024, 213, 108792. [Google Scholar] [CrossRef]
- Laimbeer, F.P.E.; Bargmann, B.O.; Holt, S.H.; Pratt, T.; Peterson, B.; Doulis, A.G.; Buell, C.R.; Veilleux, R.E. Characterization of the F locus responsible for floral anthocyanin production in potato. G3 Genes Genomes Genet. 2020, 10, 3871–3879. [Google Scholar] [CrossRef] [PubMed]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Castellano Moreno, M.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN 1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, S.; Hu, J.; Jin, L.; Xu, J. Transcriptome Profiling Identifies Key Regulators of Tuber Skin Color in Potato. Plants 2025, 14, 1544. [Google Scholar] [CrossRef]
- Lin, Y.-C.J.; Chen, H.; Li, Q.; Li, W.; Wang, J.P.; Shi, R.; Tunlaya-Anukit, S.; Shuai, P.; Wang, Z.; Ma, H.; et al. Reciprocal cross-regulation of VND and SND multigene TF families for wood formation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2017, 114, E9722–E9729. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Guo, Z.; Zhang, X.; Guo, H. Molecular Characterization of Tissue-Specific Anthocyanin Biosynthesis in Potato Stamens. Plants 2025, 14, 3260. https://doi.org/10.3390/plants14213260
Li S, Guo Z, Zhang X, Guo H. Molecular Characterization of Tissue-Specific Anthocyanin Biosynthesis in Potato Stamens. Plants. 2025; 14(21):3260. https://doi.org/10.3390/plants14213260
Chicago/Turabian StyleLi, Sunjin, Zongming Guo, Xing Zhang, and Huachun Guo. 2025. "Molecular Characterization of Tissue-Specific Anthocyanin Biosynthesis in Potato Stamens" Plants 14, no. 21: 3260. https://doi.org/10.3390/plants14213260
APA StyleLi, S., Guo, Z., Zhang, X., & Guo, H. (2025). Molecular Characterization of Tissue-Specific Anthocyanin Biosynthesis in Potato Stamens. Plants, 14(21), 3260. https://doi.org/10.3390/plants14213260





