The Structure, Evolution, and Expression Patterns Analysis Reveals the bHLH Members Associated with Powdery Mildew Resistance in Rubber Tree
Abstract
1. Introduction
2. Results
2.1. Identification and Structural Analysis of Members of the bHLH Family
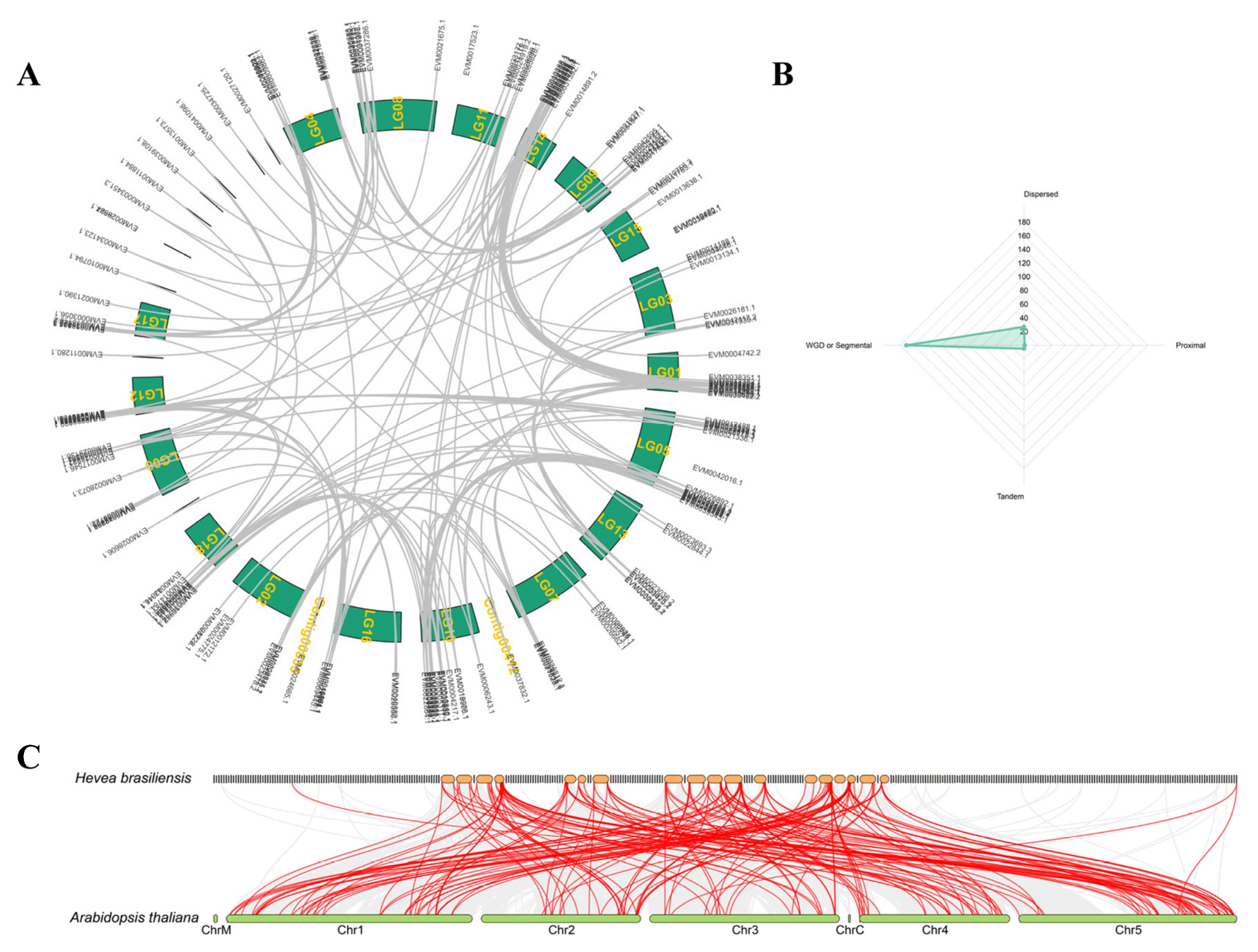
2.2. Evolution and Collinearity Analysis of the bHLH Family
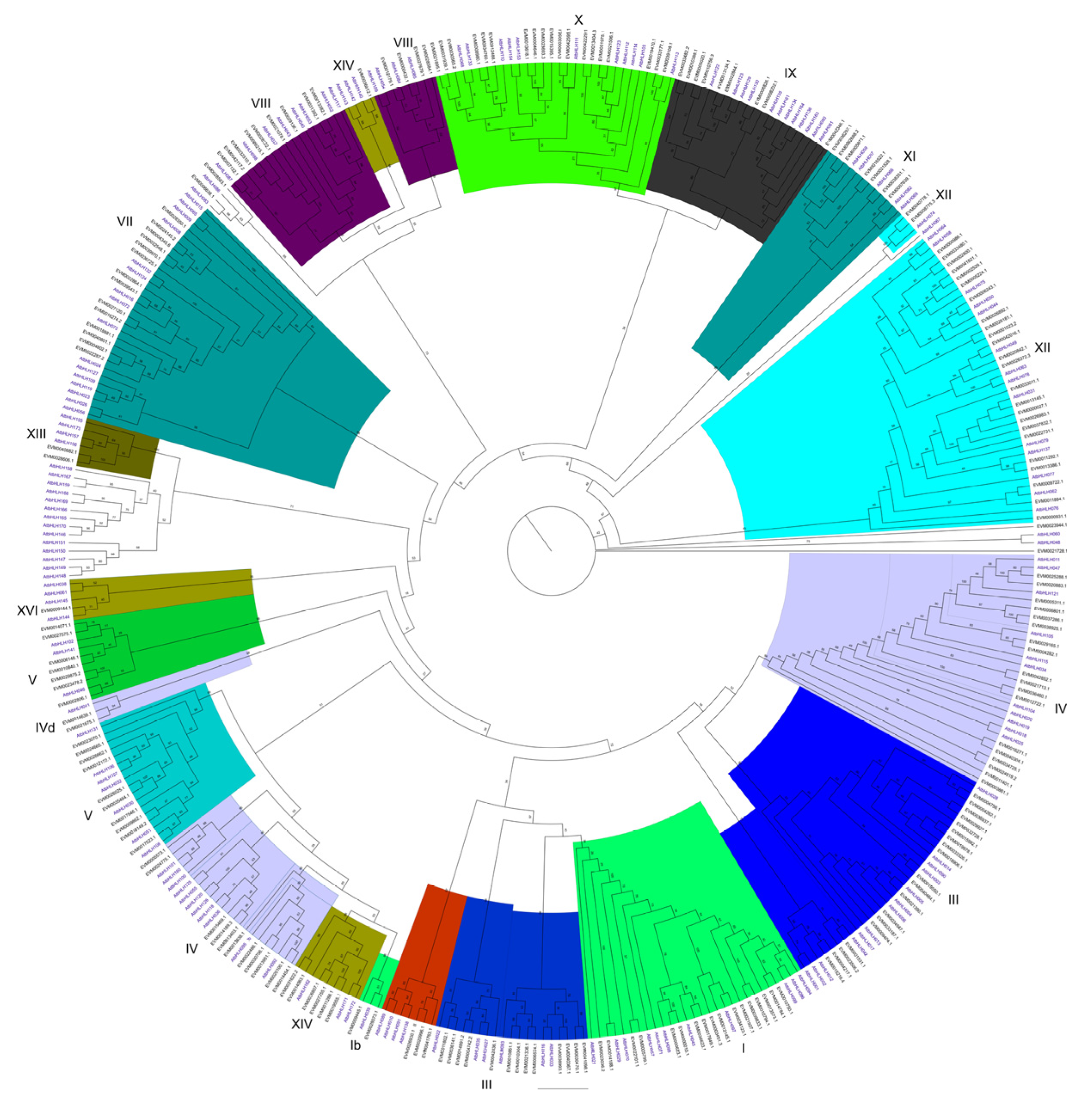
2.3. Systematic Evolutionary Relationships Among Members of the bHLH Family
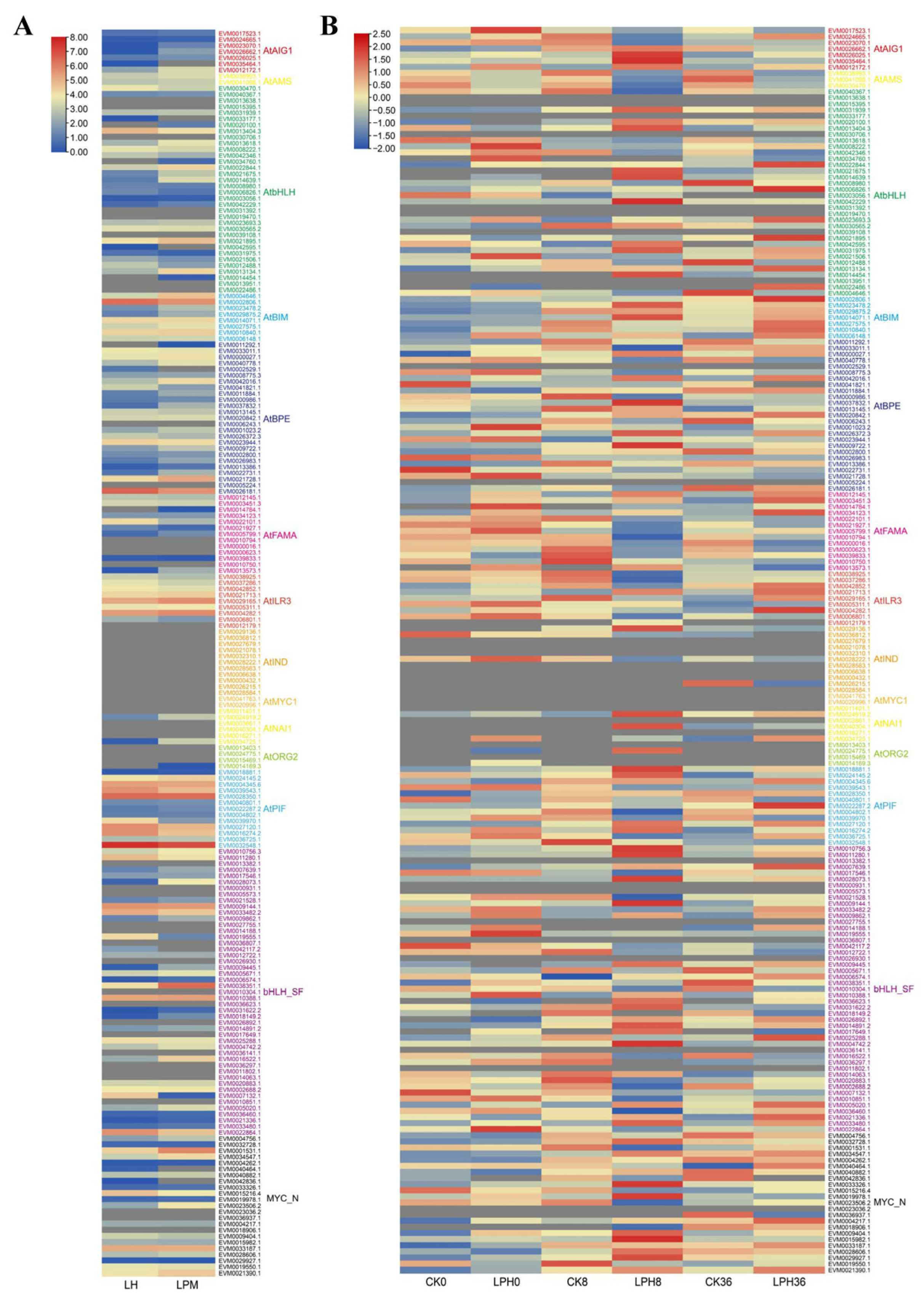
2.4. RNA Sequencing Analysis of bHLH Family Member Expression Patterns Following Powdery Mildew Treatment
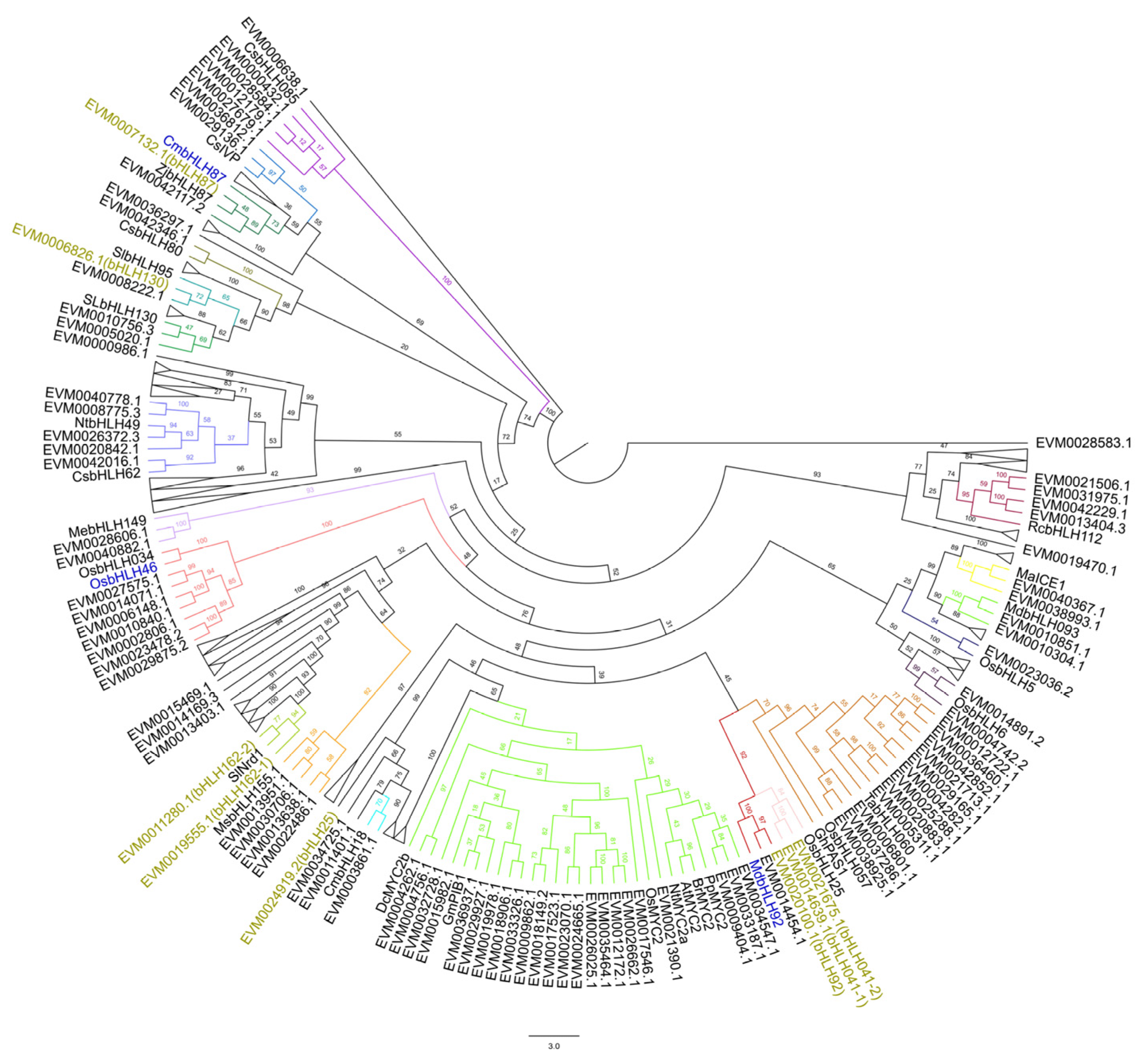
2.5. Phylogenetic Relationships Between Plant Disease-Related bHLH Proteins and bHLH Family Members in Rubber Trees
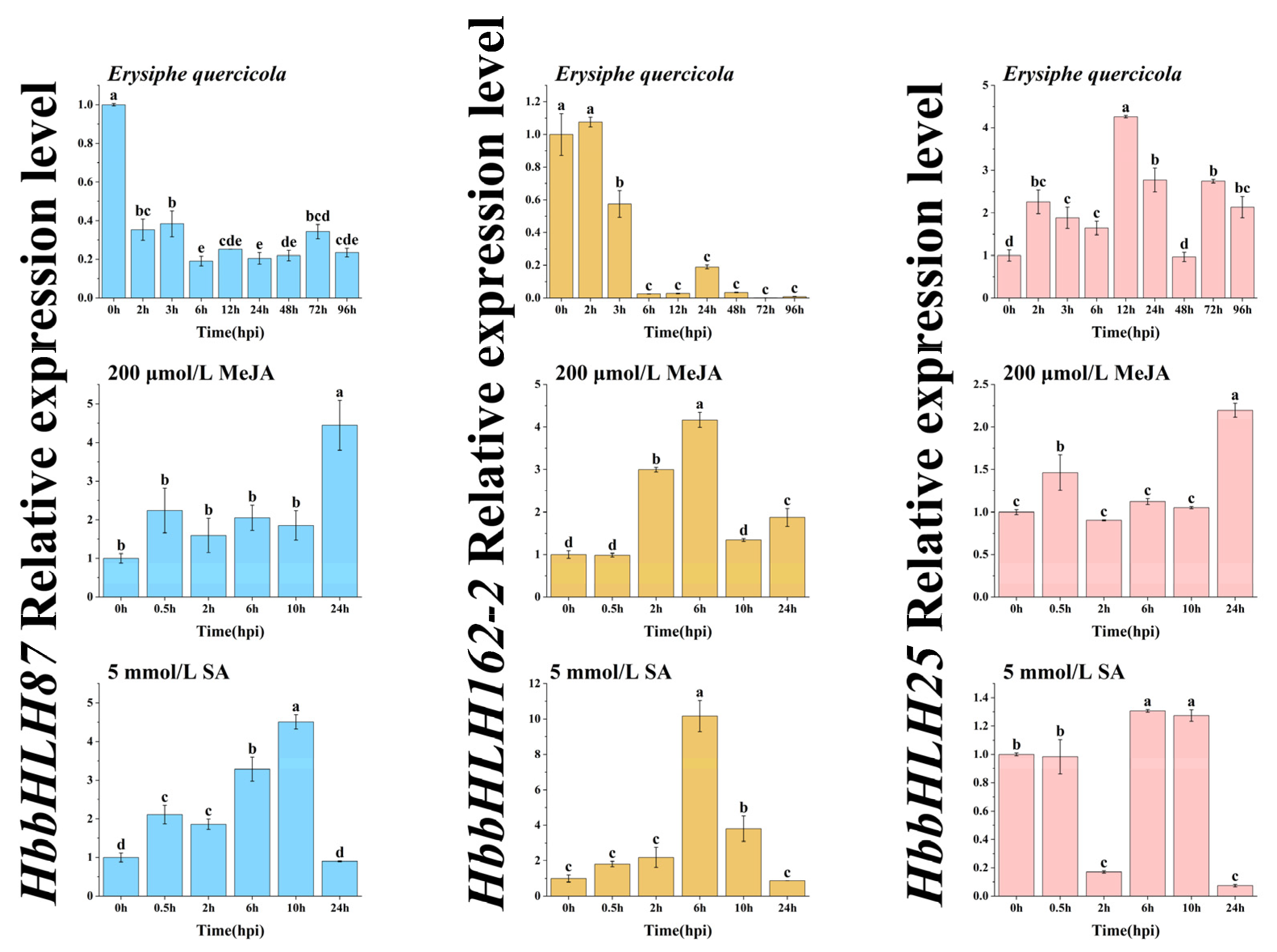
2.6. Expression Patterns of Key bHLH Genes Under Erysiphe quercicola and Hormone Treatments
3. Discussion
4. Materials and Methods
4.1. Data Downloading and Identification of the bHLH Gene Family in the Whole Genome
4.2. bHLH Gene Structure, MEME Clustering Analysis and Collinearity Analysis
4.3. Construction of the bHLH System Evolutionary Tree
4.4. Analysis of Cis-Acting Elements in the Promoter Regions of bHLH Family Members
4.5. RNA Sequencing Analysis
4.6. Relative Fluorescence Quantitative Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liyanage, K.K.; Khan, S.; Brooks, S.; Mortimer, P.E.; Karunarathna, S.C.; Xu, J.C.; Hyde, K.D. Morph-Molecular Characterization of two Ampelomyces spp. (Pleosporales) Strains Mycoparasites of Powdery Mildew of Hevea brasiliensis. Front. Microbiol. 2018, 9, 12. [Google Scholar] [CrossRef]
- Cao, X.R.; Han, Q.H.; Xiao, Y.; He, J.J.; Chuan, X.; Jiang, G.Z.; West, J.S.; Xu, X.M. Population Genetic Structure of the Rubber Tree Powdery Mildew Pathogen (Erysiphe quercicola) from China. Plant Dis. 2024, 108, 62–70. [Google Scholar] [CrossRef]
- Zhai, D.L.; Thaler, P.; Worthy, F.R.; Xu, J.C. Rubber latex yield is affected by interactions between antecedent temperature, rubber phenology, and powdery mildew disease. Int. J. Biometeorol. 2023, 67, 1569–1579. [Google Scholar] [CrossRef]
- Liyanage, K.K.; Khan, S.; Mortimer, P.E.; Hyde, K.D.; Xu, J.; Brooks, S.; Ming, Z. Powdery mildew disease of rubber tree. For. Pathol. 2016, 46, 90–103. [Google Scholar] [CrossRef]
- Solpot, T.C.; Borja, B.T.; Prado, M.M.; Abubakar, J.V.; Cabasan, M.T.N. Leaf diseases of Hevea brasiliensis Müll. Arg. in major rubber growing areas of Cotabato, Philippines. J. Rubber Res. 2024, 27, 11–20. [Google Scholar] [CrossRef]
- Liyanage, K.K.; Khan, S.; Herath, V.; Brooks, S.; Mortimer, P.E.; Nadir, S.; Hyde, K.D.; Xu, J. Genome Wide Identification of the MLO Gene Family Associated with Powdery Mildew Resistance in Rubber Trees (Hevea brasiliensis). Trop. Plant. Biol. 2020, 13, 331–342. [Google Scholar] [CrossRef]
- Qin, B.; Wang, M.; He, H.X.; Xiao, H.X.; Zhang, Y.; Wang, L.F. Identification and Characterization of a Potential Candidate Mlo Gene Conferring Susceptibility to Powdery Mildew in Rubber Tree. Phytopathology 2019, 109, 1236–1245. [Google Scholar] [CrossRef]
- Li, X.; Zhao, W.Y.; Zhang, Z.M.; Fang, Y.X.; Dong, L.P.; Yin, J.Y.; Liu, Y.H.; Chen, D.P.; Li, Z.G.; Liu, W.B.; et al. The Rubber Tree (Heveae brasiliensis) MLO Protein HbMLO12 Promotes Plant Susceptibility to Sustain Infection by a Powdery Mildew Fungus. Mol. Plant Microbe Interact. 2023, 36, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef]
- Liang, X.Y.; Ma, Z.; Ke, Y.H.; Wang, J.L.; Wang, L.F.; Qin, B.; Tang, C.R.; Liu, M.Y.; Xian, X.M.; Yang, Y.; et al. Single-cell transcriptomic analyses reveal cellular and molecular patterns of rubber tree response to early powdery mildew infection. Plant Cell Environ. 2023, 46, 2222–2237. [Google Scholar] [CrossRef]
- Wang, M.; Xiao, H.; Li, X.; Wan, S.; Yang, Y.; Yu, H.; Zhang, Y.; Qin, B. Functional characterization of powdery mildew resistance-related genes HbSGT1a and HbSGT1b in Hevea brasiliensis Muell. Arg. Eur. J. Plant Pathol. 2023, 165, 153–161. [Google Scholar] [CrossRef]
- Li, X.L.; He, Q.G.; Liu, Y.H.; Xu, X.Z.; Xie, Q.B.; Li, Z.G.; Lin, C.H.; Liu, W.B.; Chen, D.P.; Li, X.; et al. Ectopic Expression of HbRPW8-a from Hevea brasiliensis Improves Arabidopsis thaliana Resistance to Powdery Mildew Fungi (Erysiphe cichoracearum UCSC1). Int. J. Mol. Sci. 2022, 23, 12588. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, H.; Mei, Q.; Yang, J.; Ma, F.; Mao, K. Characteristics of bHLH transcription factors and their roles in the abiotic stress responses of horticultural crops. Sci. Hortic. 2023, 310, 111710. [Google Scholar] [CrossRef]
- Wei, K.F.; Chen, H.Q. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 2018, 18, 309. [Google Scholar] [CrossRef]
- Wang, Z.; Yuan, Y.; Rehman, F.; Wang, X.; Wu, T.K.; Deng, Z.; Cheng, H. Genome-Wide Identification and Characterization of bHLH Gene Family in Hevea brasiliensis. Forests 2024, 15, 2027. [Google Scholar] [CrossRef]
- Huang, X.; Su, L.Y.; Xian, B.H.; Yu, Q.Y.; Zhang, M.; Fan, J.; Zhang, C.X.; Liu, Y.Q.; He, H.Z.; Zhong, X.; et al. Genome-wide identification and characterization of the sweet orange (Citrus sinensis) basic helix-loop-helix (bHLH) family reveals a role for CsbHLH085 as a regulator of citrus bacterial canker resistance. Int. J. Biol. Macromol. 2024, 267, 131442. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.W.; Yang, C.; Cao, J.D.; Chen, H.; Pang, J.H.; Zhao, Q.Q.; Wang, Z.Y.; Fu, Z.Q.; Liu, J. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef]
- Yu, D.; Wei, W.; Fan, Z.Q.; Chen, J.Y.; You, Y.L.; Huang, W.D.; Zhan, J.C. VabHLH137 promotes proanthocyanidin and anthocyanin biosynthesis and enhances resistance to Colletotrichum gloeosporioides in grapevine. Hortic. Res. 2023, 10, 12. [Google Scholar] [CrossRef]
- Ma, H.; Zou, F.Y.; Li, D.M.; Wan, Y.; Zhang, Y.P.; Zhao, Z.Y.; Wang, X.P.; Gao, H. Transcription Factor MdbHLH093 Enhances Powdery Mildew Resistance by Promoting Salicylic Acid Signaling and Hydrogen Peroxide Accumulation. Int. J. Mol. Sci. 2023, 24, 9390. [Google Scholar] [CrossRef]
- Guo, W.L.; Chen, B.H.; Guo, Y.Y.; Chen, X.J.; Li, Q.F.; Yang, H.L.; Li, X.Z.; Zhou, J.G.; Wang, G.Y. Expression of Pumpkin CmbHLH87 Gene Improves Powdery Mildew Resistance in Tobacco. Front. Plant Sci. 2020, 11, 163. [Google Scholar] [CrossRef]
- Wu, H.; Mori, A.; Jiang, X.; Wang, Y.; Yang, M. The INDEHISCENT protein regulates unequal cell divisions in Arabidopsis fruit. Planta 2006, 224, 971–979. [Google Scholar] [CrossRef]
- van Gelderen, K.; van Rongen, M.; Liu, A.a.; Otten, A.; Offringa, R. An INDEHISCENT-controlled auxin response specifies the separation layer in early Arabidopsis fruit. Mol. Plant 2016, 9, 857–869. [Google Scholar] [CrossRef]
- Gremski, K.; Ditta, G.; Yanofsky, M.F. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 2007, 134, 3593–3601. [Google Scholar] [CrossRef] [PubMed]
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.W.; Huang, S.; Zeng, Y.; Liu, C.; Ma, Q.Y.; Pruneda-Paz, J.; Kay, S.A.; Li, L. Two bHLH transcription factors, bHLH48 and bHLH60, associate with phytochrome interacting factor 7 to regulate hypocotyl elongation in Arabidopsis. Cell Rep. 2021, 35, 109054. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.T.; Deng, C.Y.; Feng, G.Z.; Liu, D. Genome-wide analysis of phytochrome-interacting factor (PIF) families and their potential roles in light and gibberellin signaling in Chinese pine. BMC Genom. 2024, 25, 1017. [Google Scholar] [CrossRef]
- Gao, F.; Dubos, C. The arabidopsis bHLH transcription factor family. Trends Plant Sci. 2024, 29, 668–680. [Google Scholar] [CrossRef]
- Zhang, J.L.; Liu, X.Y.; Yin, Z.Z.; Zhao, T.T.; Du, D.; Li, J.; Zhu, M.K.; Sun, Y.Y.; Pan, Y. Genome- and Transcriptome-Wide Characterization and Expression Analyses of bHLH Transcription Factor Family Reveal Their Relevance to Salt Stress Response in Tomato. Plants 2025, 14, 200. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Heim, M.A. The Basic Helix-Loop-Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Zhang, L.; Ma, H.; Zhang, Y.C.; Zhang, X.M.; Ji, M.M.; van Nocker, S.; Ahmad, B.; Zhao, Z.Y.; Wang, X.P.; et al. Overexpression of the apple (Malus× domestica) MdERF100 in Arabidopsis increases resistance to powdery mildew. Int. J. Mol. Sci. 2021, 22, 5713. [Google Scholar] [CrossRef]
- Liao, H.; Fang, Y.; Yin, J.; He, M.; Wei, Y.; Zhang, J.; Yong, S.; Cha, J.; Song, L.; Zhu, X.; et al. Rice transcription factor bHLH25 confers resistance to multiple diseases by sensing H2O2. Cell Res. 2025, 35, 205–219. [Google Scholar] [CrossRef]
- Yang, S.; Lovelace, A.H.; Yuan, Y.; Nie, H.Z.; Chen, W.K.; Gao, Y.; Bo, W.H.; Nagel, D.H.; Pang, X.M.; Ma, W.B. A witches’ broom phytoplasma effector induces stunting by stabilizing a bHLH transcription factor in Ziziphus jujuba plants. New Phytol. 2025, 247, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Ali, J.; Mukarram, M.; Ojo, J.; Dawam, N.; Riyazuddin, R.; Ghramh, H.A.; Khan, K.A.; Chen, R.Z.; Kurjak, D.; Bayram, A. Harnessing Phytohormones: Advancing Plant Growth and Defence Strategies for Sustainable Agriculture. Physiol. Plant. 2024, 176, e14307. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Xu, R.R.; Wang, X.N.; Zhang, X.W.; You, C.X.; Han, Y.P. MdbHLH162 connects the gibberellin and jasmonic acid signals to regulate anthocyanin biosynthesis in apple. J. Integr. Plant Biol. 2024, 66, 265–284. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Fang, Y.; Mei, H.; Zhou, B.; Xiao, X.; Yang, M.; Huang, Y.; Long, X.; Hu, S.; Tang, C. De novo Transcriptome Analysis Reveals Distinct Defense Mechanisms by Young and Mature Leaves of Hevea brasiliensis (Para Rubber Tree). Sci. Rep. 2016, 6, 33151. [Google Scholar] [CrossRef]
- Ødum, M.T.; Teufel, F.; Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Winther, O.; Nielsen, H. DeepLoc 2.1: Multi-label membrane protein type prediction using protein language models. Nucleic Acids Res. 2024, 52, W215–W220. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xiao, X.; Lin, J.; Lin, Q.; Wang, J.; Liu, K.; Li, Z.; Xing, J.; Liu, Z.; Wang, B.; et al. Pan-genome and phylogenomic analyses highlight Hevea species delineation and rubber trait evolution. Nat. Commun. 2024, 15, 7232. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, X.; Tang, X.; Lu, Y.; Zhang, Y.; Wang, C.; Zhang, Y.; Wang, L. The Structure, Evolution, and Expression Patterns Analysis Reveals the bHLH Members Associated with Powdery Mildew Resistance in Rubber Tree. Plants 2025, 14, 3244. https://doi.org/10.3390/plants14213244
Fan X, Tang X, Lu Y, Zhang Y, Wang C, Zhang Y, Wang L. The Structure, Evolution, and Expression Patterns Analysis Reveals the bHLH Members Associated with Powdery Mildew Resistance in Rubber Tree. Plants. 2025; 14(21):3244. https://doi.org/10.3390/plants14213244
Chicago/Turabian StyleFan, Xiaokang, Xiaoling Tang, Yiying Lu, Yan Zhang, Cuicui Wang, Yu Zhang, and Lifeng Wang. 2025. "The Structure, Evolution, and Expression Patterns Analysis Reveals the bHLH Members Associated with Powdery Mildew Resistance in Rubber Tree" Plants 14, no. 21: 3244. https://doi.org/10.3390/plants14213244
APA StyleFan, X., Tang, X., Lu, Y., Zhang, Y., Wang, C., Zhang, Y., & Wang, L. (2025). The Structure, Evolution, and Expression Patterns Analysis Reveals the bHLH Members Associated with Powdery Mildew Resistance in Rubber Tree. Plants, 14(21), 3244. https://doi.org/10.3390/plants14213244





