Overexpression of GmNAC03 in Soybean Enhances Salt Tolerance
Abstract
1. Introduction
2. Result
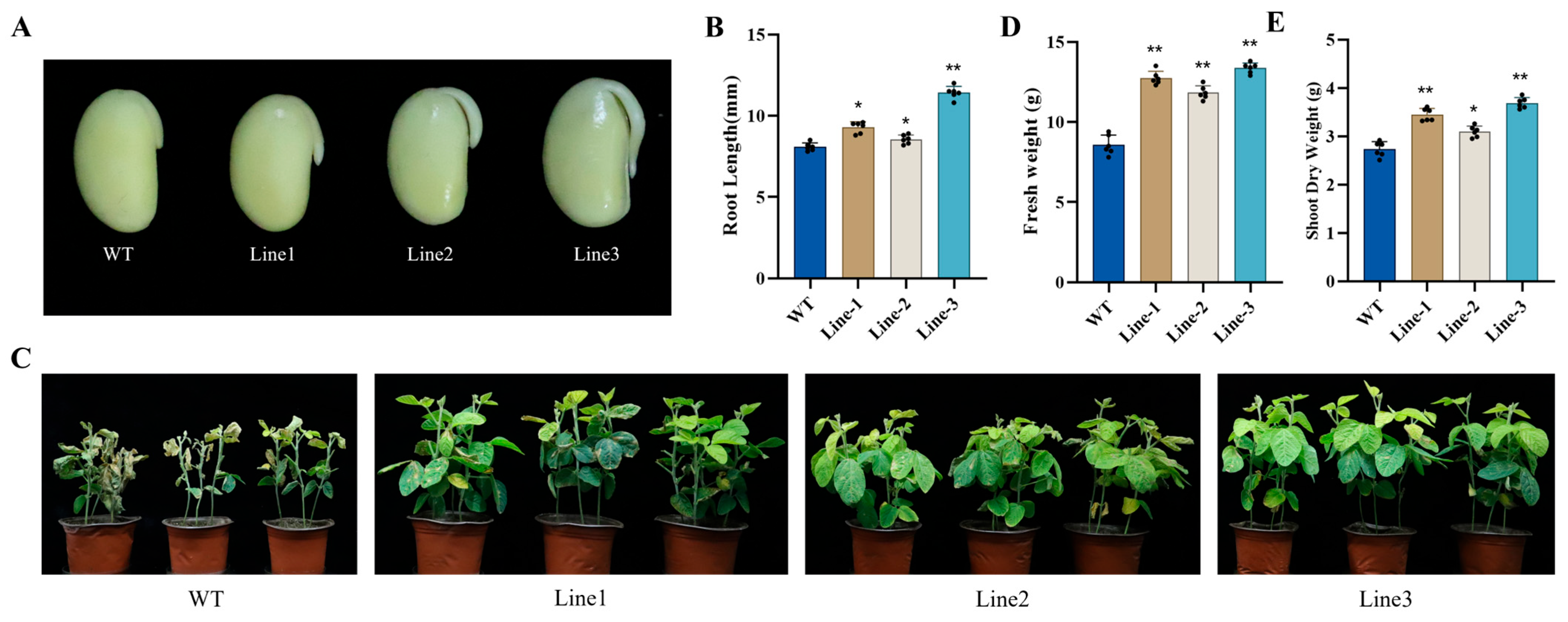
2.1. Soybean Transformation and Positive Line Identification
2.2. Overexpression of the GmNAC03 Gene Improves Soybean Salt Tolerance
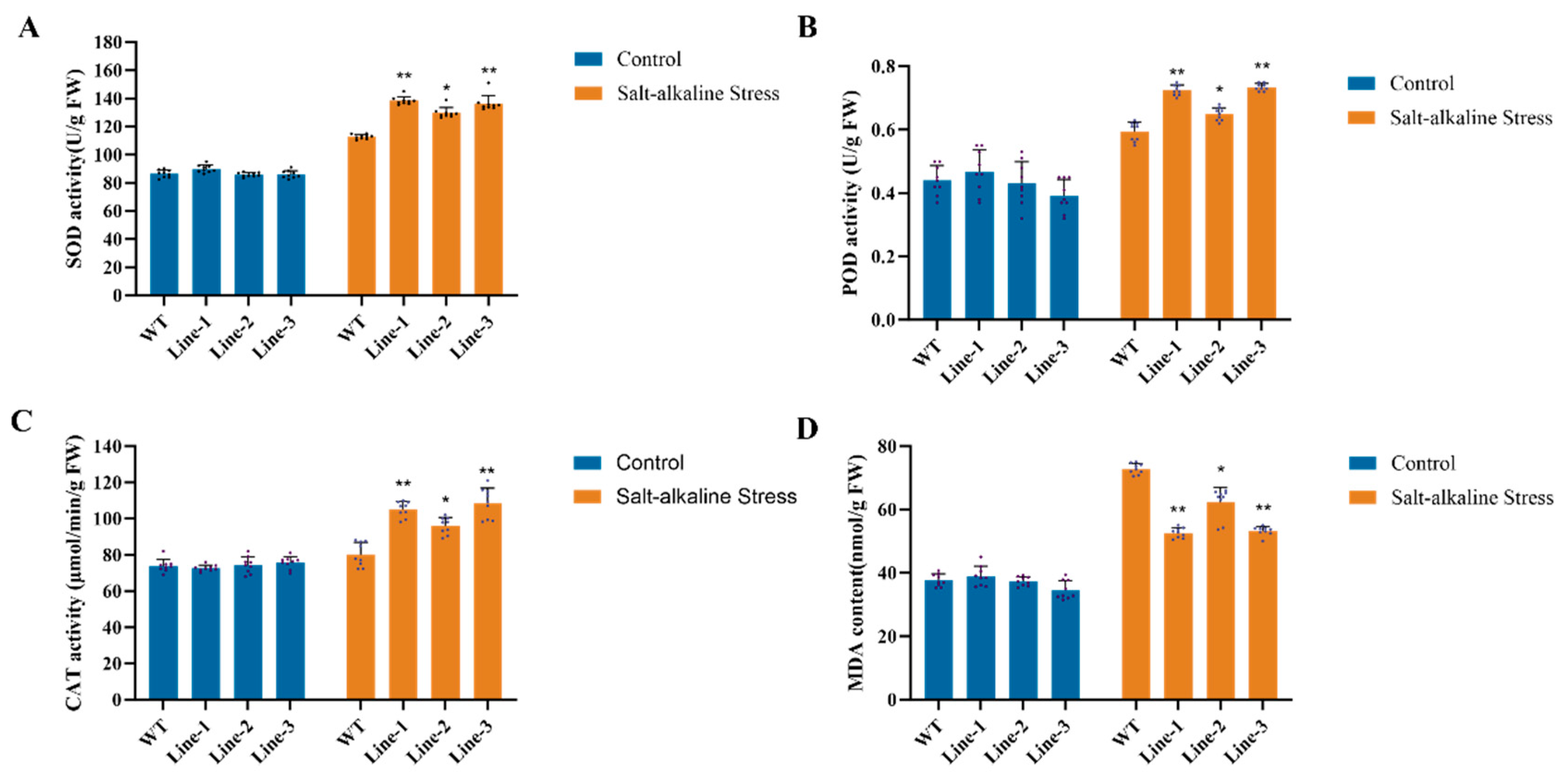
2.3. Physiological Evaluation of Salt Tolerance
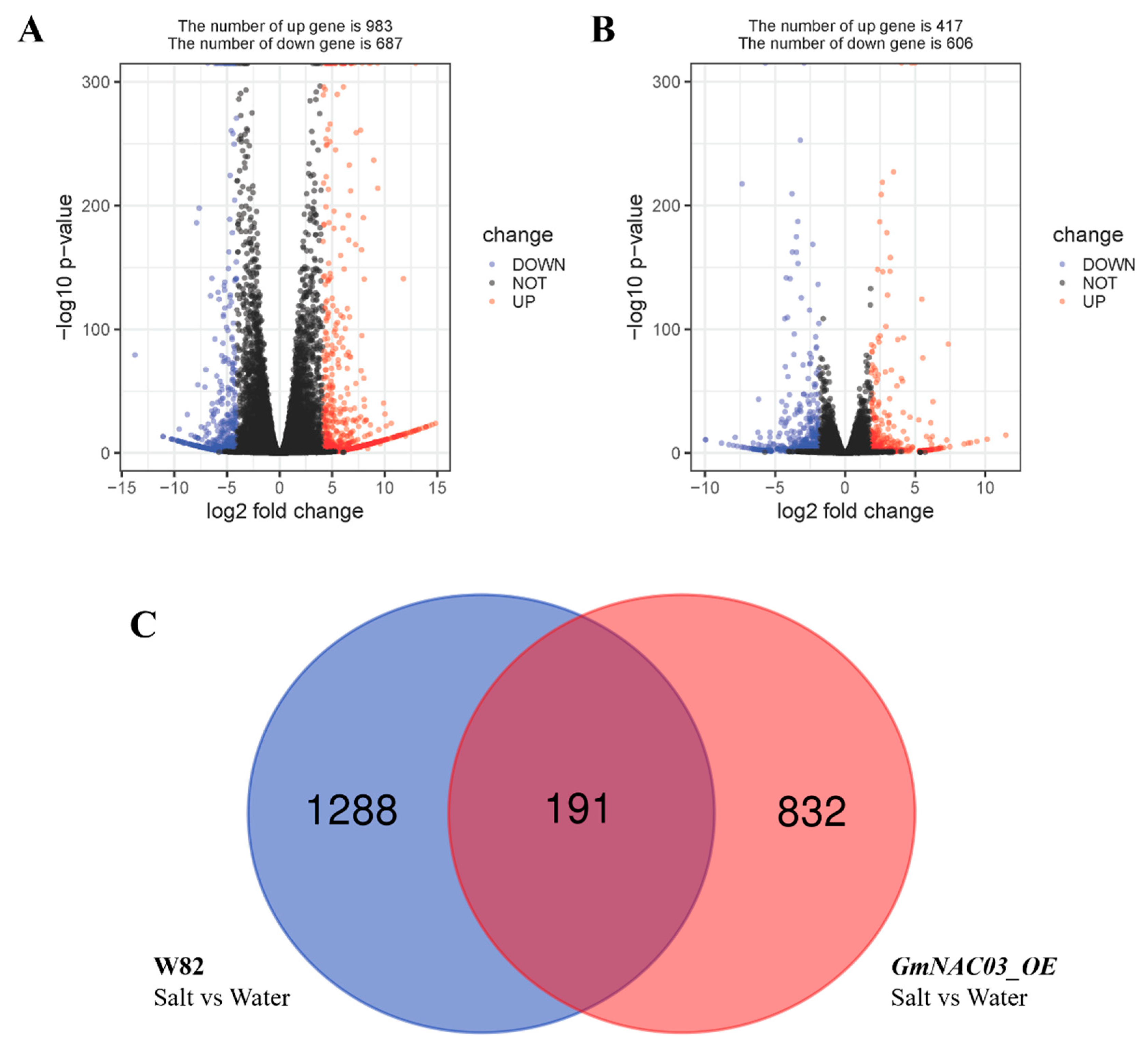
2.4. RNA-Seq Analysis of GmNAC03 Overexpression Lines and WT
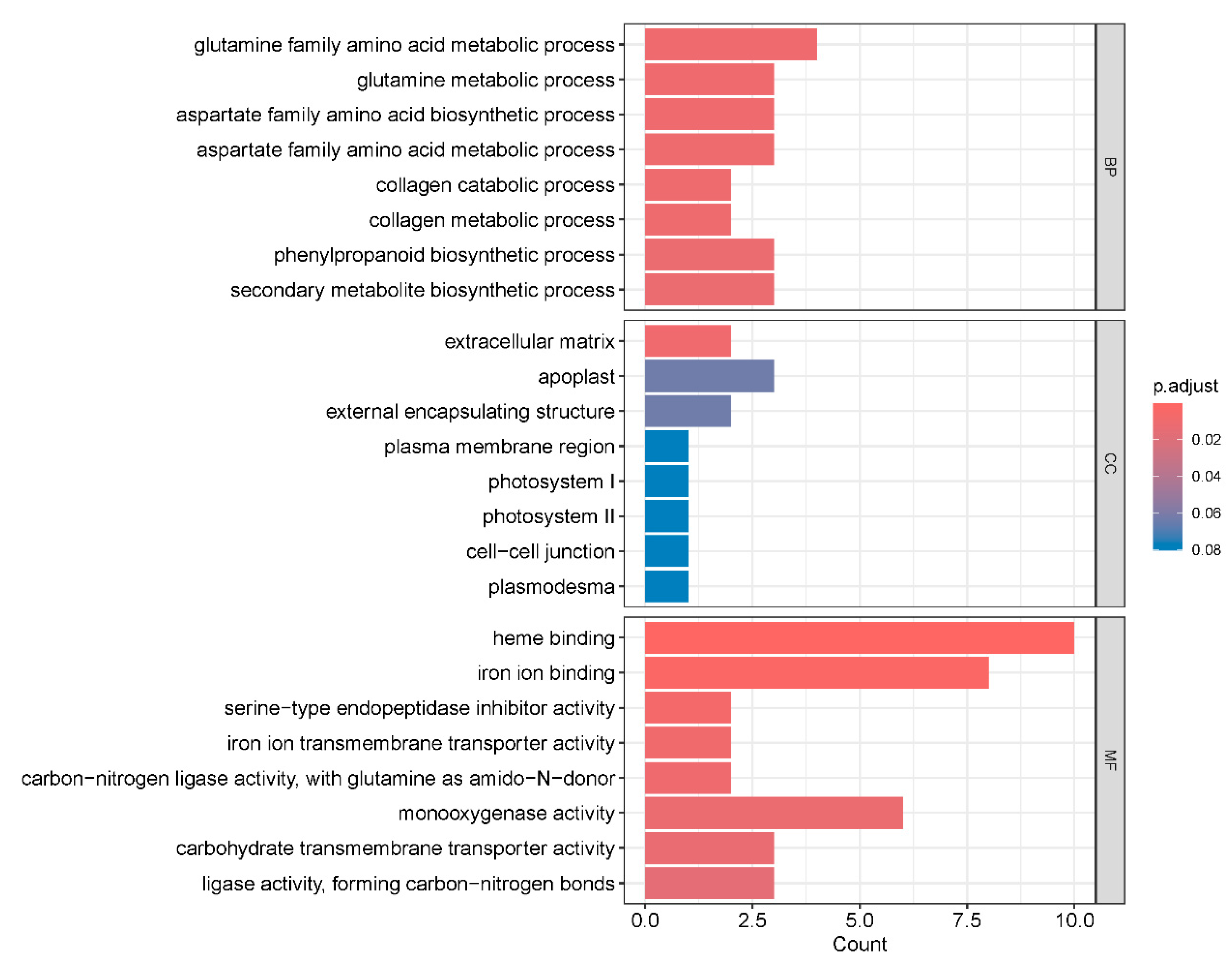
2.5. Gene Ontology Enrichment Analysis of Differentially Expressed Genes
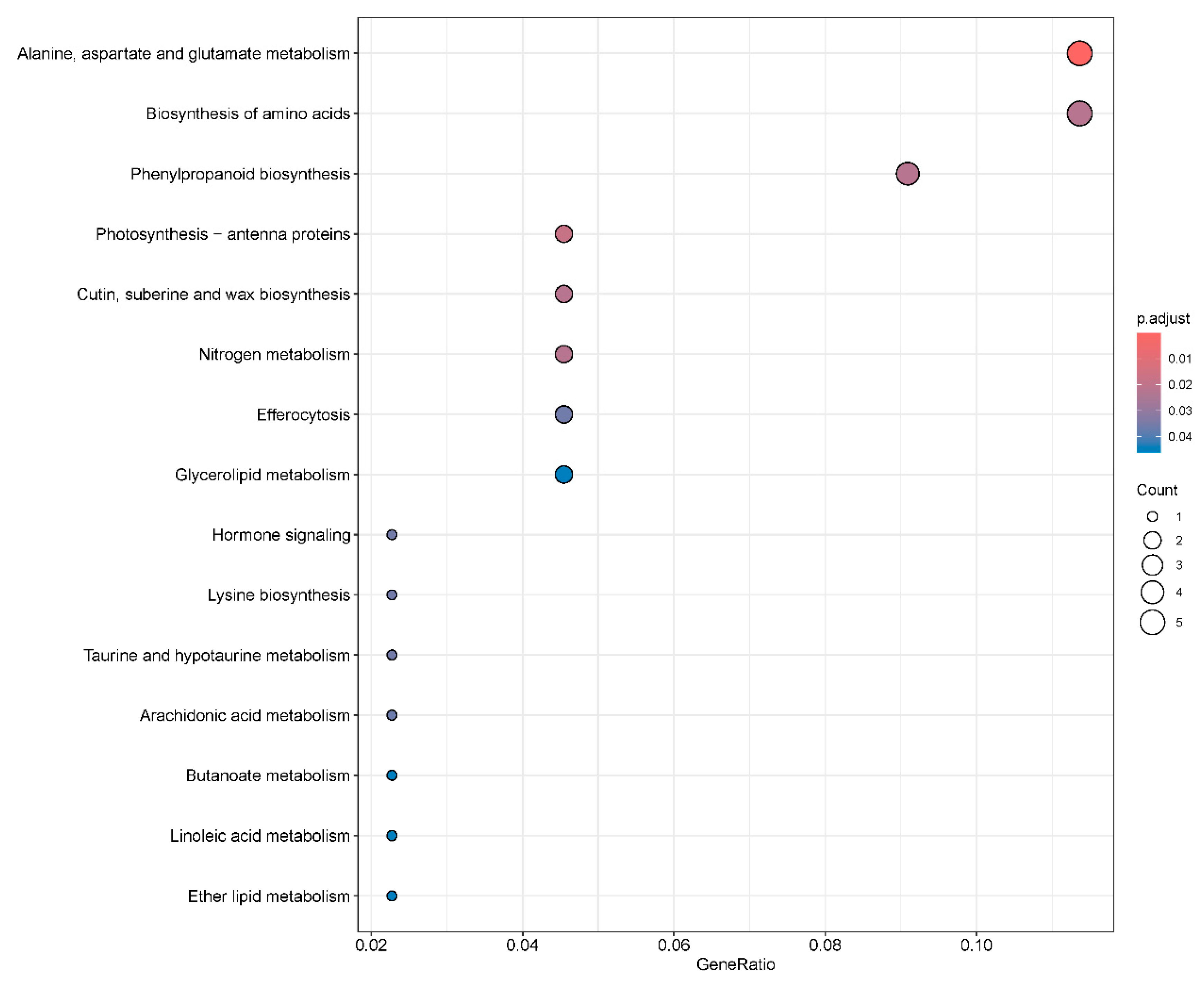
2.6. Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis of Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Vector Construction
4.3. Genetic Transformation and Progeny Identification in Soybean
4.4. Salt Stress Treatment
4.5. Determination of Salt Tolerance Parameters
4.6. RT-qPCR Validation of Gene Expression
4.7. RNA-seq Analysis
4.8. Gene Ontology and KEGG Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qin, P.; Wang, T.; Luo, Y. A review on plant-based proteins from soybean: Health benefits and soy product development. J. Agric. Food Res. 2022, 7, 100265. [Google Scholar] [CrossRef]
- Cuiyu, L.; Ming, Y.; Xianbin, H.; Zhaohe, Y. Effects of NaCl stress on growth and ion homeostasis in pomegranate tissues. Eur. J. Hortic. Sci. 2020, 85, 42–50. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Okur, B.; Örçen, N. Soil salinization and climate change. In Climate Change and Soil Interactions; Elsevier: Amsterdam, The Netherlands, 2020; pp. 331–350. [Google Scholar]
- Afzal, M.; Hindawi, S.E.S.; Alghamdi, S.S.; Migdadi, H.H.; Khan, M.A.; Hasnain, M.U.; Arslan, M.; Habib ur Rahman, M.; Sohaib, M. Potential breeding strategies for improving salt tolerance in crop plants. J. Plant Growth Regul. 2023, 42, 3365–3387. [Google Scholar] [CrossRef]
- Demidchik, V.; Maathuis, F.J. Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 2007, 175, 387–404. [Google Scholar] [CrossRef]
- Demidchik, V.; Tester, M. Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol. 2002, 128, 379–387. [Google Scholar] [CrossRef]
- Hao, S.; Wang, Y.; Yan, Y.; Liu, Y.; Wang, J.; Chen, S. A review on plant responses to salt stress and their mechanisms of salt resistance. Horticulturae 2021, 7, 132. [Google Scholar] [CrossRef]
- Feng, G.; Zhang, F.; Li, X.; Tian, C.; Tang, C.; Rengel, Z. Improved tolerance of maize plants to salt stress by arbuscular mycorrhiza is related to higher accumulation of soluble sugars in roots. Mycorrhiza 2002, 12, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.M. Synthesis versus degradation: Directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol. Biol. 2018, 98, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Wang, J.; Li, K.; Wang, S.; Qin, J.; Zhang, G.; Na, X.; Wang, X.; Bi, Y. Integrated physiological, transcriptomic, and metabolomic analyses revealed molecular mechanism for salt resistance in soybean roots. Int. J. Mol. Sci. 2021, 22, 12848. [Google Scholar] [CrossRef]
- Hu, J.; Zhuang, Y.; Li, X.; Li, X.; Sun, C.; Ding, Z.; Xu, R.; Zhang, D. Time-series transcriptome comparison reveals the gene regulation network under salt stress in soybean (Glycine max) roots. BMC Plant Biol. 2022, 22, 157. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Asif, S.; Jang, Y.-H.; Park, J.-R.; Zhao, D.-D.; Kim, E.-G.; Kim, K.-M. Effect of different salts on nutrients uptake, gene expression, antioxidant, and growth pattern of selected rice genotypes. Front. Plant Sci. 2022, 13, 895282. [Google Scholar] [CrossRef] [PubMed]
- Aizaz, M.; Lubna; Jan, R.; Asaf, S.; Bilal, S.; Kim, K.-M.; AL-Harrasi, A. Regulatory dynamics of plant hormones and transcription factors under salt stress. Biology 2024, 13, 673. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Yang, M.; Teng, Y.; Yue, T.; Wang, Z.; Feng, G.; Ruan, J.; Yan, S.; Zheng, Y.; Zhang, L.; Chen, Q.; et al. The overexpression of peanut (Arachis hypogaea L.) AhALDH2B6 in soybean enhances cold resistance. Plants 2023, 12, 2928. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Li, Y.; Zhao, Y.; Yang, M.; Zhang, X.; Teng, Y.; Jing, L.; Kong, D.; Liu, T.; Li, S.; et al. Heterologous expression of arabidopsis AtARA6 in soybean enhances salt tolerance. Front. Genet. 2022, 13, 849357. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, N.; Yang, J.; Guo, H.; Liu, Z.; Zheng, X.; Li, S.; Xiang, F. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020, 291, 110326. [Google Scholar] [CrossRef]
- Ni, X.; Wang, Y.; Dai, L.; Jiang, K.; Zeng, S.; Huang, Y.; Zhou, Y.; Duan, L.; Bian, C.; Liu, Q.; et al. The transcription factor GmbZIP131 enhances soybean salt tolerance by regulating flavonoid biosynthesis. Plant Physiol. 2025, 197, kiaf092. [Google Scholar] [CrossRef]
- Ma, J.; Wang, L.-Y.; Dai, J.-X.; Wang, Y.; Lin, D. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 11. [Google Scholar] [CrossRef]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef]
- Hussey, S.G.; Mizrachi, E.; Spokevicius, A.V.; Bossinger, G.; Berger, D.K.; Myburg, A.A. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 2011, 11, 173. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, M.R.; Silva, P.A.; Mendes, G.C.; Alves, J.R.; Caetano, H.D.N.; Machado, J.P.B.; Brustolini, O.J.B.; Carpinetti, P.A.; Melo, B.P.; Silva, J.C.F.; et al. The stress-induced soybean NAC transcription factor GmNAC81 plays a positive role in developmentally programmed leaf senescence. Plant Cell Physiol. 2016, 57, 1098–1114. [Google Scholar] [CrossRef]
- Li, S.; Wang, N.; Ji, D.; Zhang, W.; Wang, Y.; Yu, Y.; Zhao, S.; Lyu, M.; You, J.; Zhang, Y.; et al. A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 2019, 31, 2107–2130. [Google Scholar] [CrossRef]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.Y.; Walley, J.; Yin, Y. GSK 3-like kinase BIN 2 phosphorylates RD 26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef]
- Kamranfar, I.; Xue, G.P.; Tohge, T.; Sedaghatmehr, M.; Fernie, A.R.; Balazadeh, S.; Mueller-Roeber, B. Transcription factor RD 26 is a key regulator of metabolic reprogramming during dark-induced senescence. New Phytol. 2018, 218, 1543–1557. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Gao, H.; Zhou, Y.; Jing, Y.; Li, S.; Yan, Z.; Xu, K.; Zhou, F.; Zhang, W.; Yang, X.; et al. Unfolding molecular switches for salt stress resilience in soybean: Recent advances and prospects for salt-tolerant smart plant production. Front. Plant Sci. 2023, 14, 1162014. [Google Scholar] [CrossRef] [PubMed]
- Pruthi, R.; Chaudhary, C.; Sharma, J.; Rana, P.; Kondi, R.K.R.; Richards, J.; Nguyen, H.T.; Subudhi, P.K. A comparative transcriptomic analysis provides insights into molecular mechanisms driving salt tolerance in soybean. Sci. Rep. 2025, 15, 31869. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Jia, B.; Sun, M.; Sun, X. Insights into the regulation of wild soybean tolerance to salt-alkaline stress. Front. Plant Sci. 2022, 13, 1002302. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Nowroz, F.; Fujita, M. Insight into the mechanism of salt-induced oxidative stress tolerance in soybean by the application of Bacillus subtilis: Coordinated actions of osmoregulation, ion homeostasis, antioxidant defense, and methylglyoxal detoxification. Antioxidants 2022, 11, 1856. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; Chen, Y.; Yang, X.; Jiao, S.; Zhang, H.; Ma, X.; Zhai, H.; Bai, X. GmSTK12 participates in salt stress resistance in soybean. Agronomy 2023, 13, 613. [Google Scholar] [CrossRef]
- El Moukhtari, A.; Cabassa-Hourton, C.; Farissi, M.; Savouré, A. How does proline treatment promote salt stress tolerance during crop plant development? Front. Plant Sci. 2020, 11, 1127. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Wang, L.; Tan, M.; Ogutu, C.O.; Yin, Z.; Zhou, J.; Wang, J.; Wang, L.; Yan, X. Transcriptome analysis and molecular mechanism of linseed (Linum usitatissimum L.) drought tolerance under repeated drought using single-molecule long-read sequencing. BMC Genom. 2021, 22, 109. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, W.; Chen, H.; Chen, J.; Liu, X.; Chen, X.; Yang, S. Transcriptomic analysis of salt tolerance-associated genes and diversity analysis using indel markers in yardlong bean (Vigna unguiculata ssp. sesquipedialis). BMC Genom. Data 2021, 22, 34. [Google Scholar] [CrossRef]
- Zhang, L.; Li, T.; Wang, Y.; Zhang, Y.; Dong, Y.-s. FvC5SD overexpression enhances drought tolerance in soybean by reactive oxygen species scavenging and modulating stress-responsive gene expression. Plant Cell Rep. 2019, 38, 1039–1051. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Chen, J.; Tao, L.; An, Y.; Cai, H.; Guo, C. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS ONE 2018, 13, e0192382. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Reuveni, R.; Shimoni, M.; Karchi, Z.; Kuc, J. Peroxidase activity as a biochemical marker for resistance of muskmelon (Cucumis melo) to Pseudoperonospora cubensis. Phytopathology 1992, 82, 749–753. [Google Scholar] [CrossRef]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Research 2021, 8, 1874. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, D.; Zhang, W.; Li, Y.; Li, W.; Meng, F.; Lu, W. Overexpression of GmNAC03 in Soybean Enhances Salt Tolerance. Plants 2025, 14, 3235. https://doi.org/10.3390/plants14213235
Han D, Zhang W, Li Y, Li W, Meng F, Lu W. Overexpression of GmNAC03 in Soybean Enhances Salt Tolerance. Plants. 2025; 14(21):3235. https://doi.org/10.3390/plants14213235
Chicago/Turabian StyleHan, Dezhi, Wu Zhang, Yang Li, Wei Li, Fanli Meng, and Wencheng Lu. 2025. "Overexpression of GmNAC03 in Soybean Enhances Salt Tolerance" Plants 14, no. 21: 3235. https://doi.org/10.3390/plants14213235
APA StyleHan, D., Zhang, W., Li, Y., Li, W., Meng, F., & Lu, W. (2025). Overexpression of GmNAC03 in Soybean Enhances Salt Tolerance. Plants, 14(21), 3235. https://doi.org/10.3390/plants14213235





