Understanding Callus Types in Maize by Genetic Mapping and Transcriptional Profiling
Abstract
1. Introduction
2. Results
2.1. Genetic Mapping of the Callus Type
2.2. Alleles Contributing to Type II Callus Formation
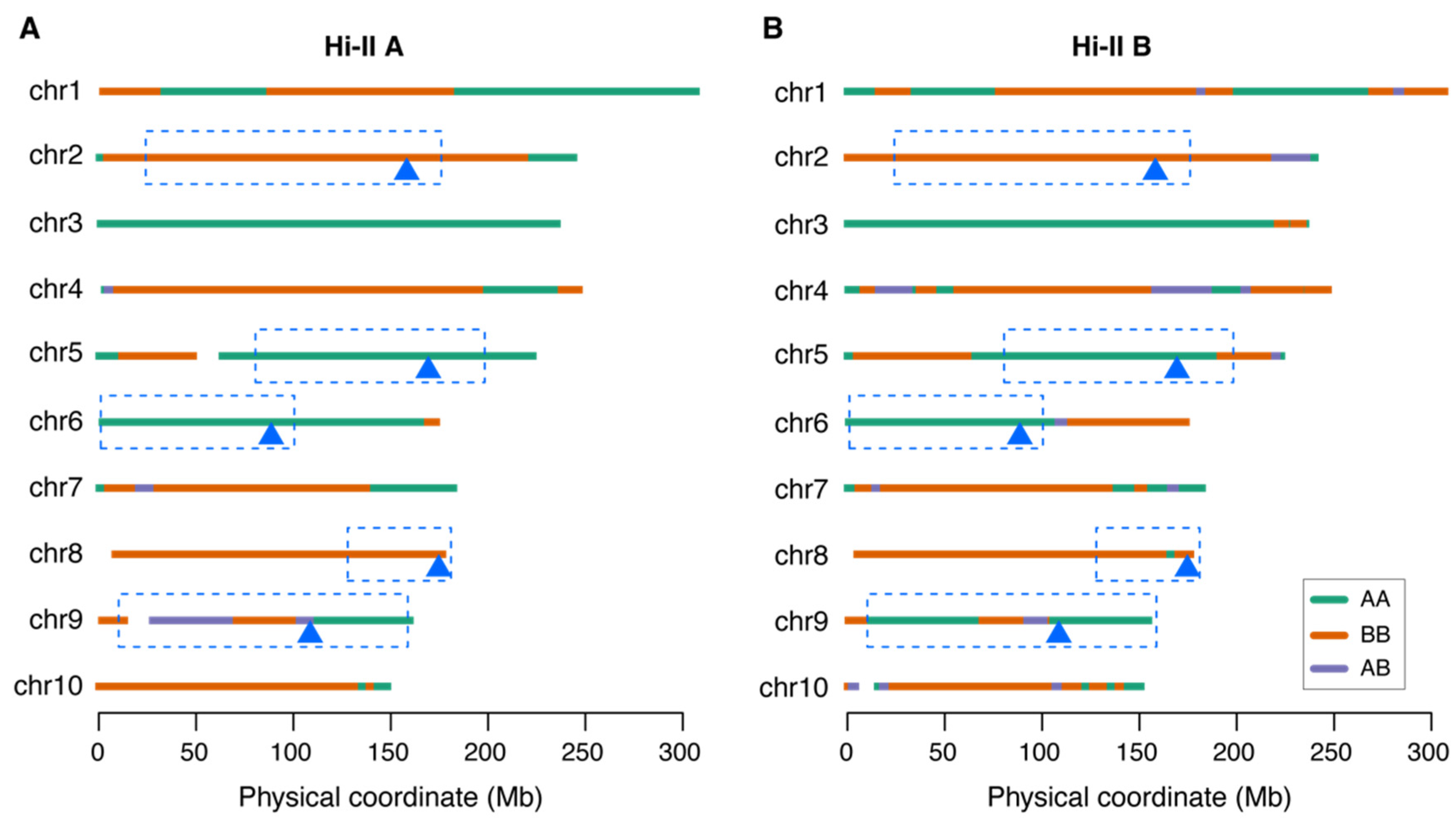
2.3. Type II Callus Favorable Alleles of Many QTLs Were Selected in Both Hi-II A and B
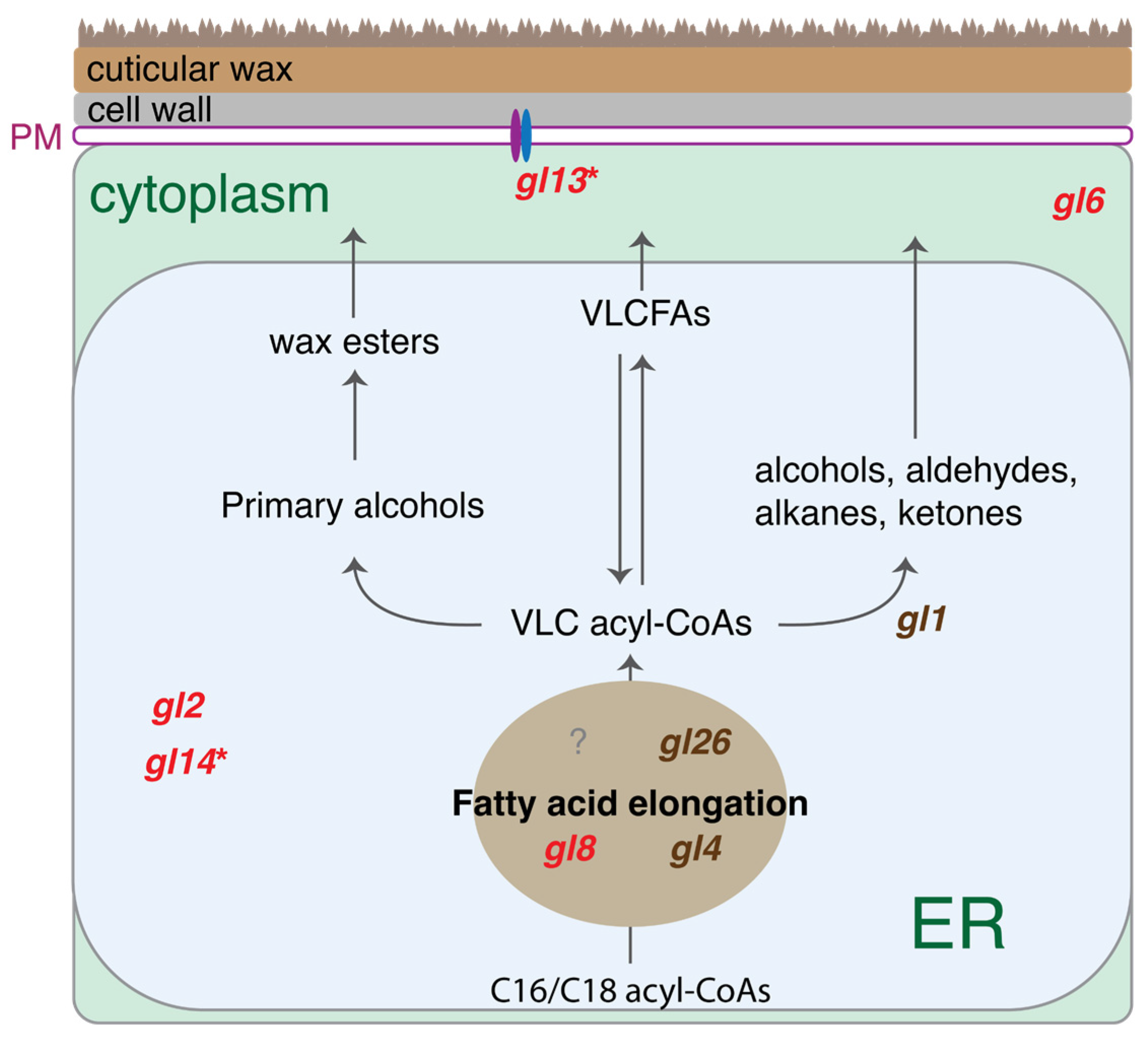
2.4. Differential Expression Between Different Types of Calli
2.5. Integration of Genetic Mapping and DEGs
3. Discussion
4. Materials and Methods
4.1. Establishment of B73xA188 F2 and A188 Calli
4.2. DNA Isolation and GBS Sequencing
4.3. Genotypes of GBS Segment Markers
4.4. Genetic Mapping Using Logistic Regression
4.5. QTL Mapping Using R/qtl
4.6. RNA Extraction and Sequencing of Bulked Samples
4.7. BSR-seq Analysis
4.8. Identification of Callus Type-Associated Genes
4.9. Gene Ontology (GO) Enrichment Analysis
4.10. Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Que, Q.; Elumalai, S.; Li, X.; Zhong, H.; Nalapalli, S.; Schweiner, M.; Fei, X.; Nuccio, M.; Kelliher, T.; Gu, W.; et al. Maize Transformation Technology Development for Commercial Event Generation. Front. Plant Sci. 2014, 5, 379. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.L.; Green, C.E.; Phillips, R.L. Development and Availability of Germplasm with High Type II Culture Formation Response. Maize Genet. Coop. Newsl. 1991, 65, 92–93. [Google Scholar]
- Duncan, D.R.; Williams, M.E.; Zehr, B.E.; Widholm, J.M. The Production of Callus Capable of Plant Regeneration from Immature Embryos of Numerous Zea mays Genotypes. Planta 1985, 165, 322–332. [Google Scholar] [CrossRef]
- Kotchoni, S.O.; Noumavo, P.A.; Adjanohoun, A.; Russo, D.P.; Dell’Angelo, J.; Gachomo, E.W.; Baba-Moussa, L. A Simple and Efficient Seed-Based Approach to Induce Callus Production from B73 Maize Genotype. Am. J. Mol. Biol. 2012, 2, 380–385. [Google Scholar] [CrossRef]
- Cho, M.-J.; Wu, E.; Kwan, J.; Yu, M.; Banh, J.; Linn, W.; Anand, A.; Li, Z.; TeRonde, S.; Register, J.C., 3rd; et al. Agrobacterium-Mediated High-Frequency Transformation of an Elite Commercial Maize (Zea mays L.) Inbred Line. Plant Cell Rep. 2014, 33, 1767–1777. [Google Scholar] [CrossRef]
- Cho, M.-J.; Banh, J.; Yu, M.; Kwan, J.; Jones, T.J. Improvement of Agrobacterium-Mediated Transformation Frequency in Multiple Modern Elite Commercial Maize (Zea mays L.) Inbreds by Media Modifications. Plant Cell Tissue Organ. Cult. 2015, 121, 519–529. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby Boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The Gene TaWOX5 Overcomes Genotype Dependency in Wheat Genetic Transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF–GIF Chimeric Protein Improves the Regeneration Efficiency of Transgenic Plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef]
- Tomes, D.T.; Smith, O.S. The Effect of Parental Genotype on Initiation of Embryogenic Callus from Elite Maize (Zea mays L.) Germplasm. Theor. Appl. Genet. 1985, 70, 505–509. [Google Scholar] [CrossRef]
- Hodges, T.K.; Kamo, K.K.; Imbrie, C.W.; Becwar, M.R. Genotype Specificity of Somatic Embryogenesis and Regeneration in Maize. Nat. Biotechnol. 1986, 4, 219–223. [Google Scholar] [CrossRef]
- Welter, M.E.; Clayton, D.S.; Miller, M.A.; EPetolino, J. Morphotypes of Friable Embryogenic Maize Callus. Plant Cell Rep. 1995, 14, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Frame, B.R.; Zhang, H.; Cocciolone, S.M.; Sidorenko, L.V.; Dietrich, C.R.; Pegg, S.E.; Zhen, S.; Schnable, P.S.; Wang, K. Production of Transgenic Maize from Bombarded Type II Callus: Effect of Gold Particle Size and Callus Morphology on Transformation Efficiency. Vitr. Cell. Dev. Biol. Plant 2000, 36, 21–29. [Google Scholar] [CrossRef]
- D’Halluin, K.; Bonne, E.; Bossut, M.; De Beuckeleer, M.; Leemans, J. Transgenic Maize Plants by Tissue Electroporation. Plant Cell 1992, 4, 1495–1505. [Google Scholar] [PubMed]
- Armstrong, C.L.; Green, C.E. Establishment and Maintenance of Friable, Embryogenic Maize Callus and the Involvement of L-Proline. Planta 1985, 164, 207–214. [Google Scholar] [CrossRef]
- Kausch, A.P.; Wang, K.; Kaeppler, H.F.; Gordon-Kamm, W. Maize Transformation: History, Progress, and Perspectives. Mol. Breed. 2021, 41, 38. [Google Scholar] [CrossRef]
- McCain, J.W.; Kamo, K.K.; Hodges, T.K. Characterization of Somatic Embryo Development and Plant Regeneration from Friable Maize Callus Cultures. Bot. Gaz. 1988, 149, 16–20. [Google Scholar] [CrossRef]
- Ishida, Y.; Saito, H.; Ohta, S.; Hiei, Y.; Komari, T.; Kumashiro, T. High Efficiency Transformation of Maize (Zea mays L.) Mediated by Agrobacterium Tumefaciens. Nat. Biotechnol. 1996, 14, 745–750. [Google Scholar] [CrossRef]
- Songstad, D.D.; Armstrong, C.L.; Petersen, W.L.; Hairston, B.; Hinchee, M.A.W. Production of Transgenic Maize Plants and Progeny by Bombardment of Hi-II Immature Embryos. Vitr. Cell. Dev. Biol. Plant 1996, 32, 179–183. [Google Scholar] [CrossRef]
- Lin, G.; He, C.; Zheng, J.; Koo, D.-H.; Le, H.; Zheng, H.; Tamang, T.M.; Lin, J.; Liu, Y.; Zhao, M.; et al. Chromosome-Level Genome Assembly of a Regenerable Maize Inbred Line A188. Genome Biol. 2021, 22, 175. [Google Scholar] [CrossRef]
- Armstrong, C.L.; Romero-Severson, J.; Hodges, T.K. Improved Tissue Culture Response of an Elite Maize Inbred through Backcross Breeding, and Identification of Chromosomal Regions Important for Regeneration by RFLP Analysis. Züchter Genet. Breed. Res. 1992, 84, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Lowe, B.A.; Way, M.M.; Kumpf, J.M.; Rout, J.; Warner, D.; Johnson, R.; Armstrong, C.L.; Spencer, M.T.; Chomet, P.S. Marker Assisted Breeding for Transformability in Maize. Mol. Breed. 2006, 18, 229–239. [Google Scholar] [CrossRef]
- Krakowsky, M.D.; Lee, M.; Garay, L.; Woodman-Clikeman, W.; Long, M.J.; Sharopova, N.; Frame, B.; Wang, K. Quantitative Trait Loci for Callus Initiation and Totipotency in Maize (Zea mays L.). Züchter Genet. Breed. Res. 2006, 113, 821–830. [Google Scholar] [CrossRef][Green Version]
- Salvo, S.; Cook, J.; Carlson, A.R.; Hirsch, C.N.; Kaeppler, S.M.; Kaeppler, H.F. Genetic Fine-Mapping of a Quantitative Trait Locus (QTL) Associated with Embryogenic Tissue Culture Response and Plant Regeneration Ability in Maize (Zea mays L.). Plant Genome 2018, 11, 170111. [Google Scholar] [CrossRef] [PubMed]
- McFarland, F.L.; Collier, R.; Walter, N.; Martinell, B.; Kaeppler, S.M.; Kaeppler, H.F. A Key to Totipotency: Wuschel-like Homeobox 2a Unlocks Embryogenic Culture Response in Maize (Zea mays L.). Plant Biotechnol. J. 2023, 21, 1860–1872. [Google Scholar] [CrossRef]
- Chu, C.C. Establishment of an Efficient Medium for Anther Culture of Rice through Comparative Experiments on the Nitrogen Sources. Sci. Sin. 1975, 18, 659–668. [Google Scholar]
- Ren, W.; Gong, X.; Li, K.; Zhang, H.; Chen, F.; Pan, Q. Recombination Pattern Characterization via Simulation Using Different Maize Populations. Int. J. Mol. Sci. 2020, 21, 2222. [Google Scholar] [CrossRef]
- Hoopes, G.M.; Hamilton, J.P.; Wood, J.C.; Esteban, E.; Pasha, A.; Vaillancourt, B.; Provart, N.J.; Buell, C.R. An Updated Gene Atlas for Maize Reveals Organ-Specific and Stress-Induced Genes. Plant J. 2019, 97, 1154–1167. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Robin Buell, C.; de Leon, N.; Kaeppler, S.M. An Expanded Maize Gene Expression Atlas Based on RNA Sequencing and Its Use to Explore Root Development. Plant Genome 2016, 9, plantgenome2015.04.0025. [Google Scholar] [CrossRef]
- Li, L.; Li, D.; Liu, S.; Ma, X.; Dietrich, C.R.; Hu, H.-C.; Zhang, G.; Liu, Z.; Zheng, J.; Wang, G.; et al. The Maize glossy13 Gene, Cloned via BSR-Seq and Seq-Walking Encodes a Putative ABC Transporter Required for the Normal Accumulation of Epicuticular Waxes. PLoS ONE 2013, 8, e82333. [Google Scholar] [CrossRef]
- Zheng, J.; He, C.; Qin, Y.; Lin, G.; Park, W.D.; Sun, M. Co-expression Analysis Aids in the Identification of Genes in the Cuticular Wax Pathway in Maize. Plant J. 2019, 97, 530–542. [Google Scholar] [CrossRef]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax Biosynthesis in Response to Danger: Its Regulation upon Abiotic and Biotic Stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Sun, W.; Chen, Z.; Shi, L.; Hong, J.; Shi, J. Plant GDSL Esterases/Lipases: Evolutionary, Physiological and Molecular Functions in Plant Development. Plants 2022, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Adesina, O.; Bika, R.; Singh, R.; Jugulam, M.; Liu, S. Homotools: A Suite of Genomic Tools for Homologous Retrieval and Comparison. Genom. Commun. 2024, 1, e002. [Google Scholar] [CrossRef]
- Ranocha, P.; Denancé, N.; Vanholme, R.; Freydier, A.; Martinez, Y.; Hoffmann, L.; Köhler, L.; Pouzet, C.; Renou, J.-P.; Sundberg, B.; et al. Walls Are Thin 1 (WAT1), an Arabidopsis Homolog of Medicago Truncatula NODULIN21, Is a Tonoplast-Localized Protein Required for Secondary Wall Formation in Fibers. Plant J. 2010, 63, 469–483. [Google Scholar] [CrossRef]
- Koseoglou, E.; Hanika, K.; Mohd Nadzir, M.M.; Kohlen, W.; van der Wolf, J.M.; Visser, R.G.F.; Bai, Y. Inactivation of Tomato WAT1 Leads to Reduced Susceptibility to Clavibacter Michiganensis through Downregulation of Bacterial Virulence Factors. Front. Plant Sci. 2023, 14, 1082094. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in Plant Growth and Potential Applications in Crop Improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef]
- Shen, Y.; Jiang, Z.; Yao, X.; Zhang, Z.; Lin, H.; Zhao, M.; Liu, H.; Peng, H.; Li, S.; Pan, G. Genome Expression Profile Analysis of the Immature Maize Embryo during Dedifferentiation. PLoS ONE 2012, 7, e32237. [Google Scholar] [CrossRef]
- Nishikubo, N.; Takahashi, J.; Roos, A.A.; Derba-Maceluch, M.; Piens, K.; Brumer, H.; Teeri, T.T.; Stålbrand, H.; Mellerowicz, E.J. Xyloglucan Endo-Transglycosylase-Mediated Xyloglucan Rearrangements in Developing Wood of Hybrid Aspen. Plant Physiol. 2011, 155, 399–413. [Google Scholar] [CrossRef]
- Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Luo, X.; Huang, X.; Zhang, Y.; He, X.; Liu, M.; Lin, H.; Peng, H.; Li, L.; Zhang, Z.; et al. Genome-Wide Analysis of Transcription Factors Involved in Maize Embryonic Callus Formation. Physiol. Plant. 2016, 158, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular Mechanisms of Plant Regeneration. Annu. Rev. Plant Biol. 2019, 70, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jung, J.H.; Lee, S.B.; Go, Y.S.; Kim, H.J.; Cahoon, R.; Markham, J.E.; Cahoon, E.B.; Suh, M.C. Arabidopsis 3-Ketoacyl-Coenzyme a synthase9 Is Involved in the Synthesis of Tetracosanoic Acids as Precursors of Cuticular Waxes, Suberins, Sphingolipids, and Phospholipids. Plant Physiol. 2013, 162, 567–580. [Google Scholar] [CrossRef]
- Liu, S.; Dietrich, C.R.; Schnable, P.S. DLA-Based Strategies for Cloning Insertion Mutants: Cloning the gl4 Locus of Maize Using Mu Transposon Tagged Alleles. Genetics 2009, 183, 1215–1225. [Google Scholar] [CrossRef]
- Shang, B.; Xu, C.; Zhang, X.; Cao, H.; Xin, W.; Hu, Y. Very-Long-Chain Fatty Acids Restrict Regeneration Capacity by Confining Pericycle Competence for Callus Formation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 5101–5106. [Google Scholar] [CrossRef]
- Nobusawa, T.; Okushima, Y.; Nagata, N.; Kojima, M.; Sakakibara, H.; Umeda, M. Synthesis of Very-Long-Chain Fatty Acids in the Epidermis Controls Plant Organ Growth by Restricting Cell Proliferation. PLoS Biol. 2013, 11, e1001531. [Google Scholar] [CrossRef]
- Qin, Y.-M.; Hu, C.-Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.-X. Saturated Very-Long-Chain Fatty Acids Promote Cotton Fiber and Arabidopsis Cell Elongation by Activating Ethylene Biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef]
- Frame, B.R.; McMurray, J.M.; Fonger, T.M.; Main, M.L.; Taylor, K.W.; Torney, F.J.; Paz, M.M.; Wang, K. Improved Agrobacterium-Mediated Transformation of Three Maize Inbred Lines Using MS Salts. Plant Cell Rep. 2006, 25, 1024–1034. [Google Scholar] [CrossRef]
- Liu, S.; Yeh, C.-T.; Tang, H.M.; Nettleton, D.; Schnable, P.S. Gene Mapping via Bulked Segregant RNA-Seq (BSR-Seq). PLoS ONE 2012, 7, e36406. [Google Scholar] [CrossRef]
- Ott, A.; Liu, S.; Schnable, J.C.; Yeh, C.T.E. tGBS® Genotyping-by-Sequencing Enables Reliable Genotyping of Heterozygous Loci. Nucleic Acids 2017, 45, e178. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, J.; Migeon, P.; Ren, J.; Hu, Y.; He, C.; Liu, H.; Fu, J.; White, F.F.; Toomajian, C.; et al. Unbiased K-Mer Analysis Reveals Changes in Copy Number of Highly Repetitive Sequences During Maize Domestication and Improvement. Sci. Rep. 2017, 7, 42444. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Peluso, P.; Shi, J.; Liang, T.; Stitzer, M.C.; Wang, B.; Campbell, M.S.; Stein, J.C.; Wei, X.; Chin, C.-S.; et al. Improved Maize Reference Genome with Single-Molecule Technologies. Nature 2017, 546, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling Accurate Genetic Variant Discovery to Tens of Thousands of Samples. bioRxiv. 2018, 201178. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Broman, K.W.; Wu, H.; Sen, S.; Churchill, G.A. R/qtl: QTL Mapping in Experimental Crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef]
- Broman, K.W. Review of Statistical Methods for QTL Mapping in Experimental Crosses. Lab Anim. 2001, 30, 44–52. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
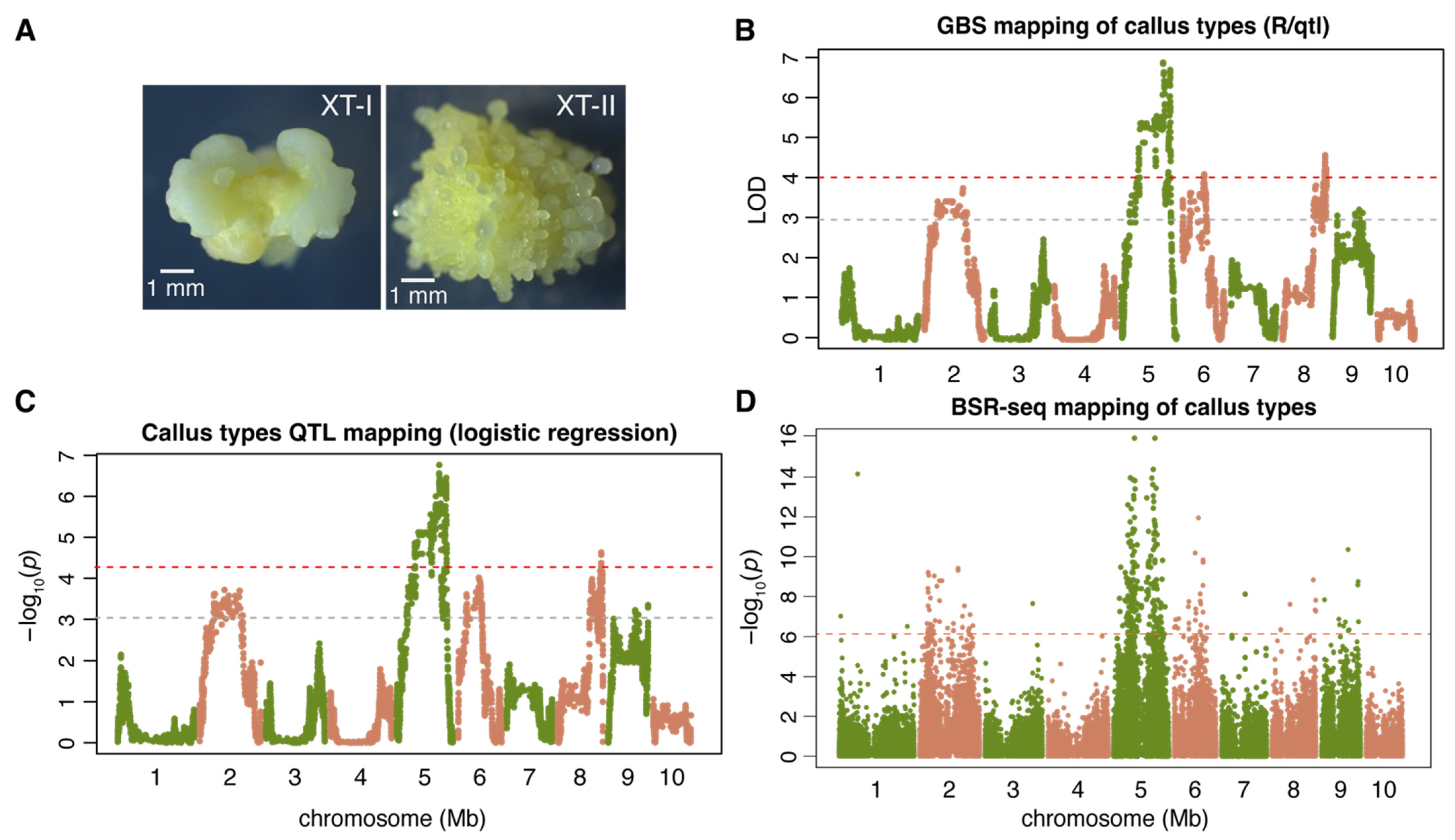

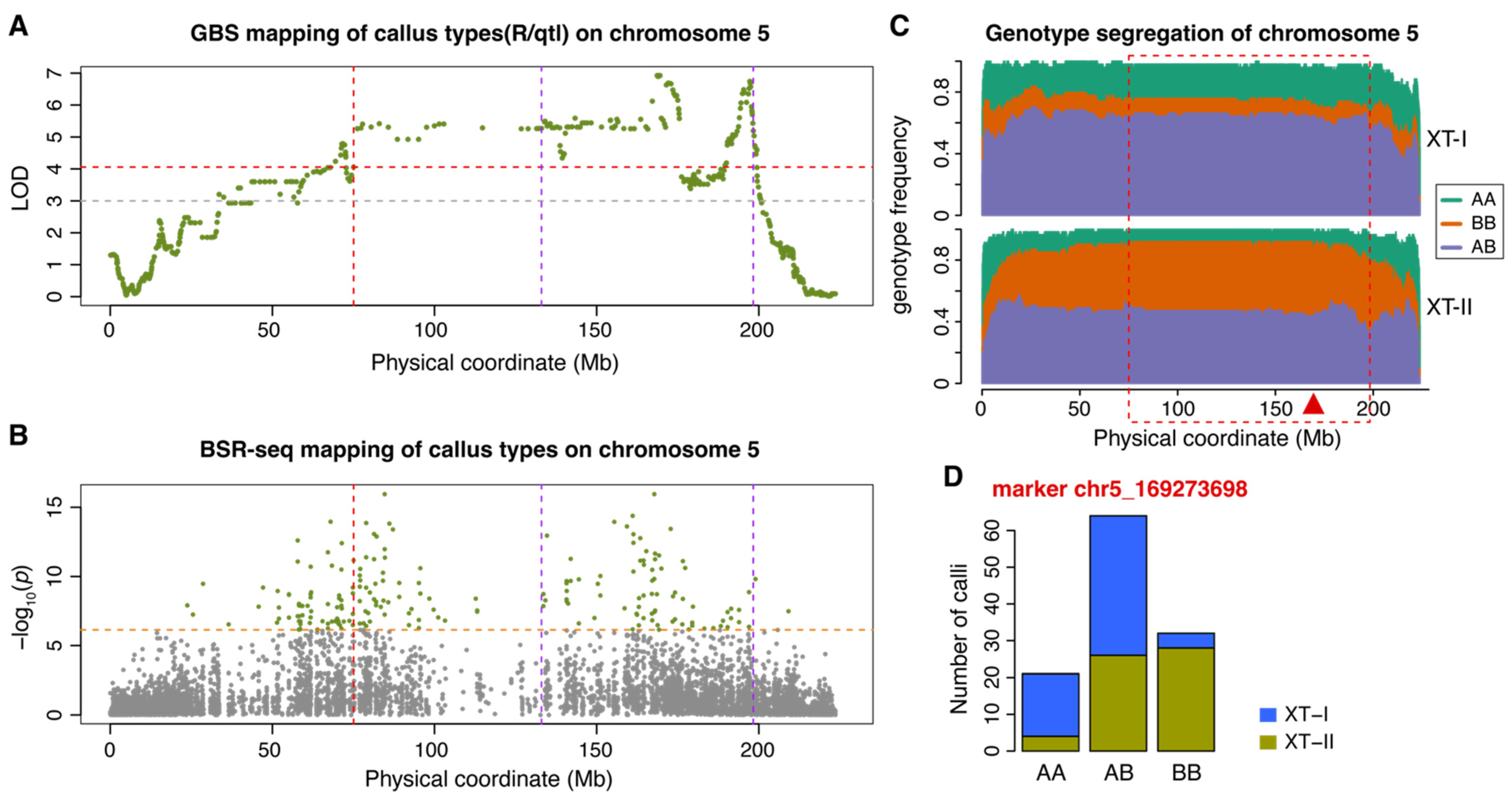

| QTL | chr | R/qtl | Logistic Regression | BSR-seq | QTL Interval (bp) * | ||||
|---|---|---|---|---|---|---|---|---|---|
| Pos (bp) | LOD | Pos (bp) | p-Value | Pos (bp) | p-Value | Start | End | ||
| ctAB5a | 5 | 169,273,698 | 6.9 | 169,273,698 | 1.7 × 10−7 | 84,632,155 | 0.0 × 100 | 80,458,296 | 198,270,432 |
| ctAB8a | 8 | 174,632,571 | 4.6 | 174,479,910 | 2.4 × 10−5 | 169,073,812 | 1.5 × 10−9 | 127,872,514 | 180,929,814 |
| ctAB6a | 6 | 88,567,616 | 4.1 | 83,296,319 | 9.6 × 10−5 | 99,960,681 | 1.1 × 10−12 | 1,006,642 | 100,419,946 |
| ctAB2a | 2 | 158,183,928 | 3.8 | 99,580,047 | 1.9 × 10−4 | 156,883,753 | 3.9 × 10−10 | 24,095,104 | 176,011,606 |
| ctAB9a | 9 | 108,566,128 | 3.3 | 159,752,864 | 4.5 × 10−4 | 108,540,812 | 4.4 × 10−11 | 10,213,832 | 158,723,248 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, G.; Liu, Y.; Tamang, T.M.; Qin, Y.; Zhao, M.; Zheng, J.; Wang, G.; Wei, H.; Park, S.; Cho, M.-J.; et al. Understanding Callus Types in Maize by Genetic Mapping and Transcriptional Profiling. Plants 2025, 14, 3168. https://doi.org/10.3390/plants14203168
Lin G, Liu Y, Tamang TM, Qin Y, Zhao M, Zheng J, Wang G, Wei H, Park S, Cho M-J, et al. Understanding Callus Types in Maize by Genetic Mapping and Transcriptional Profiling. Plants. 2025; 14(20):3168. https://doi.org/10.3390/plants14203168
Chicago/Turabian StyleLin, Guifang, Yan Liu, Tej Man Tamang, Yang Qin, Mingxia Zhao, Jun Zheng, Guoying Wang, Hairong Wei, Sunghun Park, Myeong-Je Cho, and et al. 2025. "Understanding Callus Types in Maize by Genetic Mapping and Transcriptional Profiling" Plants 14, no. 20: 3168. https://doi.org/10.3390/plants14203168
APA StyleLin, G., Liu, Y., Tamang, T. M., Qin, Y., Zhao, M., Zheng, J., Wang, G., Wei, H., Park, S., Cho, M.-J., White, F. F., Liu, Y., & Liu, S. (2025). Understanding Callus Types in Maize by Genetic Mapping and Transcriptional Profiling. Plants, 14(20), 3168. https://doi.org/10.3390/plants14203168






