Comprehensive Identification and Expression Analysis of the SWEET Gene Family in Actinidia eriantha Reveals That Two AeSWEET11 Genes Function in Sucrose and Hexose Transport
Abstract
1. Introduction
2. Results
2.1. Identification of the AeSWEET Family Genes in A. eriantha
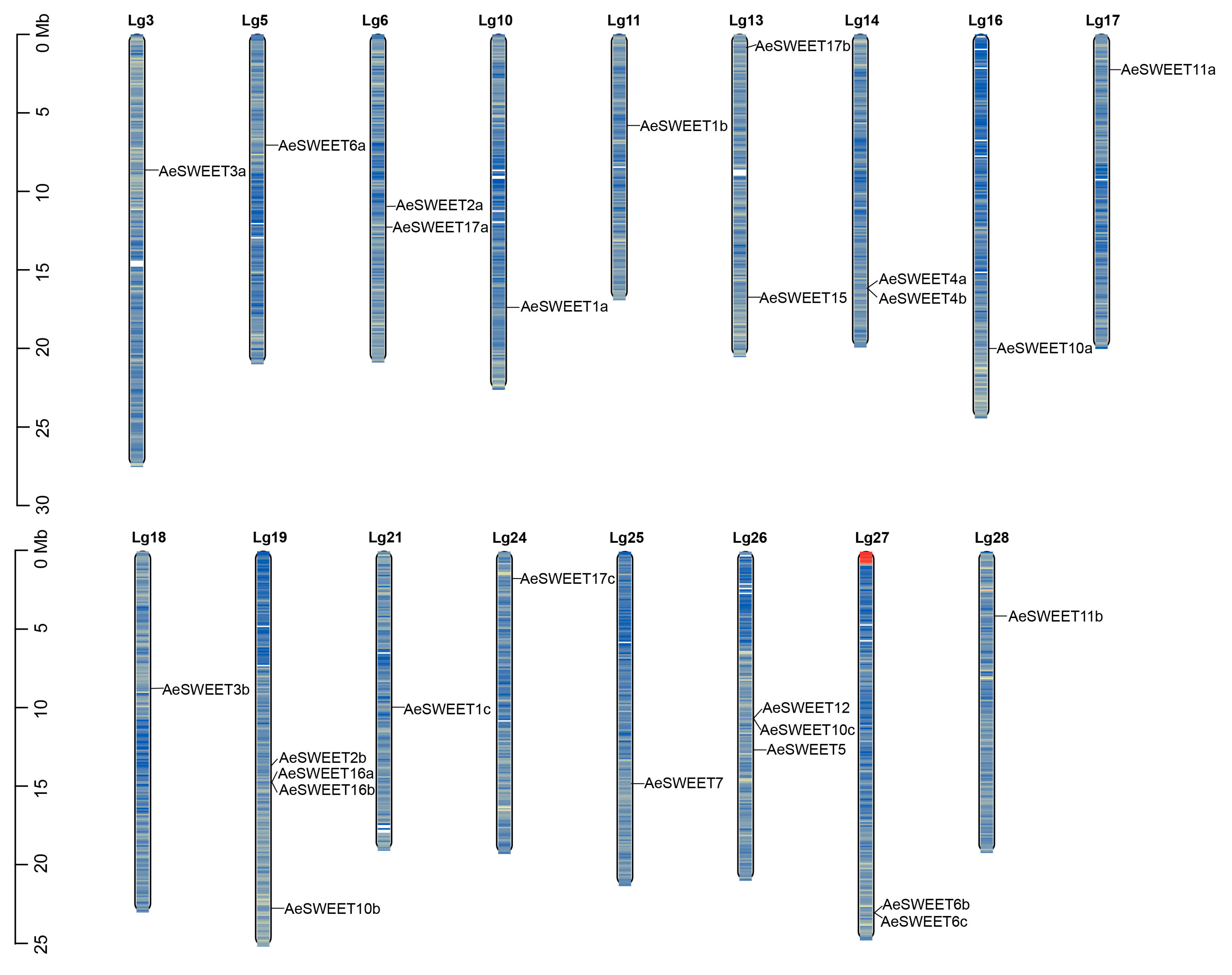
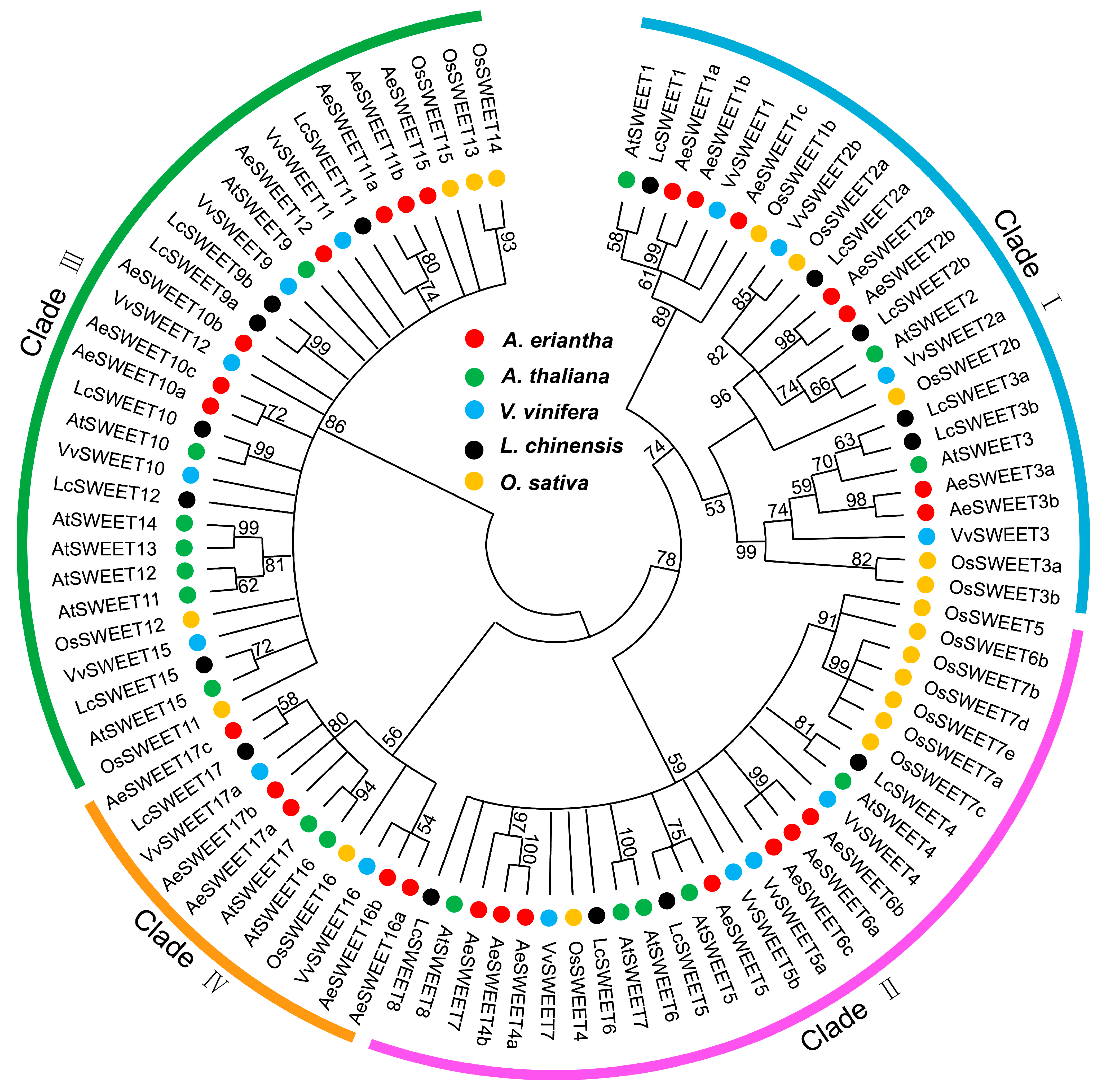
2.2. Chromosomal Mapping and Evolutionary Analysis of AeSWEETs
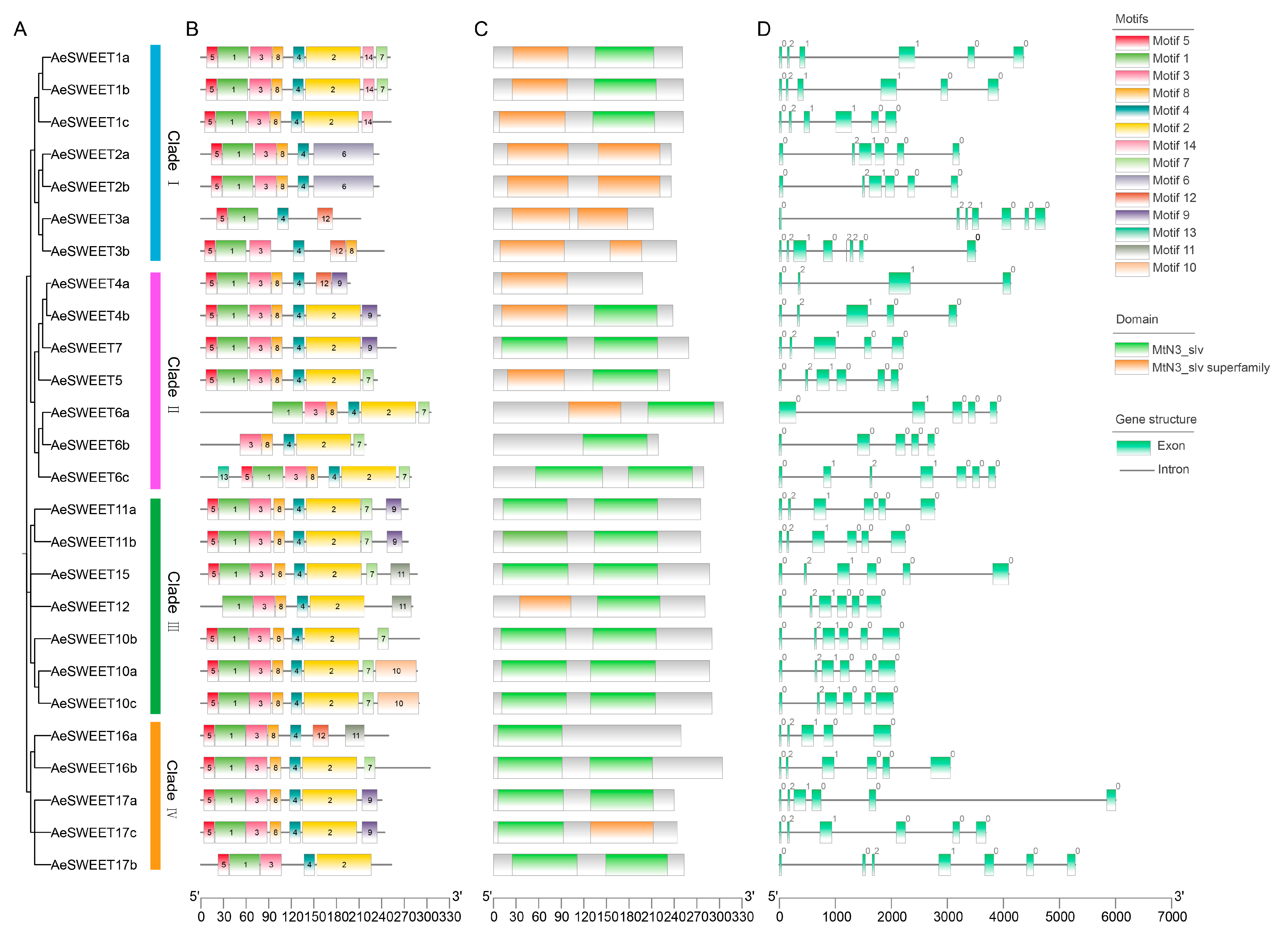
2.3. Protein Motifs, Conserved Domains and Gene Structure Features of AeSWEETs
2.4. Collinearity and Synteny Patterns of AeSWEETs
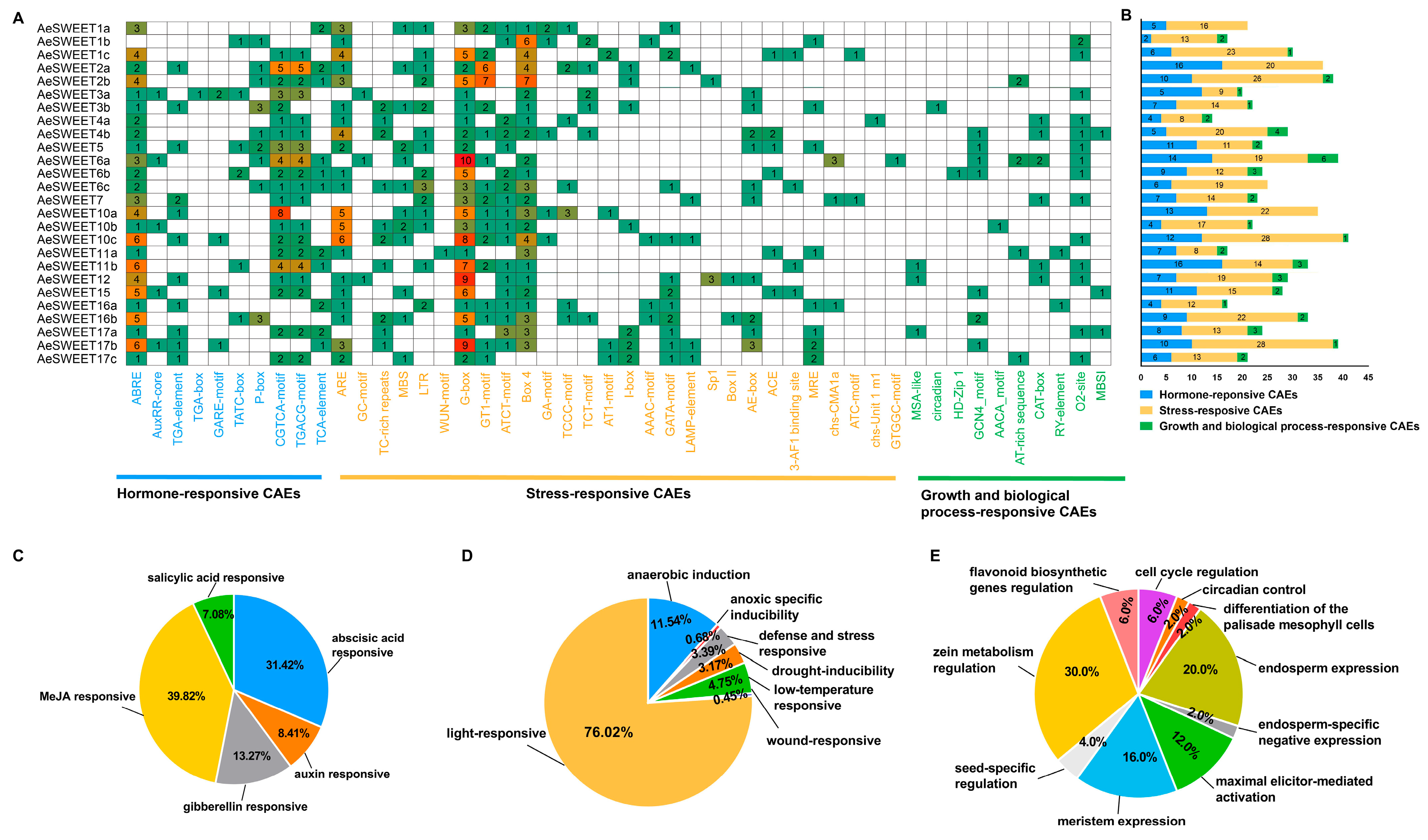
2.5. Diverse Cis-Acting Elements Present in the Promoters of AeSWEET Genes
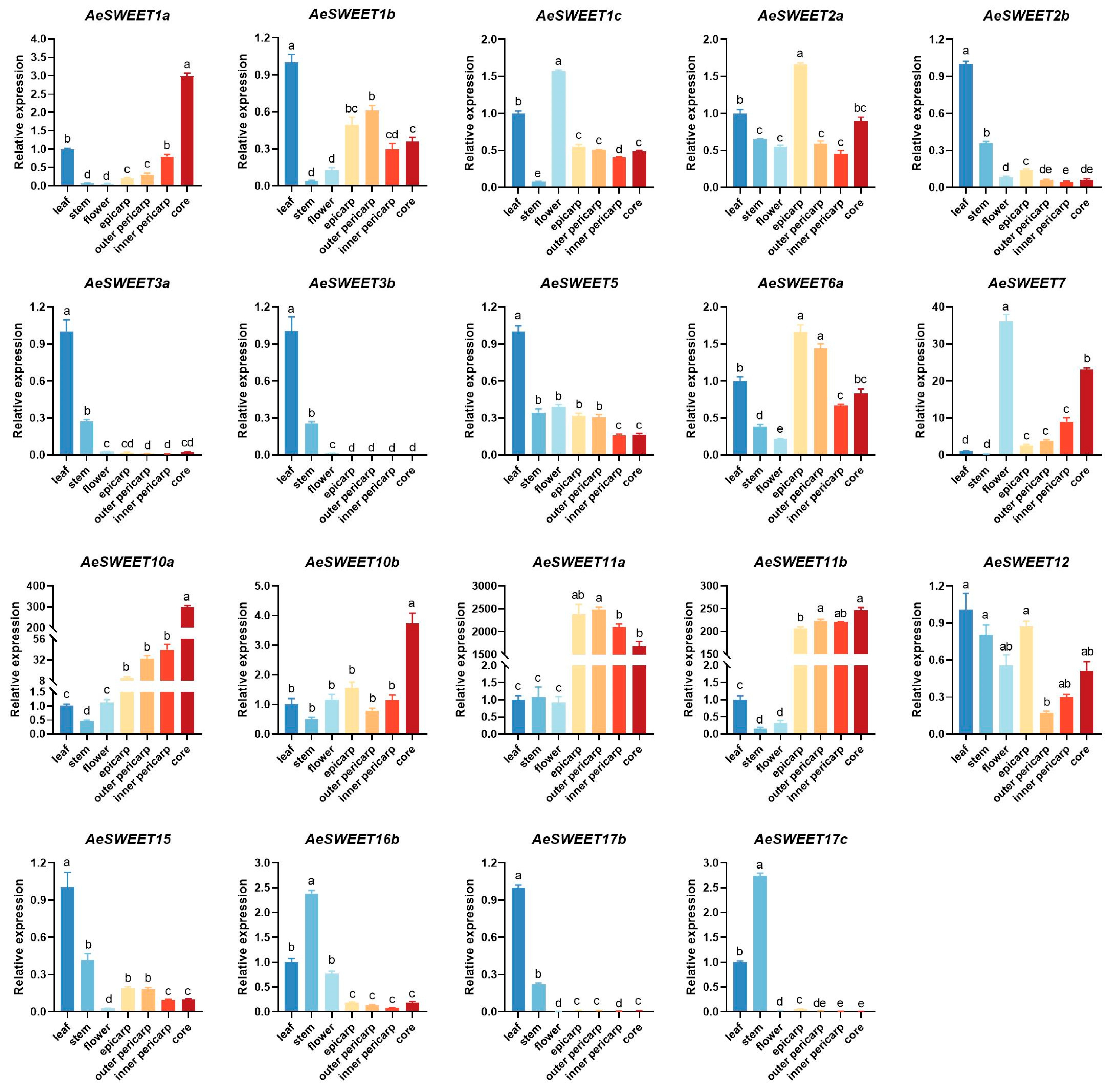
2.6. Tissue Expression Patterns of the AeSWEET Family Genes
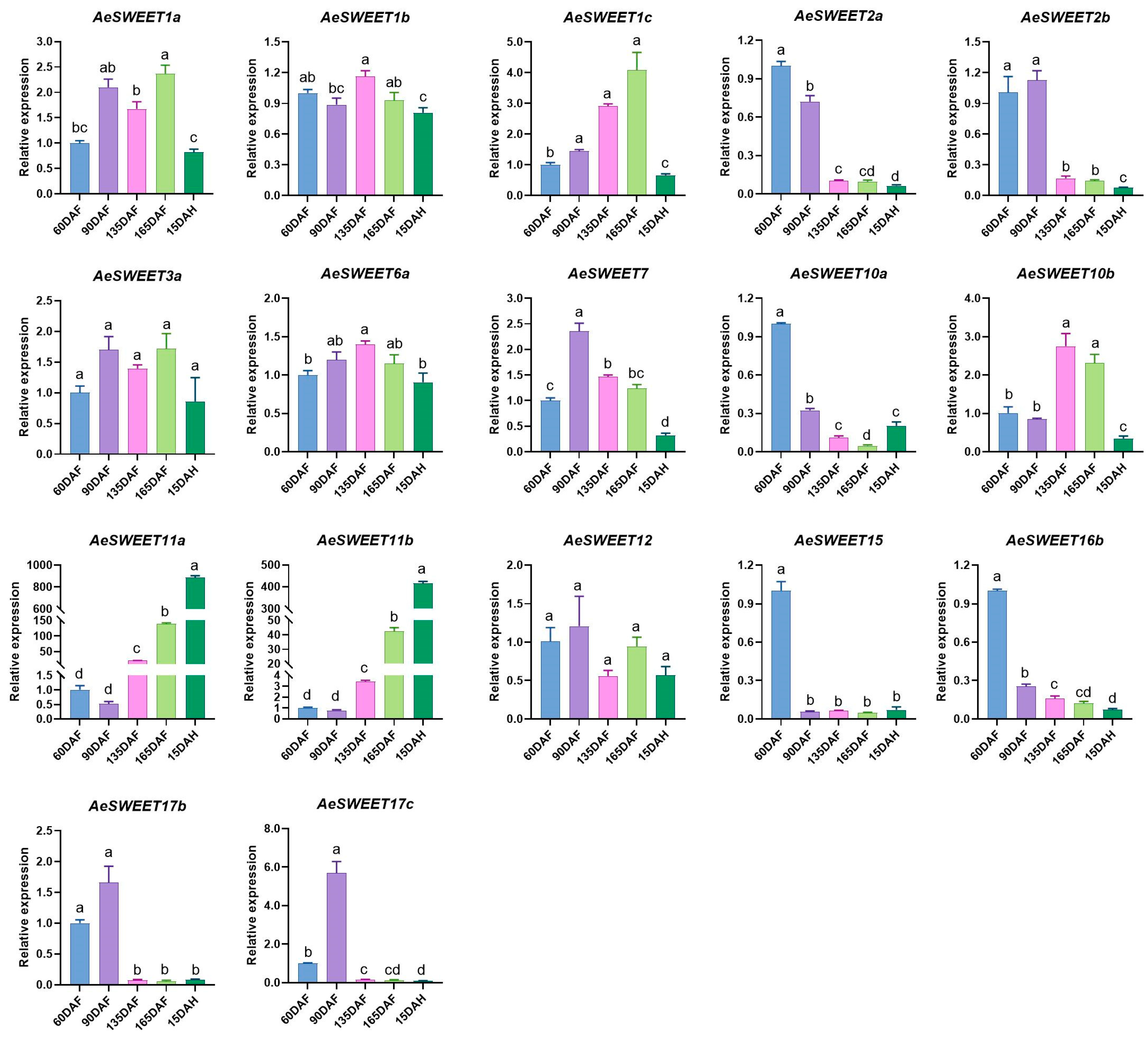
2.7. Differential Expression of AeSWEET Genes During Fruit Development and Ripening
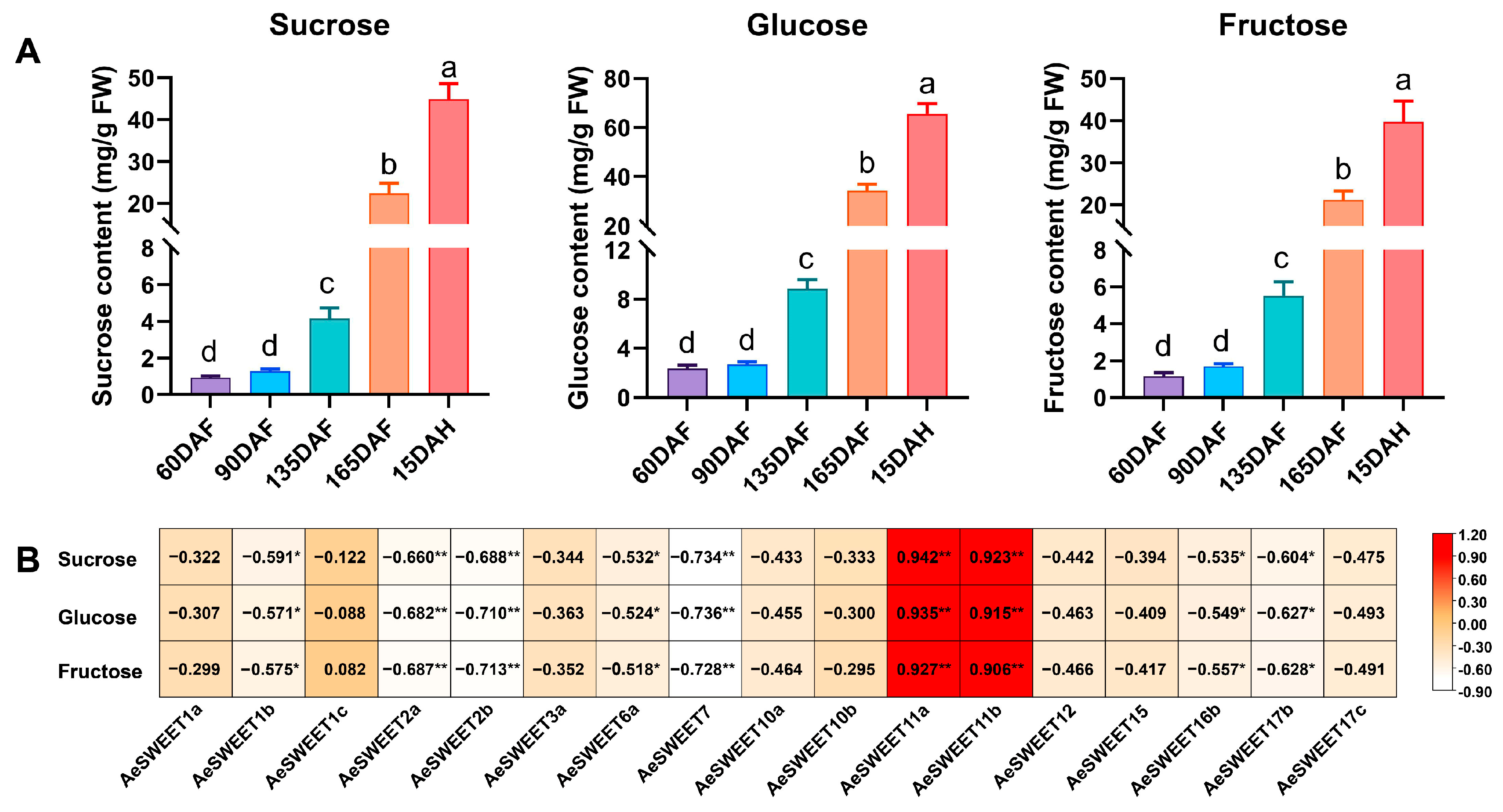
2.8. Correlation Analysis of AeSWEET Gene Expression and Sugar Contents During Fruit Development and Ripening
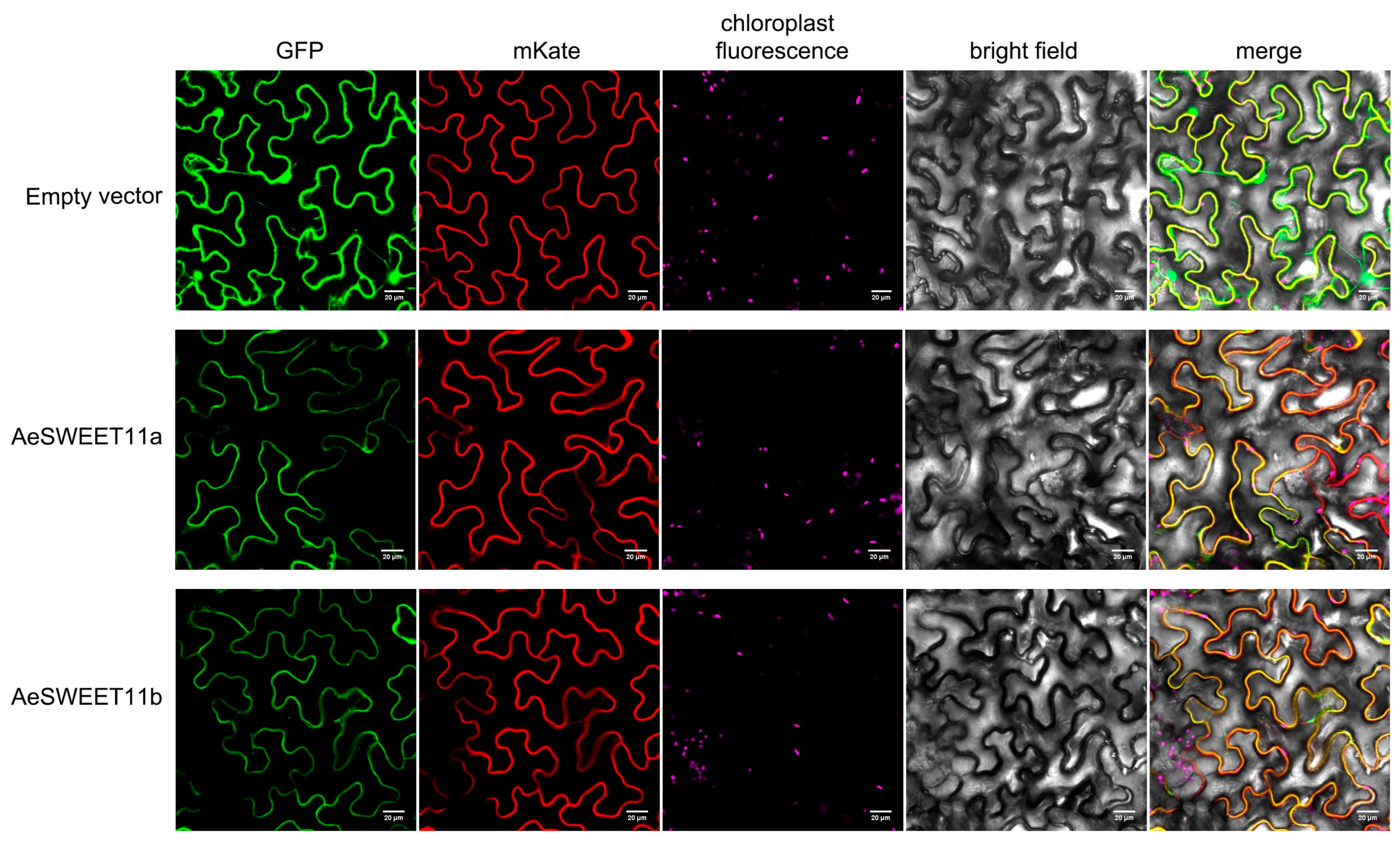
2.9. Subcellular Localization of AeSWEET11a and AeSWEET11b
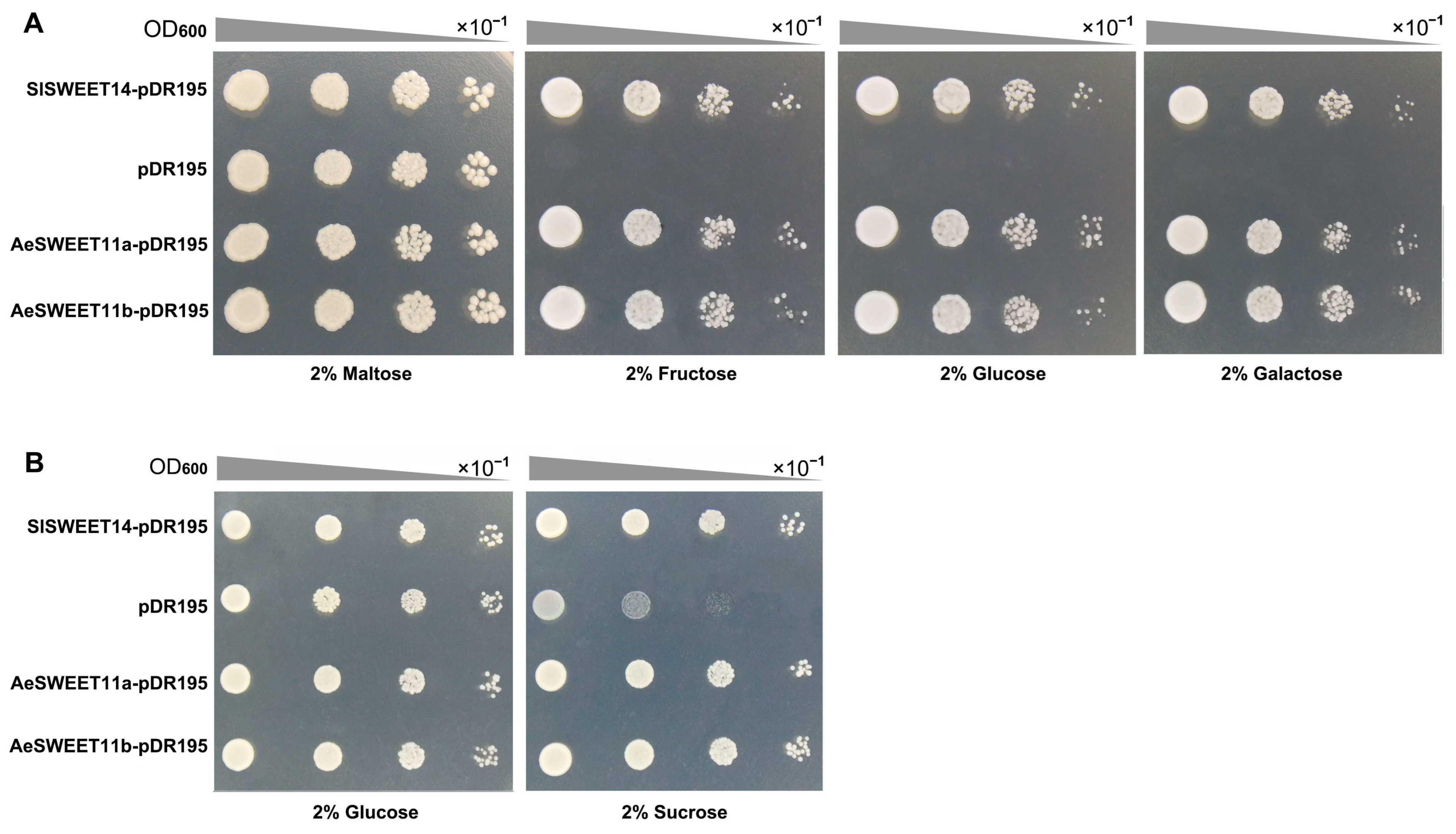
2.10. Functions of AeSWEET11a and AeSWEET11b in Sugar Transporting
3. Discussion
3.1. The Characteristics of AeSWEET Family Genes in A. eriantha
3.2. Expression and Functional Diversity of AeSWEET Genes
3.3. AeSWEET11a and AeSWEET11b Probably Participate in Sugar Transport and Accumulation During Fruit Development and Ripening
4. Materials and Methods
4.1. Identification of AeSWEET Genes Through a Genome-Wide Analysis
4.2. Chromosomal Location and Phylogenetic Relationship of AeSWEETs
4.3. Gene Structure and Protein Domains of AeSWEETs
4.4. Intraspecific and Interspecific Collinearity Analysis
4.5. Cis-Acting Element Analysis of AeSWEET Promoters
4.6. Plant Materials and Quantitative Real-Time PCR
4.7. Measurement of Sucrose, Glucose and Fructose
4.8. Subcellular Localization
4.9. Yeast Mutant Complementary Growth Assay
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeandet, P.; Formela-Luboińska, M.; Labudda, M.; Morkunas, I. The role of sugars in plant responses to stress and their regulatory function during development. Int. J. Mol. Sci. 2022, 23, 5161. [Google Scholar] [CrossRef]
- Wen, S.; Neuhaus, H.E.; Cheng, J.; Bie, Z. Contributions of sugar transporters to crop yield and fruit quality. J. Exp. Bot. 2022, 73, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Falchi, R.; Bonghi, C.; Drincovich, M.F.; Famiani, F.; Lara, M.V.; Walker, R.P.; Vizzotto, G. Sugar metabolism in stone fruit: Source-sink relationships and environmental and agronomical effects. Front. Plant Sci. 2020, 11, 573982. [Google Scholar] [CrossRef]
- Zhang, C.; Bian, Y.; Hou, S.; Li, X. Sugar transport played a more important role than sugar biosynthesis in fruit sugar accumulation during Chinese jujube domestication. Planta 2018, 248, 1187–1199. [Google Scholar] [CrossRef]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar transporters in plants: New insights and discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef]
- Xue, X.; Wang, J.; Shukla, D.; Cheung, L.S.; Chen, L.Q. When SWEETs turn tweens: Updates and perspectives. Annu. Rev. Plant Biol. 2022, 73, 379–403. [Google Scholar] [CrossRef]
- Slewinski, T.L. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol. Plant 2011, 4, 641–662. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Zhu, Y.; Tian, Y.; Han, S.; Wang, J.; Liu, Y.; Yin, J. Structure, evolution, and roles of SWEET proteins in growth and stress responses in plants. Int. J. Biol. Macromol. 2024, 263, 130441. [Google Scholar] [CrossRef]
- Singh, J.; Das, S.; Jagadis Gupta, K.; Ranjan, A.; Foyer, C.H.; Thakur, J.K. Physiological implications of SWEETs in plants and their potential applications in improving source-sink relationships for enhanced yield. Plant Biotechnol. J. 2023, 21, 1528–1541. [Google Scholar] [CrossRef] [PubMed]
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Gerós, H.; Granell, A. Plant SWEETs: From sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021, 186, 836–852. [Google Scholar] [CrossRef]
- Fakher, B.; Ashraf, M.A.; Wang, L.; Wang, X.; Zheng, P.; Aslam, M.; Qin, Y. Pineapple SWEET10 is a glucose transporter. Hortic. Res. 2023, 10, uhad175. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Shi, Y.; Zhang, P.; Xie, W.; Li, S.; Xiao, J.; Yuan, M. Deletion of the sugar importer gene OsSWEET1b accelerates sugar starvation-promoted leaf senescence in rice. Plant Physiol. 2024, 195, 2176–2194. [Google Scholar] [CrossRef]
- Fan, X.W.; Sun, J.L.; Cai, Z.; Zhang, F.; Li, Y.Z.; Palta, J.A. MeSWEET15a/b genes play a role in the resistance of cassava (Manihot esculenta Crantz) to water and salt stress by modulating sugar distribution. Plant Physiol. Biochem. 2023, 194, 394–405. [Google Scholar] [CrossRef]
- Ji, J.; Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y. Plant SWEET family of sugar transporters: Structure, evolution and biological functions. Biomolecules 2022, 12, 205. [Google Scholar] [CrossRef]
- Yamada, K.; Osakabe, Y. Sugar compartmentation as an environmental stress adaptation strategy in plants. Semi. Cell Dev. Biol. 2018, 83, 106–114. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Ren, Y.; Gan, C.; Li, B.; Fan, Y.; Zhao, X.; Yuan, Z. Identification, analysis and gene cloning of the SWEET gene family provide insights into sugar transport in Pomegranate (Punica granatum). Int. J. Mol. Sci. 2022, 23, 2471. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Li, Y.; Fu, M.; Li, J.; Wu, J.; Shua, Z.; Chen, T.; Yao, W.; Huai, D. Genome-wide identification of SWEET genes reveals their roles during seed development in peanuts. BMC Genom. 2024, 25, 259. [Google Scholar] [CrossRef]
- Li, J.; Qin, M.; Qiao, X.; Cheng, Y.; Li, X.; Zhang, H.; Wu, J. A new insight into the evolution and functional divergence of SWEET transporters in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef]
- Miao, H.; Sun, P.; Liu, Q.; Miao, Y.; Liu, J.; Zhang, K.; Hu, W.; Zhang, J.; Wang, J.; Wang, Z.; et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 2017, 7, 3536. [Google Scholar] [CrossRef]
- Liu, S.; Nie, X.; Chen, H.; Shen, X. Identification of the SWEET gene family and functional characterization of PsSWEET1a and PsSWEET17b in the regulation of sugar accumulation in ‘Fengtang’ plum (Prunus salicina Lindl.). BMC Plant Biol. 2025, 25, 407. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Piron, M.C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, K.; Wen, S.; Yang, D.; Gao, J.; Wang, Z.; Zhu, P.; Bie, Z.; Cheng, J. Phloem unloading in cultivated melon fruits follows an apoplasmic pathway during enlargement and ripening. Hortic. Res. 2023, 10, uhad123. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Yao, X.; Wang, J.; Feng, S.; Sun, L.; Ma, S.; Xu, K.; Chen, L.Q.; Sui, X. Hexose transporter CsSWEET7a in cucumber mediates phloem unloading in companion cells for fruit development. Plant Physiol. 2021, 186, 640–654. [Google Scholar] [CrossRef]
- Ko, H.Y.; Ho, L.H.; Neuhaus, H.E.; Guo, W.J. Transporter SlSWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021, 187, 2230–2245. [Google Scholar] [CrossRef]
- Nie, P.; Wang, L.; Li, M.; Lyu, D.; Qin, S.; Xue, X. MdSWEET23, a sucrose transporter from apple (Malus × domestica Borkh.), influences sugar metabolism and enhances cold tolerance in tomato. Front. Plant Sci. 2023, 14, 1266194. [Google Scholar] [CrossRef]
- Huang, S.; Ding, J.; Deng, D.; Tang, W.; Sun, H.; Liu, D.; Zhang, L.; Niu, X.; Zhang, X.; Meng, M. Draft genome of the kiwifruit Actinidia chinensis. Nature Commun. 2013, 4, 2640. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Shu, P.; Zhang, C.; Zhang, J.; Chen, Y.; Zhang, Y.; Du, K.; Xie, Y.; Li, M.; Ma, T.; et al. Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytol. 2022, 233, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zheng, H.; Fan, J.; Liu, F.; Li, J.; Zhong, C.; Zhang, Q. Comparative study on physicochemical and nutritional qualities of kiwifruit varieties. Foods 2022, 12, 108. [Google Scholar] [CrossRef]
- Tao, J.; Jia, H.; Wu, M.; Zhong, W.; Huang, Y.; Huang, L.; Xu, Y.; Huang, C. Integrated metabolome and transcriptome analysis reveals the mechanism related to the formation of peelability in Actinidia eriantha. Sci. Hortic. 2024, 330, 113072. [Google Scholar] [CrossRef]
- Fang, H.; Shi, Y.; Liu, S.; Jin, R.; Sun, J.; Grierson, D.; Li, S.; Chen, K. The transcription factor CitZAT5 modifies sugar accumulation and hexose proportion in citrus fruit. Plant Physiol. 2023, 192, 1858–1876. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Y.; Zhang, Q.; Ma, N.; Liu, X.; Tao, W.; Lou, Z.; Zhong, C.; Deng, X.W.; Li, D.; et al. Two haplotype-resolved, gap-free genome assemblies for Actinidia latifolia and Actinidia chinensis shed light on the regulatory mechanisms of vitamin C and sucrose metabolism in kiwifruit. Mol. Plant 2023, 16, 452–470. [Google Scholar] [CrossRef] [PubMed]
- Nardozza, S.; Boldingh, H.L.; Kashuba, M.P.; Feil, R.; Jones, D.; Thrimawithana, A.H.; Ireland, H.S.; Philippe, M.; Wohlers, M.W.; McGhie, T.K. Carbon starvation reduces carbohydrate and anthocyanin accumulation in red-fleshed fruit via trehalose 6-phosphate and MYB27. Plant Cell Environ. 2020, 43, 819–835. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, M.; Wu, Y.; Zhang, F.; Ren, W.; Lin, Y.; Chen, Q.; Zhang, S.; Yue, J.; Liu, Y. Telomere-to-telomere and haplotype-resolved genome of the kiwifruit Actinidia eriantha. Mol. Hortic. 2023, 3, 4. [Google Scholar] [CrossRef]
- Linster, E.; Layer, D.; Bienvenut, W.V.; Dinh, T.V.; Weyer, F.A.; Leemhuis, W.; Brünje, A.; Hoffrichter, M.; Miklankova, P.; Kopp, J.; et al. The Arabidopsis N(α) -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response. New Phytol. 2020, 228, 554–569. [Google Scholar] [CrossRef]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef]
- Riesmeier, J.W.; Willmitzer, L.; Frommer, W.B. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992, 11, 4705–4713. [Google Scholar] [CrossRef]
- Xie, H.; Wang, D.; Qin, Y.; Ma, A.; Fu, J.; Qin, Y.; Hu, G.; Zhao, J. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 2019, 19, 499. [Google Scholar] [CrossRef]
- Geng, K.; Zhan, Z.; Xue, X.; Hou, C.; Li, D.; Wang, Z. Genome--wide identification of the SWEET gene family in grape (Vitis vinifera L.) and expression analysis of VvSWEET14a in response to water stress. Physiol. Mol. Biol. Plants 2024, 30, 1565–1579. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.J.; Nagy, R.; Chen, H.Y.; Pfrunder, S.; Yu, Y.C.; Santelia, D.; Frommer, W.B.; Martinoia, E. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol. 2014, 164, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Niu, K.; Ma, H. Identification and expression analysis of the SWEET gene family from Poa pratensis under abiotic stresses. DNA Cell Biol. 2020, 39, 1606–1620. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhou, L.; Li, T.; Ruan, Y.; Zhang, A.; Dong, X.; Zhu, Y.; Li, C.; Fan, J. Genome-wide investigation and characterization of SWEET gene family with focus on their evolution and expression during hormone and abiotic stress response in Maize. Genes 2022, 13, 1682. [Google Scholar] [CrossRef]
- Li, W.; Ren, Z.; Wang, Z.; Sun, K.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Song, C.; Zhou, X.; et al. Evolution and Stress Responses of Gossypium hirsutum SWEET Genes. Int. J. Mol. Sci. 2018, 19, 769. [Google Scholar] [CrossRef]
- Li, X.; Si, W.; Qin, Q.; Wu, H.; Jiang, H. Deciphering evolutionary dynamics of SWEET genes in diverse plant lineages. Sci. Rep. 2018, 8, 13440. [Google Scholar] [CrossRef]
- Pan, Y.; Niu, K.; Miao, P.; Zhao, G.; Zhang, Y.; Ju, Z.; Chai, J.; Yang, J.; Cui, X.; Zhang, R. Genome-wide analysis of the SWEET gene family and its response to powdery mildew and leaf spot infection in the common oat (Avena sativa L.). BMC Genom. 2024, 25, 995. [Google Scholar] [CrossRef]
- Sun, Y.; Deng, T.; Zhang, A.; Moore, M.J.; Landis, J.B.; Lin, N.; Zhang, H.; Zhang, X.; Huang, J.; Zhang, X.; et al. Genome sequencing of the endangered kingdonia uniflora (Circaeasteraceae, Ranunculales) reveals potential mechanisms of evolutionary specialization. iScience 2020, 23, 101124. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Moore, B.M.; Panchy, N.L.; Meng, F.; Lehti-Shiu, M.D.; Shiu, S.H. Factors influencing gene family size variation among related species in a plant family, solanaceae. Genome Biol. Evol. 2018, 10, 2596–2613. [Google Scholar] [CrossRef]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J.S. SWEET11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef]
- Ge, Y.X.; Angenent, G.C.; Wittich, P.E.; Peters, J.; Franken, J.; Busscher, M.; Zhang, L.M.; Dahlhaus, E.; Kater, M.M.; Wullems, G.J.; et al. NEC1, a novel gene, highly expressed in nectary tissue of Petunia hybrida. Plant J. 2000, 24, 725–734. [Google Scholar] [CrossRef]
- Ge, Y.X.; Angenent, G.C.; Dahlhaus, E.; Franken, J.; Peters, J.; Wullems, G.J.; Creemers-Molenaar, J. Partial silencing of the NEC1 gene results in early opening of anthers in Petunia hybrida. Mol. Genet. Genom. 2001, 265, 414–423. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.Q.; Gase, K.; Kim, S.G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.H.; Qu, X.Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Cao, L.; Wang, J.; Wang, L.; Liu, H.; Wu, W.; Hou, F.; Liu, Y.; Gao, Y.; Cheng, X.; Li, S.; et al. Genome-wide analysis of the SWEET gene family in Hemerocallis citrina and functional characterization of HcSWEET4a in response to salt stress. BMC Plant Biol. 2024, 24, 661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, Y.N.; Wu, B.H.; Yuan, Y.Y.; Zhao, Z.Y. Plasma membrane-localized transporter MdSWEET12 is involved in sucrose unloading in apple fruit. J. Agric. Food Chem. 2022, 70, 15517–15530. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Lin, I.W.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef]
- Haberer, G.; Hindemitt, T.; Meyers, B.C.; Mayer, K.F. Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 2004, 136, 3009–3022. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef]
- Yue, W.; Cai, K.; Xia, X.; Liu, L.; Wang, J. Genome-wide identification, expression pattern and genetic variation analysis of SWEET gene family in barley reveal the artificial selection of HvSWEET1a during domestication and improvement. Front. Plant Sci. 2023, 14, 1137434. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, M.; Guo, S.; Sun, H.; Zhao, J.; Zhang, J.; Liu, G.; He, H.; Tian, S.; Yu, Y.; et al. Evolutionary gain of oligosaccharide hydrolysis and sugar transport enhanced carbohydrate partitioning in sweet watermelon fruits. Plant Cell 2021, 33, 1554–1573. [Google Scholar] [CrossRef]
- Tao, J.; Wu, M.; Jiao, X.; Chen, S.; Jia, D.; Xu, X.; Huang, C. Dynamic changes of fruit physiological quality and sugar components during fruit growth and development of Actinidia eriantha. Horticulturae 2022, 8, 529. [Google Scholar] [CrossRef]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Weigle, A.T.; Shukla, D. The Arabidopsis AtSWEET13 transporter discriminates sugars by selective facial and positional substrate recognition. Commun. Biol. 2024, 7, 764. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, C.; Wang, M.; Li, T.; Liu, X.; Jiang, J. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic. Res. 2021, 8, 186. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.-Q.; Sosso, D.; Ducat, D.C.; Hou, B.-H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef]
- Luo, M.; Jia, M.; Pan, L.; Chen, W.; Zhou, K.; Xi, W. Sugar transporters PpSWEET11a and PpSWEET14 synergistically mediate peach sucrose allocation from source leaves to fruit. Commun. Biol. 2024, 7, 1068. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Chini, V.; Foka, A.; Dimitracopoulos, G.; Spiliopoulou, I. Absolute and relative real-time PCR in the quantification of tst gene expression among methicillin-resistant Staphylococcus aureus: Evaluation by two mathematical models. Lett. Appl. Microbiol. 2007, 45, 479–484. [Google Scholar] [CrossRef]
- Shcherbo, D.; Murphy, C.S.; Ermakova, G.V.; Solovieva, E.A.; Chepurnykh, T.V.; Shcheglov, A.S.; Verkhusha, V.V.; Pletnev, V.Z.; Hazelwood, K.L.; Roche, P.M.; et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009, 418, 567–574. [Google Scholar] [CrossRef]

| Protein Name | Amino Acids (aa) | Molecular Weight (Da) | Isoelectric Point | Instability Index | GRAVY | TMH | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| AeSWEET1a | 251 | 27,425.63 | 9.35 | 33.64 | 0.596 | 7 | PM |
| AeSWEET1b | 252 | 27,780.02 | 9.47 | 33.1 | 0.508 | 7 | PM |
| AeSWEET1c | 252 | 27,485.73 | 9.68 | 28.76 | 0.636 | 7 | PM |
| AeSWEET2a | 236 | 25,895.9 | 9.24 | 38.06 | 0.883 | 7 | PM |
| AeSWEET2b | 236 | 26,221.08 | 8.94 | 41 | 0.783 | 7 | PM |
| AeSWEET3a | 212 | 23,971.52 | 9.7 | 38.24 | 0.250 | 7 | PM |
| AeSWEET3b | 243 | 27,228.28 | 9.73 | 24.97 | 0.519 | 7 | PM |
| AeSWEET4a | 198 | 21,746.05 | 9.72 | 33.39 | 0.703 | 7 | PM |
| AeSWEET4b | 238 | 26,245.35 | 9.52 | 34.27 | 0.713 | 7 | PM |
| AeSWEET5 | 234 | 26,451.54 | 8.66 | 36.72 | 0.65 | 7 | PM |
| AeSWEET6a | 305 | 34,066.5 | 9.62 | 39.75 | 0.518 | 7 | PM |
| AeSWEET6b | 219 | 24,547.16 | 8.92 | 42.66 | 0.616 | 6 | PM |
| AeSWEET6c | 279 | 31,541.67 | 9.48 | 36.55 | 0.569 | 7 | PM |
| AeSWEET7 | 259 | 28,591.22 | 9.71 | 45.36 | 0.59 | 7 | PM |
| AeSWEET10a | 287 | 32,095.65 | 9.39 | 39.72 | 0.802 | 7 | PM |
| AeSWEET10b | 290 | 32,615.36 | 8.15 | 39.24 | 0.85 | 7 | PM |
| AeSWEET10c | 290 | 32,593.16 | 9.03 | 39.01 | 0.751 | 7 | PM |
| AeSWEET11a | 275 | 30,940.75 | 8.47 | 37.23 | 0.627 | 7 | PM |
| AeSWEET11b | 275 | 30,917.98 | 8.37 | 38.51 | 0.719 | 7 | PM |
| AeSWEET12 | 281 | 31,991.57 | 9.25 | 40.11 | 0.29 | 7 | PM |
| AeSWEET15 | 287 | 32,096.05 | 7.04 | 36.89 | 0.671 | 7 | PM |
| AeSWEET16a | 249 | 27,717.86 | 9.26 | 40.34 | 0.417 | 5 | PM |
| AeSWEET16b | 304 | 33,148.24 | 9.65 | 33.44 | 0.377 | 7 | PM |
| AeSWEET17a | 240 | 26,569.19 | 6.5 | 48.66 | 0.588 | 7 | PM |
| AeSWEET17b | 253 | 27,794.08 | 9.15 | 36.01 | 0.734 | 7 | PM |
| AeSWEET17c | 244 | 26,962.85 | 7.77 | 35.7 | 0.674 | 7 | PM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Huang, Q.; Gao, M.; Lai, R.; Chen, Y. Comprehensive Identification and Expression Analysis of the SWEET Gene Family in Actinidia eriantha Reveals That Two AeSWEET11 Genes Function in Sucrose and Hexose Transport. Plants 2025, 14, 3140. https://doi.org/10.3390/plants14203140
Feng X, Huang Q, Gao M, Lai R, Chen Y. Comprehensive Identification and Expression Analysis of the SWEET Gene Family in Actinidia eriantha Reveals That Two AeSWEET11 Genes Function in Sucrose and Hexose Transport. Plants. 2025; 14(20):3140. https://doi.org/10.3390/plants14203140
Chicago/Turabian StyleFeng, Xin, Qingqing Huang, Minxia Gao, Ruilian Lai, and Yiting Chen. 2025. "Comprehensive Identification and Expression Analysis of the SWEET Gene Family in Actinidia eriantha Reveals That Two AeSWEET11 Genes Function in Sucrose and Hexose Transport" Plants 14, no. 20: 3140. https://doi.org/10.3390/plants14203140
APA StyleFeng, X., Huang, Q., Gao, M., Lai, R., & Chen, Y. (2025). Comprehensive Identification and Expression Analysis of the SWEET Gene Family in Actinidia eriantha Reveals That Two AeSWEET11 Genes Function in Sucrose and Hexose Transport. Plants, 14(20), 3140. https://doi.org/10.3390/plants14203140






