How Do Arabidopsis Seedlings Sense and React to Increasing Ambient Temperatures?
Abstract
1. Introduction
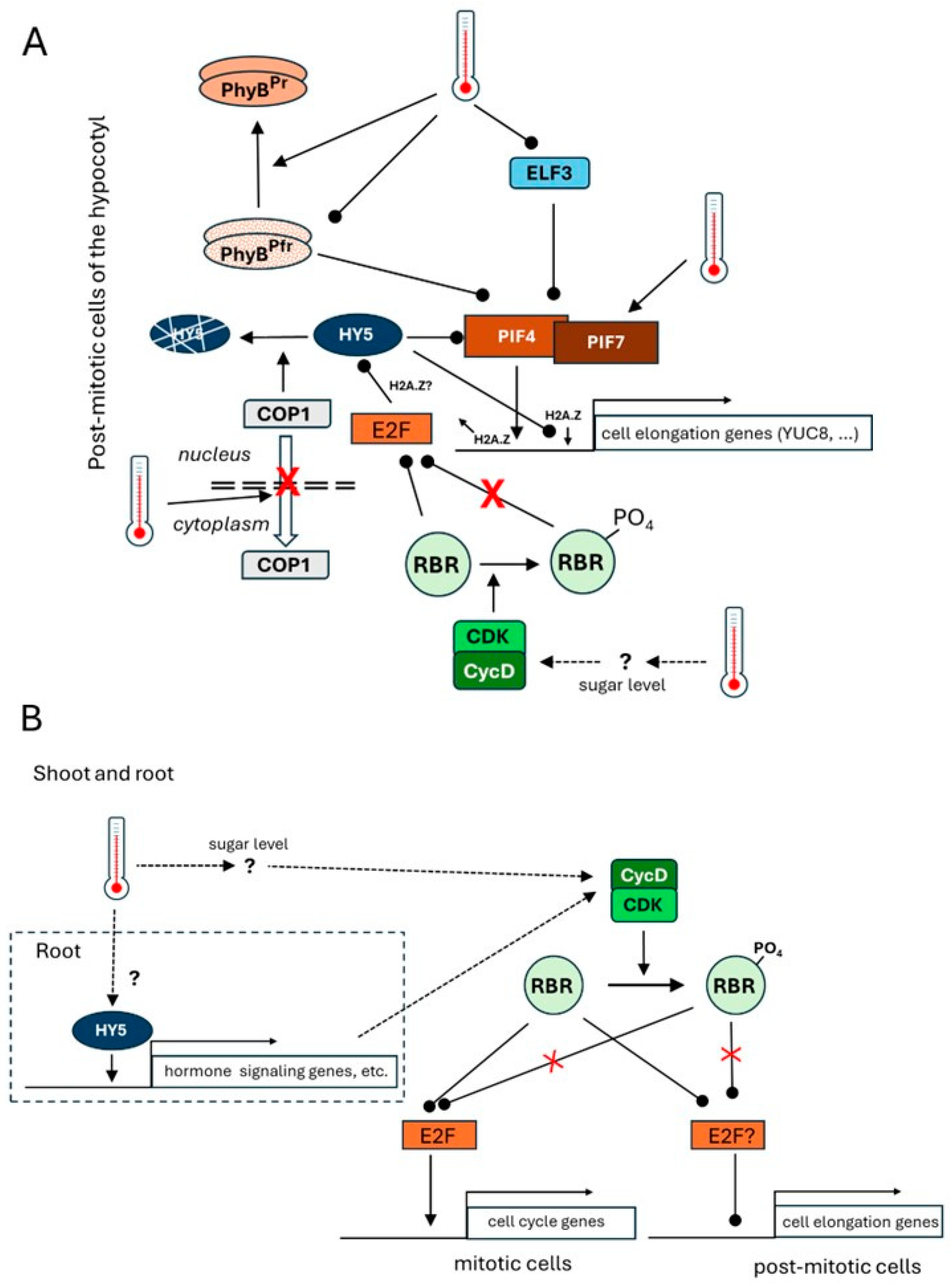
2. Thermal Response of the Hypocotyl—A Long and Bright Story
3. The Thermal Response of the Root—Hidden in the Dark
4. Summary and Conclusions
Funding
Conflicts of Interest
References
- Ray, D.K.; West, P.C.; Clark, M.; Gerber, J.S.; Prishchepov, A.V.; Chatterjee, S. Climate Change Has Likely Already Affected Global Food Production. PLoS ONE 2019, 14, e0217148. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Bobea, A.; Ault, T.R.; Carrillo, C.M.; Chambers, R.G.; Lobell, D.B. Anthropogenic Climate Change Has Slowed Global Agricultural Productivity Growth. Nat. Clim. Chang. 2021, 11, 306–312. [Google Scholar] [CrossRef]
- Mirón, I.J.; Linares, C.; Díaz, J. The Influence of Climate Change on Food Production and Food Safety. Environ. Res. 2023, 216, 114674. [Google Scholar] [CrossRef] [PubMed]
- Arnell, N.W.; Lowe, J.A.; Challinor, A.J.; Osborn, T.J. Global and Regional Impacts of Climate Change at Different Levels of Global Temperature Increase. Clim. Chang. 2019, 155, 377–391. [Google Scholar] [CrossRef]
- Lippmann, R.; Babben, S.; Menger, A.; Delker, C.; Quint, M. Development of Wild and Cultivated Plants under Global Warming Conditions. Curr. Biol. 2019, 29, R1326–R1338. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.E.; Meacham-Hensold, K.; Lemonnier, P.; Slattery, R.A.; Benjamin, C.; Bernacchi, C.J.; Lawson, T.; Cavanagh, A.P. The Effect of Increasing Temperature on Crop Photosynthesis: From Enzymes to Ecosystems. J. Exp. Bot. 2021, 72, 2822–2844. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.; Bayer, P.E.; Edwards, D. Climate Change and the Need for Agricultural Adaptation. Curr. Opin. Plant Biol. 2020, 56, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J.; Balasubramanian, S. Thermomorphogenesis. Annu. Rev. Plant Biol. 2019, 70, 321–346. [Google Scholar] [CrossRef]
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; van Zanten, M. Molecular and Genetic Control of Plant Thermomorphogenesis. Nat. Plants 2016, 2, 15190. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhu, Z. Light Signaling-Mediated Growth Plasticity in Arabidopsis Grown under High-Temperature Conditions. Stress Biol. 2022, 2, 53. [Google Scholar] [CrossRef]
- Crawford, A.J.; McLachlan, D.H.; Hetherington, A.M.; Franklin, K.A. High Temperature Exposure Increases Plant Cooling Capacity. Curr. Biol. 2012, 22, R396–R397. [Google Scholar] [CrossRef] [PubMed]
- Bridge, L.J.; Franklin, K.A.; Homer, M.E. Impact of Plant Shoot Architecture on Leaf Cooling: A Coupled Heat and Mass Transfer Model. J. R. Soc. Interface 2013, 10, 20130326. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Lee, H.-J.; Gil, K.-E.; Kim, J.Y.; Lee, J.-H.; Lee, H.; Cho, H.-T.; Vu, L.D.; De Smet, I.; Park, C.-M. Developmental Programming of Thermonastic Leaf Movement. Plant Physiol. 2019, 180, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J.; Qüesta, J.I. Light and Temperature Cues: Multitasking Receptors and Transcriptional Integrators. New Phytol. 2018, 217, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Romero-Montepaone, S.; Sellaro, R.; Esteban Hernando, C.; Costigliolo-Rojas, C.; Bianchimano, L.; Ploschuk, E.L.; Yanovsky, M.J.; Casal, J.J. Functional Convergence of Growth Responses to Shade and Warmth in Arabidopsis. New Phytol. 2021, 231, 1890–1905. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Shi, Y.; Terzaghi, W.; Yang, S.; Li, J. Integration of Light and Temperature Signaling Pathways in Plants. J. Integr. Plant Biol. 2022, 64, 393–411. [Google Scholar] [CrossRef]
- Legris, M. Light and Temperature Regulation of Leaf Morphogenesis in Arabidopsis. New Phytol. 2023, 240, 2191–2196. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, P.; Liu, Y. Thermosensing and Thermal Responses in Plants. Trends Biochem. Sci. 2023, 48, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes Function as Thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Pardi, S.A.; Nusinow, D.A. Out of the Dark and Into the Light: A New View of Phytochrome Photobodies. Front. Plant Sci. 2021, 12, 732947. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Kwon, Y.; Jeong, J.; Kang, M.; Lee, G.S.; Moon, J.H.; Lee, H.-J.; Park, Y.-I.; Choi, G. Phytochrome B Photobodies Are Comprised of Phytochrome B and Its Primary and Secondary Interacting Proteins. Nat. Commun. 2023, 14, 1708. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Lyu, M.; Kou, X.; Li, J.; Yang, Z.; Gao, L.; Li, Y.; Fan, L.-M.; Shi, H.; Zhong, S. Integration of Light and Temperature Sensing by Liquid-Liquid Phase Separation of Phytochrome B. Mol. Cell 2022, 82, 3015–3029.e6. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 Interacts with PIF4 to Regulate High Temperature-Mediated Hypocotyl Elongation in Response to Blue Light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Sharma, A.; Fraser, D.P.; Trevisan, M.; Cragg-Barber, C.K.; Tavridou, E.; Fankhauser, C.; Jenkins, G.I.; Franklin, K.A. UV-B Perceived by the UVR8 Photoreceptor Inhibits Plant Thermomorphogenesis. Curr. Biol. 2017, 27, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Schachtschabel, J.; Mishkind, M.; Munnik, T.; Arisz, S.A. Hot Topic: Thermosensing in Plants. Plant Cell Environ. 2021, 44, 2018–2033. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Takahashi, Y.; Fujii, Y.; Sasaki, K.; Hirano, S.; Okajima, K.; Kodama, Y. The Photo-Thermochemical Properties and Functions of Marchantia Phototropin Encoded by an Unduplicated Gene in Land Plant Evolution. J. Photochem. Photobiol. B 2021, 224, 112305. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, A.; Takase, T.; Abe, H.; Watahiki, M.; Hirakawa, Y.; Kiyosue, T. ZEITLUPE Enhances Expression of PIF4 and YUC8 in the Upper Aerial Parts of Arabidopsis Seedlings to Positively Regulate Hypocotyl Elongation. Plant Cell Rep. 2021, 40, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Seo, P.J.; Oh, E.; Kim, J. Temperature Perception by Plants. Trends Plant Sci. 2023, 28, 924–940. [Google Scholar] [CrossRef] [PubMed]
- Koini, M.A.; Alvey, L.; Allen, T.; Tilley, C.A.; Harberd, N.P.; Whitelam, G.C.; Franklin, K.A. High Temperature-Mediated Adaptations in Plant Architecture Require the bHLH Transcription Factor PIF4. Curr. Biol. 2009, 19, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Stavang, J.A.; Gallego-Bartolomé, J.; Gómez, M.D.; Yoshida, S.; Asami, T.; Olsen, J.E.; García-Martínez, J.L.; Alabadí, D.; Blázquez, M.A. Hormonal Regulation of Temperature-Induced Growth in Arabidopsis. Plant J. 2009, 60, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Chu, L.; Lin, H.; Qi, Y.; Fang, Z.; Xu, D. PIFs- and COP1-HY5-Mediated Temperature Signaling in Higher Plants. Stress Biol. 2022, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.; Nieto, C.; Prat, S. Convergent Regulation of PIFs and the E3 Ligase COP1/SPA1 Mediates Thermosensory Hypocotyl Elongation by Plant Phytochromes. Curr. Opin. Plant Biol. 2018, 45, 188–203. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yu, H.; Yuan, R.; Yang, Y.; An, F.; Qin, G. Arabidopsis Transcription Factor TCP5 Controls Plant Thermomorphogenesis by Positively Regulating PIF4 Activity. iScience 2019, 15, 611–622. [Google Scholar] [CrossRef]
- Lee, S.; Paik, I.; Huq, E. SPAs Promote Thermomorphogenesis by Regulating the phyB-PIF4 Module in Arabidopsis. Development 2020, 147, dev189233. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y. Regulation of PIF4-Mediated Thermosensory Growth. Plant Sci. 2020, 297, 110541. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Pasoreck, E.K.; Yoo, C.Y.; He, J.; Wang, H.; Bajracharya, A.; Li, M.; Larsen, H.D.; Cheung, S.; Chen, M. RCB Initiates Arabidopsis Thermomorphogenesis by Stabilizing the Thermoregulator PIF4 in the Daytime. Nat. Commun. 2021, 12, 2042. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Z. PIF4 and PIF4-Interacting Proteins: At the Nexus of Plant Light, Temperature and Hormone Signal Integrations. Int. J. Mol. Sci. 2021, 22, 10304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Bao, Y. PIF4: Integrator of Light and Temperature Cues in Plant Growth. Plant Sci. 2021, 313, 111086. [Google Scholar] [CrossRef]
- Verma, N.; Singh, D.; Mittal, L.; Banerjee, G.; Noryang, S.; Sinha, A.K. MPK4 Mediated Phosphorylation of PIF4 Controls Thermosensing by Regulation of H2A.Z Deposition in Arabidopsis. Plant Cell 2023, 36, 4535–4556. [Google Scholar]
- Sun, Q.; Wang, S.; Xu, G.; Kang, X.; Zhang, M.; Ni, M. SHB1 and CCA1 Interaction Desensitizes Light Responses and Enhances Thermomorphogenesis. Nat. Commun. 2019, 10, 3110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-L.; Luo, A.; Davis, S.J.; Liu, J.-X. Timing to Grow: Roles of Clock in Thermomorphogenesis. Trends Plant Sci. 2021, 26, 1248–1257. [Google Scholar] [CrossRef]
- Hendrix, S. Remembering a Warm Day: Daytime Temperature Influences Nighttime Hypocotyl Growth in Arabidopsis. Plant Cell 2022, 34, 2110–2111. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.; Park, J.; Park, J.; Hwang, G.; Seo, P.J.; Oh, E. ZTL Regulates Thermomorphogenesis through TOC1 and PRR5. Plant Cell Environ. 2023, 46, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hwang, G.; Kim, S.; Thi, T.N.; Kim, H.; Jeong, J.; Kim, J.; Kim, J.; Choi, G.; Oh, E. The Epidermis Coordinates Thermoresponsive Growth through the phyB-PIF4-Auxin Pathway. Nat. Commun. 2020, 11, 1053. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.-Y.; Bai, M.-Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.-Y. Cell Elongation Is Regulated through a Central Circuit of Interacting Transcription Factors in the Arabidopsis Hypocotyl. eLife 2014, 3, e03031. [Google Scholar] [CrossRef] [PubMed]
- Bouré, N.; Kumar, S.V.; Arnaud, N. The BAP Module: A Multisignal Integrator Orchestrating Growth. Trends Plant Sci. 2019, 24, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-Interacting Factor 4 (PIF4) Regulates Auxin Biosynthesis at High Temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Qi, L.; Li, Y.; Chu, J.; Li, C. PIF4-Mediated Activation of YUCCA8 Expression Integrates Temperature into the Auxin Pathway in Regulating Arabidopsis Hypocotyl Growth. PLoS Genet. 2012, 8, e1002594. [Google Scholar] [CrossRef] [PubMed]
- Bianchimano, L.; De Luca, M.B.; Borniego, M.B.; Iglesias, M.J.; Casal, J.J. Temperature Regulation of Auxin-Related Gene Expression and Its Implications for Plant Growth. J. Exp. Bot. 2023, 74, 7015–7033. [Google Scholar] [CrossRef]
- Ibañez, C.; Delker, C.; Martinez, C.; Bürstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310.e3. [Google Scholar] [CrossRef]
- Martínez, C.; Espinosa-Ruíz, A.; Lucas, M.; Bernardo-García, S.; Franco-Zorrilla, J.M.; Prat, S. PIF4-Induced BR Synthesis Is Critical to Diurnal and Thermomorphogenic Growth. EMBO J 2018, 37, e99552. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Luengo, L.M.; Prat, S. Regulation of COP1 Function by Brassinosteroid Signaling. Front. Plant Sci. 2020, 11, 1151. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.-Y.; Wang, Z.-Y. Interaction between BZR1 and PIF4 Integrates Brassinosteroid and Environmental Responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef]
- Box, M.S.; Huang, B.E.; Domijan, M.; Jaeger, K.E.; Khattak, A.K.; Yoo, S.J.; Sedivy, E.L.; Jones, D.M.; Hearn, T.J.; Webb, A.A.R.; et al. ELF3 Controls Thermoresponsive Growth in Arabidopsis. Curr. Biol. 2015, 25, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Raschke, A.; Ibañez, C.; Ullrich, K.K.; Anwer, M.U.; Becker, S.; Glöckner, A.; Trenner, J.; Denk, K.; Saal, B.; Sun, X.; et al. Natural Variants of ELF3 Affect Thermomorphogenesis by Transcriptionally Modulating PIF4-Dependent Auxin Response Genes. BMC Plant. Biol. 2015, 15, 197. [Google Scholar] [CrossRef] [PubMed]
- Press, M.O.; Lanctot, A.; Queitsch, C. PIF4 and ELF3 Act Independently in Arabidopsis Thaliana Thermoresponsive Flowering. PLoS ONE 2016, 11, e0161791. [Google Scholar] [CrossRef]
- Chung, B.Y.W.; Balcerowicz, M.; Di Antonio, M.; Jaeger, K.E.; Geng, F.; Franaszek, K.; Marriott, P.; Brierley, I.; Firth, A.E.; Wigge, P.A. An RNA Thermoswitch Regulates Daytime Growth in Arabidopsis. Nat. Plants 2020, 6, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, A.-S.; Galvão, V.C.; Ince, Y.Ç.; Boccaccini, A.; Goyal, A.; Allenbach Petrolati, L.; Trevisan, M.; Fankhauser, C. PHYTOCHROME INTERACTING FACTOR 7 Is Important for Early Responses to Elevated Temperature in Arabidopsis Seedlings. New Phytol. 2020, 226, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Burko, Y.; Willige, B.C.; Seluzicki, A.; Novák, O.; Ljung, K.; Chory, J. PIF7 Is a Master Regulator of Thermomorphogenesis in Shade. Nat. Commun. 2022, 13, 4942. [Google Scholar] [CrossRef]
- Murcia, G.; Nieto, C.; Sellaro, R.; Prat, S.; Casal, J.J. Hysteresis in PHYTOCHROME-INTERACTING FACTOR 4 and EARLY-FLOWERING 3 Dynamics Dominates Warm Daytime Memory in Arabidopsis. Plant Cell 2022, 34, 2188–2204. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A Prion-like Domain in ELF3 Functions as a Thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; López-Salmerón, V.; Davière, J.-M.; Prat, S. ELF3-PIF4 Interaction Regulates Plant Growth Independently of the Evening Complex. Curr. Biol. 2015, 25, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Perrella, G.; Bäurle, I.; van Zanten, M. Epigenetic Regulation of Thermomorphogenesis and Heat Stress Tolerance. New Phytol. 2022, 234, 1144–1160. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Zheng, Y.; Lu, Y.; Issakidis-Bourguet, E.; Zhou, D.-X. Metabolic Control of Histone Demethylase Activity Involved in Plant Response to High Temperature. Plant Physiol. 2021, 185, 1813–1828. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Jung, J.-H.; Cortés Llorca, L.; Kim, S.-G.; Lee, S.; Baldwin, I.T.; Park, C.-M. FCA Mediates Thermal Adaptation of Stem Growth by Attenuating Auxin Action in Arabidopsis. Nat. Commun. 2014, 5, 5473. [Google Scholar] [CrossRef] [PubMed]
- Tasset, C.; Singh Yadav, A.; Sureshkumar, S.; Singh, R.; van der Woude, L.; Nekrasov, M.; Tremethick, D.; van Zanten, M.; Balasubramanian, S. POWERDRESS-Mediated Histone Deacetylation Is Essential for Thermomorphogenesis in Arabidopsis Thaliana. PLoS Genet. 2018, 14, e1007280. [Google Scholar] [CrossRef]
- van der Woude, L.C.; Perrella, G.; Snoek, B.L.; van Hoogdalem, M.; Novák, O.; van Verk, M.C.; van Kooten, H.N.; Zorn, L.E.; Tonckens, R.; Dongus, J.A.; et al. HISTONE DEACETYLASE 9 Stimulates Auxin-Dependent Thermomorphogenesis in Arabidopsis Thaliana by Mediating H2A.Z Depletion. Proc. Natl. Acad. Sci. USA 2019, 116, 25343–25354. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Zhang, H.; Zhao, F.; Zhao, T.; Li, H.; Jiang, D. The INO80 Chromatin Remodeling Complex Promotes Thermomorphogenesis by Connecting H2A.Z Eviction and Active Transcription in Arabidopsis. Mol. Plant 2021, 14, 1799–1813. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Wigge, P.A. H2A.Z-Containing Nucleosomes Mediate the Thermosensory Response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, S.; Charoensawan, V.; Brestovitsky, A.; Buning, R.; Ravarani, C.; Rhodes, D. Transcriptional Regulation of the Ambient Temperature Response by H2A.Z-Nucleosomes and HSF1 Transcription Factors in Arabidopsis. Mol. Plant 2017, 10, 1258–1273. [Google Scholar] [CrossRef]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted Destabilization of HY5 during Light-Regulated Development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, A.; Singh, S.; Khurana, J.P.; Burman, N. HY5-COP1: The Central Module of Light Signaling Pathway. J. Plant Biochem. Biotechnol. 2020, 29, 590–610. [Google Scholar] [CrossRef]
- Park, Y.-J.; Lee, H.-J.; Ha, J.-H.; Kim, J.Y.; Park, C.-M. COP1 Conveys Warm Temperature Information to Hypocotyl Thermomorphogenesis. New Phytol. 2017, 215, 269–280. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Kumar, S.V. DET1 and HY5 Control PIF4-Mediated Thermosensory Elongation Growth through Distinct Mechanisms. Cell Rep. 2017, 18, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Hamid, R.S.B.; Nagy, F.; Kaszler, N.; Domonkos, I.; Gombos, M.; Marton, A.; Vizler, C.; Molnár, E.; Pettkó-Szandtner, A.; Bögre, L.; et al. RETINOBLASTOMA-RELATED Has Both Canonical and Noncanonical Regulatory Functions During Thermo-Morphogenic Responses in Arabidopsis Seedlings. Plant Cell Environ. 2025, 48, 1217–1231. [Google Scholar] [CrossRef] [PubMed]
- Lang, L.; Pettkó-Szandtner, A.; Tunçay Elbaşı, H.; Takatsuka, H.; Nomoto, Y.; Zaki, A.; Dorokhov, S.; De Jaeger, G.; Eeckhout, D.; Ito, M.; et al. The DREAM Complex Represses Growth in Response to DNA Damage in Arabidopsis. Life Sci. Alliance 2021, 4, e202101141. [Google Scholar] [CrossRef] [PubMed]
- Magyar, Z.; Bögre, L.; Ito, M. DREAMs Make Plant Cells to Cycle or to Become Quiescent. Curr. Opin. Plant Biol. 2016, 34, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Desvoyes, B.; Gutierrez, C. Roles of Plant Retinoblastoma Protein: Cell Cycle and Beyond. EMBO J. 2020, 39, e105802. [Google Scholar] [CrossRef] [PubMed]
- Gombos, M.; Raynaud, C.; Nomoto, Y.; Molnár, E.; Brik-Chaouche, R.; Takatsuka, H.; Zaki, A.; Bernula, D.; Latrasse, D.; Mineta, K.; et al. The Canonical E2Fs Together with RETINOBLASTOMA-RELATED Are Required to Establish Quiescence during Plant Development. Commun. Biol. 2023, 6, 903. [Google Scholar] [CrossRef]
- Latorre, I.; Chesney, M.A.; Garrigues, J.M.; Stempor, P.; Appert, A.; Francesconi, M.; Strome, S.; Ahringer, J. The DREAM Complex Promotes Gene Body H2A.Z for Target Repression. Genes Dev. 2015, 29, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Wei, X.; Li, L.; Xu, P.; Zhang, J.; Wang, W.; Guo, T.; Kou, S.; Wang, W.; Miao, L.; et al. Arabidopsis Cryptochrome 1 Controls Photomorphogenesis through Regulation of H2A.Z Deposition. Plant Cell 2021, 33, 1961–1979. [Google Scholar] [CrossRef] [PubMed]
- Do, B.H.; Nguyen, N.H. H2A.Z Removal Mediates the Activation of Genes Accounting for Cell Elongation under Light and Temperature Stress. Plant Cell Rep. 2024, 43, 286. [Google Scholar] [CrossRef] [PubMed]
- Delker, C.; Quint, M.; Wigge, P.A. Recent Advances in Understanding Thermomorphogenesis Signaling. Curr. Opin. Plant Biol. 2022, 68, 102231. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Wang, W.; Huq, E. Spatial Regulation of Thermomorphogenesis by HY5 and PIF4 in Arabidopsis. Nat. Commun. 2021, 12, 3656. [Google Scholar] [CrossRef] [PubMed]
- Fonseca de Lima, C.F.; Kleine-Vehn, J.; De Smet, I.; Feraru, E. Getting to the Root of Belowground High Temperature Responses in Plants. J. Exp. Bot. 2021, 72, 7404–7413. [Google Scholar] [CrossRef]
- Ai, H.; Bellstaedt, J.; Bartusch, K.S.; Eschen-Lippold, L.; Babben, S.; Balcke, G.U.; Tissier, A.; Hause, B.; Andersen, T.G.; Delker, C.; et al. Auxin-Dependent Regulation of Cell Division Rates Governs Root Thermomorphogenesis. EMBO J. 2023, 42, e111926. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.; Montiel-Jorda, A.; Cayrel, A.; Huguet, S.; Roux, C.P.-L.; Ljung, K.; Vert, G. Brassinosteroid Signaling-Dependent Root Responses to Prolonged Elevated Ambient Temperature. Nat. Commun. 2017, 8, 309. [Google Scholar] [CrossRef]
- Bellstaedt, J.; Trenner, J.; Lippmann, R.; Poeschl, Y.; Zhang, X.; Friml, J.; Quint, M.; Delker, C. A Mobile Auxin Signal Connects Temperature Sensing in Cotyledons with Growth Responses in Hypocotyls. Plant Physiol. 2020, 180, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Gaillochet, C.; Burko, Y.; Platre, M.P.; Zhang, L.; Simura, J.; Kumar, S.V.; Ljung, K.; Chory, J.; Busch, W. HY5 and Phytochrome Activity Modulate Shoot-to-Root Coordination during Thermomorphogenesis in Arabidopsis. Development 2020, 147, dev192625. [Google Scholar] [CrossRef] [PubMed]
- Borniego, M.B.; Costigliolo-Rojas, C.; Casal, J.J. Shoot Thermosensors Do Not Fulfil the Same Function in the Root. New Phytol. 2022, 236, 9–14. [Google Scholar] [CrossRef]
- Yang, X.; Dong, G.; Palaniappan, K.; Mi, G.; Baskin, T.I. Temperature-compensated Cell Production Rate and Elongation Zone Length in the Root of Arabidopsis Thaliana. Plant Cell Environ. 2017, 40, 264–276. [Google Scholar] [CrossRef]
- Hanzawa, T.; Shibasaki, K.; Numata, T.; Kawamura, Y.; Gaude, T.; Rahman, A. Cellular Auxin Homeostasis under High Temperature Is Regulated through a SORTING NEXIN1–Dependent Endosomal Trafficking Pathway. Plant Cell 2013, 25, 3424–3433. [Google Scholar] [CrossRef] [PubMed]
- Feraru, E.; Feraru, M.I.; Barbez, E.; Waidmann, S.; Sun, L.; Gaidora, A.; Kleine-Vehn, J. PILS6 Is a Temperature-Sensitive Regulator of Nuclear Auxin Input and Organ Growth in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2019, 116, 3893–3898. [Google Scholar] [CrossRef] [PubMed]
- Sablowski, R.; Gutierrez, C. Cycling in a Crowd: Coordination of Plant Cell Division, Growth, and Cell Fate. Plant Cell 2022, 34, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Soni, R.; Carmichael, J.P.; Shah, Z.H.; Murray, J.A. A Family of Cyclin D Homologs from Plants Differentially Controlled by Growth Regulators and Containing the Conserved Retinoblastoma Protein Interaction Motif. Plant Cell 1995, 7, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Boniotti, M.B.; Gutierrez, C. A Cell-Cycle-Regulated Kinase Activity Phosphorylates Plant Retinoblastoma Protein and Contains, in Arabidopsis, a CDKA/Cyclin D Complex. Plant J. 2001, 28, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Shimotohno, A.; Aki, S.S.; Takahashi, N.; Umeda, M. Regulation of the Plant Cell Cycle in Response to Hormones and the Environment. Annu. Rev. Plant Biol. 2021, 72, 273–296. [Google Scholar] [CrossRef] [PubMed]
- Meijer, M.; Murray, J.A.H. The Role and Regulation of D-Type Cyclins in the Plant Cell Cycle. Plant Mol. Biol. 2000, 43, 621–633. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fehér, A.; Hamid, R.S.B.; Magyar, Z. How Do Arabidopsis Seedlings Sense and React to Increasing Ambient Temperatures? Plants 2025, 14, 248. https://doi.org/10.3390/plants14020248
Fehér A, Hamid RSB, Magyar Z. How Do Arabidopsis Seedlings Sense and React to Increasing Ambient Temperatures? Plants. 2025; 14(2):248. https://doi.org/10.3390/plants14020248
Chicago/Turabian StyleFehér, Attila, Rasik Shiekh Bin Hamid, and Zoltán Magyar. 2025. "How Do Arabidopsis Seedlings Sense and React to Increasing Ambient Temperatures?" Plants 14, no. 2: 248. https://doi.org/10.3390/plants14020248
APA StyleFehér, A., Hamid, R. S. B., & Magyar, Z. (2025). How Do Arabidopsis Seedlings Sense and React to Increasing Ambient Temperatures? Plants, 14(2), 248. https://doi.org/10.3390/plants14020248









