Genome-Wide Identification and In Silico Expression Analysis of CCO Gene Family in Citrus clementina (Citrus) in Response to Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. Identification of CCO Genes in Citrus Clementine
2.2. Phylogenetic Analysis
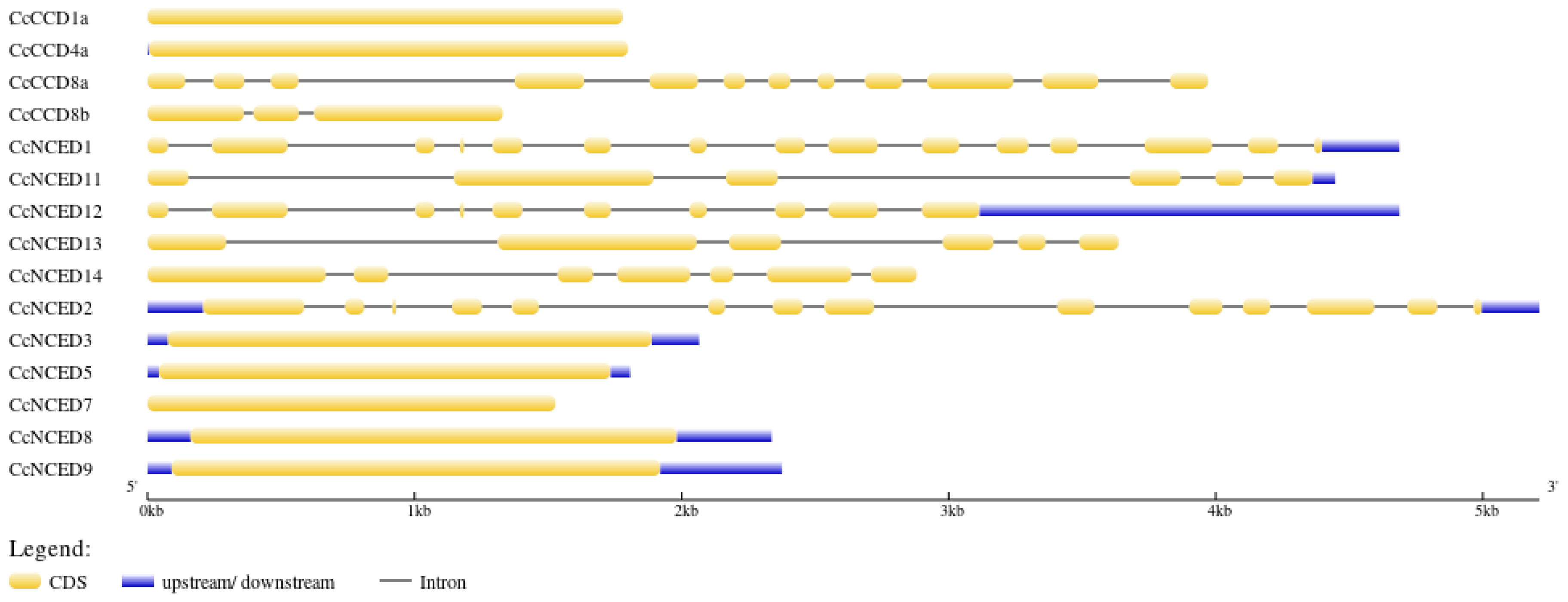
2.3. Gene Structure, Motif, and Domain Analysis
2.4. Evaluation of Duplication Event of C. clementine
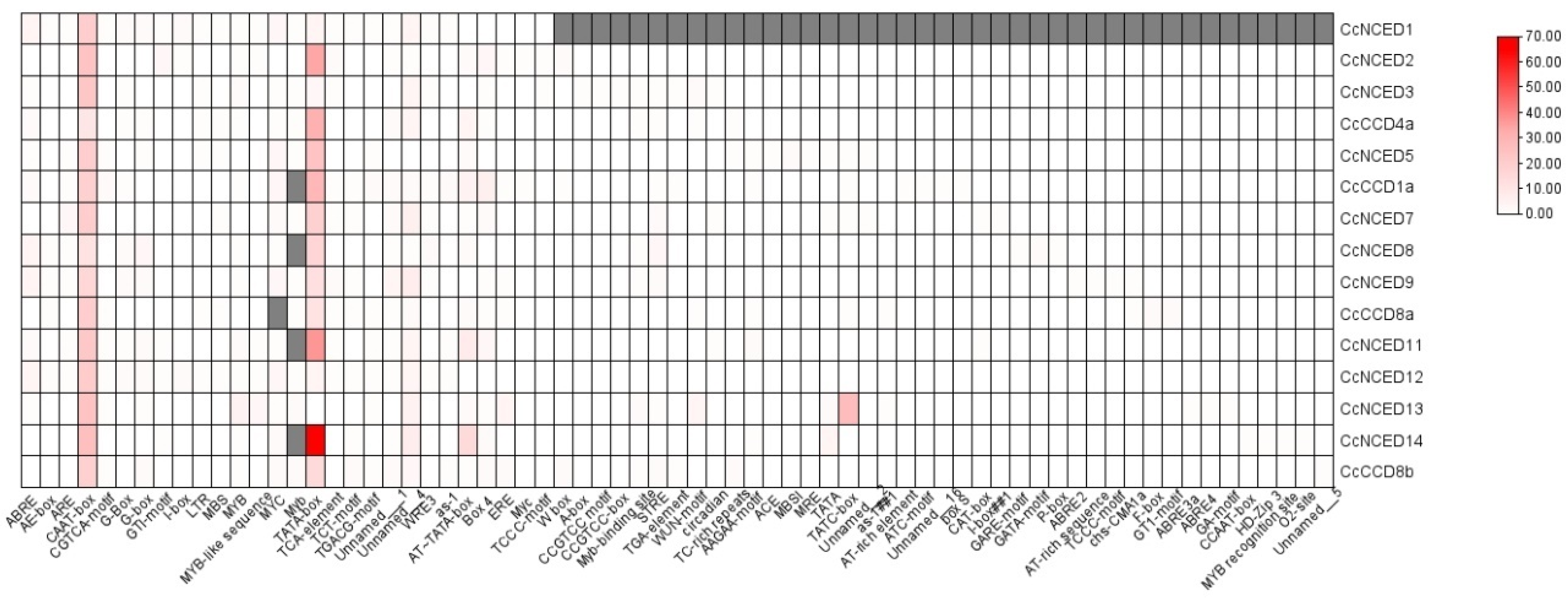
2.5. Analysis of CcCCO Cis-Regulatory Elements
2.6. Analysis of Protein–Protein Interaction Network
2.7. MicroRNA Target Site Analysis
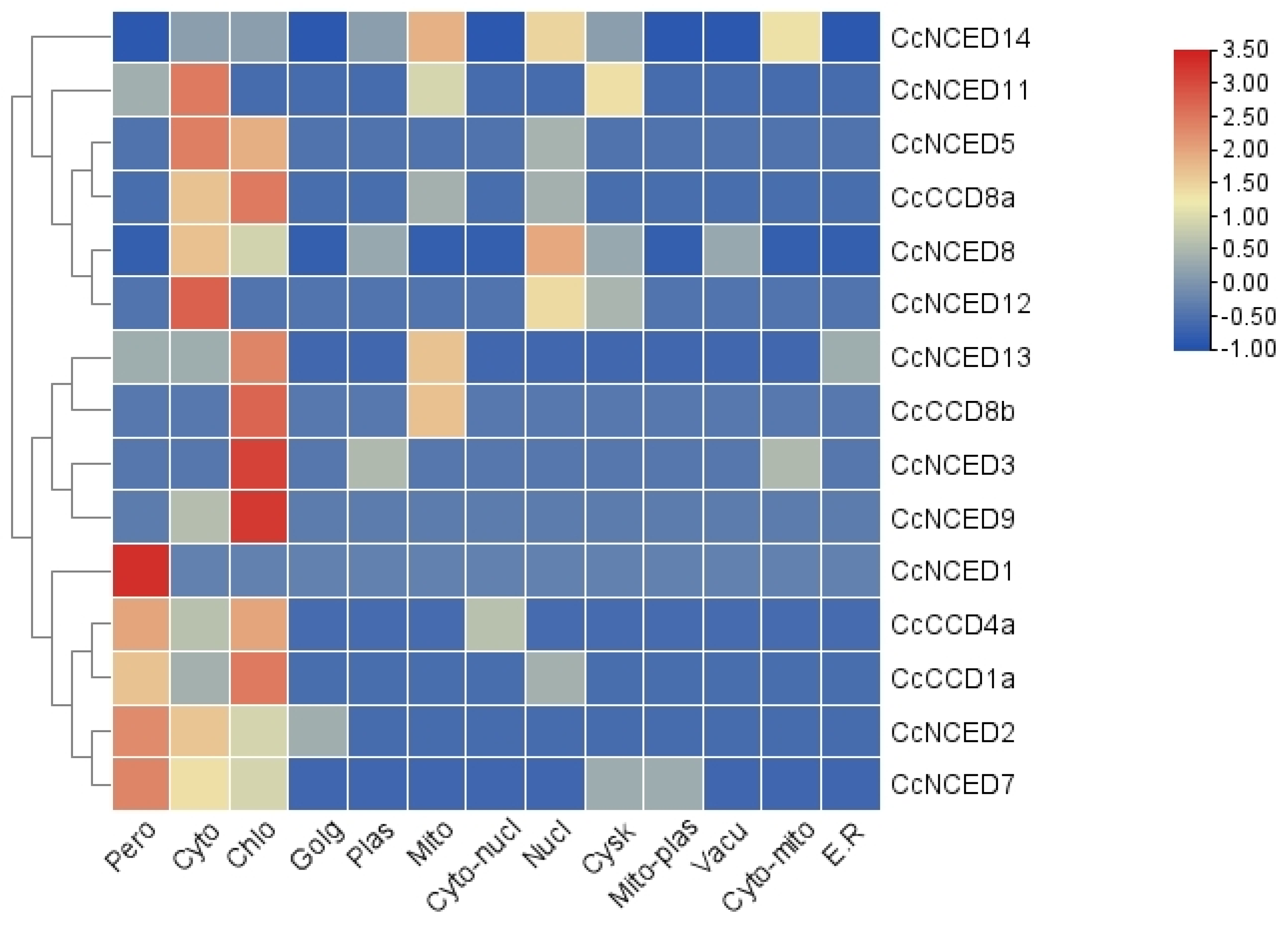
2.8. Gene Expression
3. Materials and Methods
3.1. Sequence Retrieval
3.2. Determination of Physiochemical Properties and Subcellular Localization of CCO Genes
3.3. Gene Structure, Cis-Regulatory Analysis and Motif Analysis
3.4. The Analysis of Phylogenetic
3.5. Gene Duplication and Synteny Analysis
3.6. Gene Ontology Analysis and Protein–Protein Interaction
3.7. Putative miRNA Analysis
3.8. Chromosomal Mapping
3.9. Expression Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declarations
References
- Johnson, J.D. Do carotenoids serve as transmembrane radical channels? Free Radic. Biol. Med. 2009, 47, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Guo, Y.; Zhang, Y.; Guo, J.; Li, K.; Fu, W.; Jia, Z.; Li, W.; Tran, L.-S.P.; Jia, K.-P.; et al. Genome-wide identification, characterization and expression profiles of the CCD gene family in Gossypium species. 3 Biotech 2021, 11, 249. [Google Scholar] [CrossRef]
- Yahia, E.M.; de Jesús Ornelas-Paz, J.; Emanuelli, T.; Jacob-Lopes, E.; Zepka, L.Q.; Cervantes-Paz, B. Chemistry, stability, and biological actions of carotenoids. In Fruit and Vegetable Phytochemicals: Chemistry and Human Health, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 285–346. [Google Scholar]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, G.; Al-Babili, S.; Von Lintig, J. Carotenoid oxygenases: Cleave it or leave it. Trends Plant Sci. 2003, 8, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney-Smith, C.; Mila, I.; Bouzayen, M.; Magallanes-Lundback, M.; DellaPenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef]
- Borowski, T.; Blomberg, M.R.; Siegbahn, P.E. Reaction mechanism of apocarotenoid oxygenase (ACO): A DFT study. Chem. A Eur. J. 2008, 14, 2264–2276. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Chen, H.; Zuo, X.; Shao, H.; Fan, S.; Ma, J.; Zhang, D.; Zhao, C.; Yan, X.; Liu, X.; Han, M. Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica). Plant Physiol. Biochem. 2018, 123, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Gómez-Gómez, L.; Rodrigo, M.J.; Avalos, J.; Limón, M.C. Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: Features and functions. Int. J. Mol. Sci. 2016, 17, 1781. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.D. Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 2001, 276, 25208–25211. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhu, K.; Sun, Q.; Zhang, W.; Wang, X.; Cao, H.; Tan, M.; Xie, Z.; Zeng, Y.; Ye, J. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol. Plant 2019, 12, 1294–1307. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zheng, H.; Zhao, J.; Xu, Y.; Li, X. ZmCCD7/ZpCCD7 encodes a carotenoid cleavage dioxygenase mediating shoot branching. Planta 2016, 243, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.-J.; Zhang, J.-J.; Xue, H.-W. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PLoS ONE 2012, 7, e31081. [Google Scholar] [CrossRef]
- Chernys, J.T.; Zeevaart, J.A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–354. [Google Scholar] [CrossRef]
- Thompson, A.J.; Jackson, A.C.; Symonds, R.C.; Mulholland, B.J.; Dadswell, A.R.; Blake, P.S.; Burbidge, A.; Taylor, I.B. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 2000, 23, 363–374. [Google Scholar] [CrossRef]
- Ji, K.; Kai, W.; Zhao, B.; Sun, Y.; Yuan, B.; Dai, S.; Li, Q.; Chen, P.; Wang, Y.; Pei, Y. SlNCED1 and SlCYP707A2: Key genes involved in ABA metabolism during tomato fruit ripening. J. Exp. Bot. 2014, 65, 5243–5255. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chen, L.-L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.-B.; Hao, B.-H.; Lyon, M.P. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Cittadino, G.M.; Andrews, J.; Purewal, H.; Estanislao Acuña Avila, P.; Arnone, J.T. Functional clustering of metabolically related genes is conserved across Dikarya. J. Fungi 2023, 9, 523. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yu, X.; Chen, L.; Zhao, G.; Li, S.; Zhou, H.; Dai, Y.; Sun, N.; Xie, Y.; Gao, J. Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum L.). PLoS ONE 2021, 16, e0246021. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660, 289660. [Google Scholar]
- Park, S.; Shi, A.; Mou, B. Genome-wide identification and expression analysis of the CBF/DREB1 gene family in lettuce. Sci. Rep. 2020, 10, 5733. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Edgar, R. Mining microarray data at NCBI’s Gene Expression Omnibus (GEO). In Gene Mapping, Discovery, and Expression: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2006; pp. 175–190. [Google Scholar]
- Duarte, A.; Fernandes, M.J.; Bernardes, J.P.; Miguel, M.G. Citrus as a component of the Mediterranean diet. J. Spat. Organ. Dyn. 2016, 4, 289–304. [Google Scholar]
- Kocira, A.; Staniak, M.; Tomaszewska, M.; Kornas, R.; Cymerman, J.; Panasiewicz, K.; Lipińska, H. Legume cover crops as one of the elements of strategic weed management and soil quality improvement. A review. Agriculture 2020, 10, 394. [Google Scholar] [CrossRef]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. 2018, 132, 1–25. [Google Scholar] [CrossRef]
- Mazhar, H.S.-U.-D.; Shafiq, M.; Ali, H.; Ashfaq, M.; Anwar, A.; Tabassum, J.; Ali, Q.; Jilani, G.; Awais, M.; Sahu, R. Genome-wide identification, and in-silico expression analysis of YABBY gene family in response to biotic and abiotic stresses in potato (Solanum tuberosum). Genes 2023, 14, 824. [Google Scholar] [CrossRef]
- Dockrall, S. Carotenoid Cleavage Dioxygenases (CCDs) of Grape. Ph.D. Thesis, Stellenbosch University, Stellenbosch, South Africa, December 2012. [Google Scholar]
- Qiao, L.; Zhang, X.; Han, X.; Zhang, L.; Li, X.; Zhan, H.; Ma, J.; Luo, P.; Zhang, W.; Cui, L. A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front. Plant Sci. 2015, 6, 770. [Google Scholar] [CrossRef] [PubMed]
- Tadeo, F.R.; Cercos, M.; Colmenero-Flores, J.M.; Iglesias, D.J.; Naranjo, M.A.; Rios, G.; Carrera, E.; Ruiz-Rivero, O.; Lliso, I.; Morillon, R. Molecular physiology of development and quality of citrus. Adv. Bot. Res. 2008, 47, 147–223. [Google Scholar]
- Flinn, B.; Rothwell, C.; Griffiths, R.; Lägue, M.; DeKoeyer, D.; Sardana, R.; Audy, P.; Goyer, C.; Li, X.-Q.; Wang-Pruski, G. Potato expressed sequence tag generation and analysis using standard and unique cDNA libraries. Plant Mol. Biol. 2005, 59, 407–433. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M.; Thomas, B.C. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006, 16, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A. Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 2014, 17, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.W.; De Bie, T.; Stajich, J.E.; Nguyen, C.; Cristianini, N. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 2005, 15, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, P.; Yuan, J.S. Plant protein-protein interaction network and interactome. Curr. Genom. 2010, 11, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Khanna, K.; Ruan, S. Expression of microRNAs and its regulation in plants. In Seminars in Cell & Developmental Biology; Academic Press: New York, NY, USA, 2010; pp. 790–797. [Google Scholar]







| Gene ID | Source Accession No. | Chromosome No. | Chromosome Location | Direction | pI | No. of AA | Mw (Da) | Size | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Phytozome ID | Scaffold | Start | End | Genome | Peptide | ||||
| CcNCED1 | Ciclev10031039m.1 | 4 | 25506445 | 25511133 | F | 6.05 | 589 | 66,677.35 | 4688 | 590 |
| CcNCED2 | Ciclev10031014m.1 | 4 | 25500600 | 25505812 | F | 6.33 | 597 | 67,203.01 | 5212 | 598 |
| CcNCED3 | Ciclev10031003m.1 | 4 | 18266261 | 18268327 | R | 6.87 | 603 | 66,452.87 | 2066 | 604 |
| CcCCD4a | Ciclev10011335m.1 | 6 | 23758343 | 23760142 | F | 8.53 | 597 | 66,347.09 | 1799 | 598 |
| CcNCED5 | Ciclev10028113m.1 | 8 | 17933011 | 17934818 | F | 8.34 | 563 | 63,060.24 | 1807 | 564 |
| CcCCD1a | Ciclev10006710m.1 | 9 | 10202438 | 10204217 | R | 7.32 | 592 | 65,292.48 | 1779 | 593 |
| CcNCED7 | Ciclev10030384m.1 | 8 | 17966529 | 17968056 | F | 6.56 | 508 | 56,910.31 | 1527 | 509 |
| CcNCED8 | Ciclev10019364m.1 | 3 | 29351853 | 29354190 | R | 6.37 | 606 | 67,013.31 | 2337 | 607 |
| CcNCED9 | Ciclev10014639m.1 | 2 | 35235516 | 35237892 | F | 6.30 | 609 | 67,794.81 | 2376 | 610 |
| CcCCD8a | Ciclev10010551m.1 | 1 | 3113082 | 3117053 | R | 5.79 | 611 | 69,176.61 | 3971 | 612 |
| CcCCD8b | Ciclev10013726m.1 | 6 | 23780381 | 23781711 | F | 7.25 | 412 | 45,860.27 | 1330 | 412 |
| CcNCED11 | Ciclev10008050m.1 | 1 | 3187977 | 3192424 | F | 5.93 | 510 | 56,800.61 | 4447 | 511 |
| CcNCED12 | Ciclev10031690m.1 | 4 | 25506445 | 25511133 | F | 8.27 | 410 | 46,734.14 | ||
| CcNCED13 | Ciclev10010609m.1 | 1 | 3208798 | 3212434 | F | 5.98 | 556 | 61,964.53 | 3636 | 557 |
| CcNCED14 | Ciclev10027500m.1 | 7 | 1348302 | 1351182 | R | 6.07 | 590 | 66,508.85 | 2880 | 591 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarwar, S.; Sami, A.; Haider, M.Z.; Tasawar, L.; Akram, J.; Ahmad, A.; Shafiq, M.; Zaki, H.E.M.; Ondrasek, G.; Shahid, M.S. Genome-Wide Identification and In Silico Expression Analysis of CCO Gene Family in Citrus clementina (Citrus) in Response to Abiotic Stress. Plants 2025, 14, 249. https://doi.org/10.3390/plants14020249
Sarwar S, Sami A, Haider MZ, Tasawar L, Akram J, Ahmad A, Shafiq M, Zaki HEM, Ondrasek G, Shahid MS. Genome-Wide Identification and In Silico Expression Analysis of CCO Gene Family in Citrus clementina (Citrus) in Response to Abiotic Stress. Plants. 2025; 14(2):249. https://doi.org/10.3390/plants14020249
Chicago/Turabian StyleSarwar, Sadaf, Adnan Sami, Muhammad Zeshan Haider, Layba Tasawar, Jannat Akram, Arsalan Ahmad, Muhammad Shafiq, Haitham E. M. Zaki, Gabrijel Ondrasek, and Muhammad Shafiq Shahid. 2025. "Genome-Wide Identification and In Silico Expression Analysis of CCO Gene Family in Citrus clementina (Citrus) in Response to Abiotic Stress" Plants 14, no. 2: 249. https://doi.org/10.3390/plants14020249
APA StyleSarwar, S., Sami, A., Haider, M. Z., Tasawar, L., Akram, J., Ahmad, A., Shafiq, M., Zaki, H. E. M., Ondrasek, G., & Shahid, M. S. (2025). Genome-Wide Identification and In Silico Expression Analysis of CCO Gene Family in Citrus clementina (Citrus) in Response to Abiotic Stress. Plants, 14(2), 249. https://doi.org/10.3390/plants14020249









