Behavior of Olive Genotypes Against Quick Decline Syndrome (QDS) Caused by Xylella fastidiosa subsp. pauca in Apulia
Abstract
1. Introduction
2. Material and Methods
2.1. Field Survey for Identification of Surviving Genotypes
2.2. Assessment of OQDS Symptoms Severity
2.3. Diagnostic Test and Estimation of Xfp Population in the Olive Tree Tissue
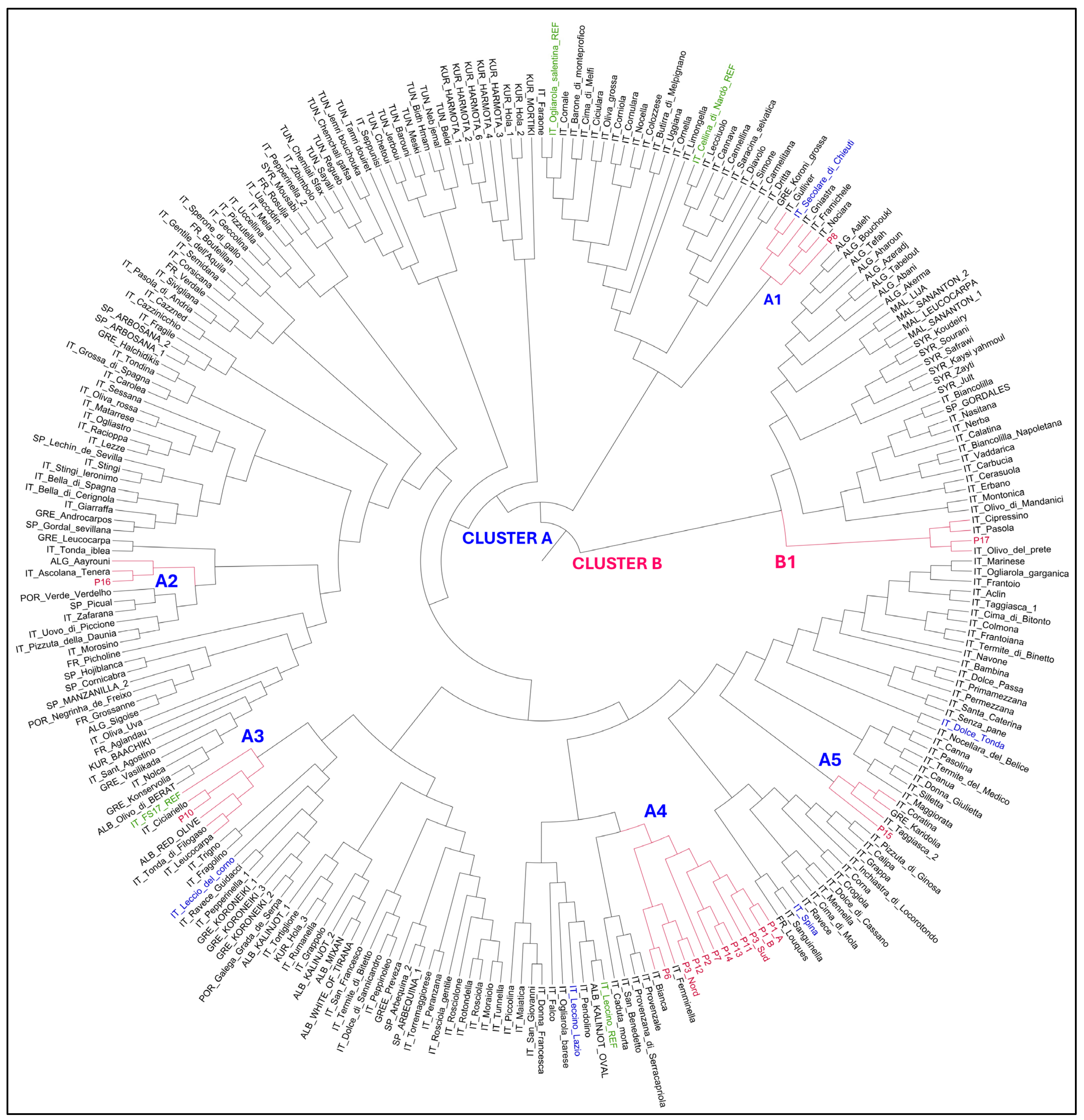
2.4. Olive Genotyping Using SSR Markers
3. Results
Field Survey for Identification of Surviving Genotypes and Assessment of Symptoms Severity and Bacterial Load
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority (EFSA); Cavalieri, V.; Fasanelli, E.; Gibin, D.; Gutierrez Linares, A.; La Notte, P.; Pasinato, L.; Delbianco, A. Update of the Xylella spp. host plant database—Systematic literature search up to 31 December 2023. EFSA J. 2024, 222, e8898. [Google Scholar] [CrossRef]
- Cornara, D.; Fereres, A.; Almeida, R. Elucidating the inoculation mechanism of Xylella fastidiosa. In Proceedings of the 15th International Symposium of Plant Virus Epidemiology (ISPVE), Madrid, Spain, 5–8 June 2022; pp. 5–8. [Google Scholar]
- Cornara, D.; Marra, M.; Morente, M.; Garzo, E.; Moreno, A.; Saponari, M.; Fereres, A. Feeding behavior in relation to spittlebug transmission of Xylella fastidiosa. J. Pest Sci. 2020, 93, 1197–1213. [Google Scholar] [CrossRef]
- Cariddi, C.; Saponari, M.; Boscia, D.; De Stradis, A.; Loconsole, G.; Nigro, F.; Porcelli, F.; Potere, O.; Martelli, G.P. Isolation of a Xylella fastidiosa strain infecting olive and oleander in Apulia, Italy. J. Plant Pathol. 2014, 96, 425–429. Available online: https://www.jstor.org/stable/24332228 (accessed on 22 October 2024).
- Schneider, K.; van der Werf, M.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Lansink, A.O. Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. USA 2020, 117, 9250–9259. [Google Scholar] [CrossRef]
- Frem, M.; Santeramo, F.G.; Lamonaca, E.; El Moujabber, M.; Choueiri, E.; La Notte, P.; Nigro, F.; Fucilli, V. Landscape restoration due to Xylella fastidiosa invasion in Italy: Assessing the hypothetical public’s preferences. NeoBiota 2021, 66, 31–54. [Google Scholar] [CrossRef]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’attoma, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D.; et al. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in Southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, J.; Ingel, B.; Blanco-Ulate, B.; Cantu, D.; Roper, C. Xylella fastidiosa: An examination of a re-emerging plant pathogen. Mol. Plant Pathol. 2018, 19, 786–800. [Google Scholar] [CrossRef]
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in olive in Apulia: Where we stand. Phytopathology 2019, 109, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Mascio, I.; Savoia, M.A.; Miazzi, M.M.; Fanelli, V.; Dellino, M.; Piarulli, L.; Montemurro, C. Insight into the European Union community trademarks olive oils traceability: The use of DNA markers as the most effective approach Trends Food Sci. Technol. 2024, 151, 104615. [Google Scholar] [CrossRef]
- European Commission. Commission Delegated Regulation (EU) 2019/1702 of 1 August 2019 supplementing Regulation (EU) 2016/2031 of the European Parliament and of the Council by establishing the list of priority pests. Off. J. Eur. Union L 2019, 260, 8–10. Available online: http://data.europa.eu/eli/reg_del/2019/1702/oj/eng (accessed on 31 October 2024).
- European Commission. Commission Implementing Regulation (EU) 2020/1201 of 14 August 2020 as regards measures to prevent the introduction into and the spread within the Union of Xylella fastidiosa (Wells et al.). Off. J. Eur. Union L 2020, 269, 2–39. Available online: http://data.europa.eu/eli/reg_impl/2020/1201/oj/eng (accessed on 31 October 2024).
- Picciotti, U.; Valverde-Urrea, M.; Garganese, F.; Lopez-Moya, F.; Foubelo, F.; Porcelli, F.; Lopez-Llorca, L.V. Brindley’s Glands Volatilome of the Predator Zelus renardii Interacting with Xylella Vectors. Insects 2023, 14, 520. [Google Scholar] [CrossRef] [PubMed]
- Savoia, M.A.; Fanelli, V.; Miazzi, M.M.; Taranto, F.; Procino, S.; Susca, L.; Montilon, V.; Potere, O.; Nigro, F.; Montemurro, C. Apulian Autochthonous Olive Germplasm: A Promising Resource to Restore Cultivation in Xylella fastidiosa-Infected Areas. Agriculture 2023, 13, 1746. [Google Scholar] [CrossRef]
- Picciotti, U.; Valverde-Urrea, M.; Sefa, V.; Ragni, M.; Garganese, F.; Porcelli, F. Performance of Artificial Diets for Zelus renardii (Hemiptera: Reduviidae) Rearing. Insects 2024, 15, 607. [Google Scholar] [CrossRef]
- Rodriguez, R.; Henson, J.; Volkenburgh, E.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.-O.; Redman, R.S. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008, 2, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Clair, J.B.S.; Lind, B.M.; Cronn, R.; Wilhelmi, N.P.; Feau, N.; Lu, M.; Vidakovic, D.O.; Hamelin, R.C.; Shaw, D.C.; et al. Genetic architecture of disease resistance and tolerance in douglas-fir trees. New Phytol. 2024, 243, 705–719. [Google Scholar] [CrossRef]
- Marchand, M.; Allery, T.; Massot, M.; Capdevielle, X.; Robin, C. Resistance, tolerance and competence for a root pathogen in six woody species. Plant Pathol. 2022, 71, 1700–1711. [Google Scholar] [CrossRef]
- Masini, L.; Grenville-Briggs, L.; Andréasson, E.; Råberg, L.; Lankinen, Å. Tolerance and overcompensation to infection by Phytophthora infestans in the wild perennial climber Solanum dulcamara. Ecol. Evol. 2019, 9, 4557–4567. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Morelli, M.; Saponari, M.; Loconsole, G.; Chiumenti, M.; Boscia, D.; Savino, V.N.; Martelli, G.P.; Saldarelli, P. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genom. 2016, 17, 475. [Google Scholar] [CrossRef] [PubMed]
- Boscia, D.; Altamura, G.; La Notte, P.; Morelli, M.; Saldarelli, P.; Saponari, M.; Savino, V.N.; Silletti, M.R.; Specchia, F.; Susca, L.; et al. Resistenza a Xylella fastidiosa in diverse cultivar di olivo. Inf. Agrar. 2017, 11, 59–63. [Google Scholar] [CrossRef]
- Sabella, E.; Buja, I.; Negro, C.; Vergine, M.; Cherubini, P.; Pavan, S.; Maruccio, G.; De Bellis, L.; Luvisi, A. The Significance of Xylem Structure and Its Chemical Components in Certain Olive Tree Genotypes with Tolerance to Xylella fastidiosa Infection. Plants 2024, 13, 930. [Google Scholar] [CrossRef]
- Montilon, V.; De Stradis, A.; Saponari, M.; Abou Kubaa, R.; Giampetruzzi, A.; D’Attoma, G.; Saldarelli, P. Xylella fastidiosa subsp. pauca ST53 exploits pit membranes of susceptible olive cultivars to spread systemically in the xylem. Plant Pathol. 2023, 72, 144–153. [Google Scholar] [CrossRef]
- Surano, A.; Abou Kubaa, R.; Nigro, F.; Altamura, G.; Losciale, P.; Saponari, M.; Saldarelli, P. Susceptible and resistant olive cultivars show differential physiological response to Xylella fastidiosa infections. Front. Plant Sci. 2022, 13, 968934. [Google Scholar] [CrossRef]
- La Notte, P.; Saponari, M.; Mousavi, S.; Mariotti, R.; Abou Kubaa, R.; Nikbakht, R.; Melcarne, G.; Specchia, F.; Altamura, G.; Ligorio, A.; et al. A survey in natural olive resources exposed to high inoculum pressure indicates the presence of traits of resistance to Xylella fastidiosa in Leccino offspring. Front. Plant Sci. 2024, 15, 1457831. [Google Scholar] [CrossRef]
- Miazzi, M.M.; Di Rienzo, V.; Mascio, I.; Montemurro, C.; Sion, S.; Sabetta, W.; Vivaldi, G.A.; Camposeo, S.; Caponio, F.; Squeo, G.; et al. Re. Ger. OP: An integrated project for the recovery of ancient and rare olive germplasm. Front. Plant Sci. 2020, 11, 73. [Google Scholar] [CrossRef]
- Saddoud Debbabi, O.; Miazzi, M.M.; Elloumi, O.; Fendri, M.; Ben Amar, F.; Savoia, M.; Sion, S.; Souabni, H.; Mnasri, S.R.; Ben Abdelaali, S.; et al. Recovery, assessment, and molecular characterization of minor olive genotypes in Tunisia. Plants 2020, 9, 382. [Google Scholar] [CrossRef]
- Falek, W.; Mascio, I.; Gadaleta, S.; Fanelli, V.; Bechkri, S.; Khelifi, D.; Miazzi, M.M.; Montemurro, C. Morphological and eco-geographic variation in Algerian wild olives. Plants 2022, 11, 1803. [Google Scholar] [CrossRef]
- Islam, A.F.; Sanders, D.; Mishra, A.K.; Joshi, V. Genetic diversity and population structure analysis of the USDA olive germplasm using genotyping-by-sequencing (GBS). Genes 2021, 12, 2007. [Google Scholar] [CrossRef]
- Sion, S.; Savoia, M.A.; Gadaleta, S.; Piarulli, L.; Mascio, I.; Fanelli, V.; Montemurro, C.; Miazzi, M.M. How to choose a good marker to analyze the olive germplasm (Olea europaea L.) and derived products. Genes 2021, 12, 1474. [Google Scholar] [CrossRef]
- Bazakos, C.; Alexiou, K.G.; Ramos-Onsins, S.; Koubouris, G.; Tourvas, N.; Xanthopoulou, A.; Mellidou, I.; Moysiadis, T.; Vourlaki, I.T.; Metzidakis, I.; et al. Whole genome scanning of a Mediterranean basin hotspot collection provides new insights into olive tree biodiversity and biology. Plant J. 2023, 116, 303–319. [Google Scholar] [CrossRef]
- Marchese, A.; Bonanno, F.; Marra, F.P.; Trippa, D.A.; Zelasco, S.; Rizzo, S.; Giovino, A.; Imperiale, V.; Ioppolo, A.; Sala, G.; et al. Recovery and genotyping ancient Sicilian monumental olive trees. Front. Conserv. Sci. 2023, 4, 1206832. [Google Scholar] [CrossRef]
- OEPP/EPPO. PM 7/24 (5) Xylella fastidiosa. OEPP/EPPO Bull. 2023, 53, 205–276. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Baptista, P.; Morelli, M.; Cameirão, C.; Lino Neto, T.; Costa, D.; D’Attoma, G.; Kubaa, A.R.; Altamura, G.; Saponari, M.; et al. Differences in the endophytic microbiome of olive cultivars infected by Xylella fastidiosa across seasons. Pathogens 2020, 9, 723. [Google Scholar] [CrossRef]
- Harper, S.J.; Ward, L.I.; Clover, G.R.G.; Marco-Noales, E.; Barbé, S.; Monterde, A.; Navarro, I.; Ferrer, A.; Dalmau, V.; Aure, C.M.; et al. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology 2010, 100, 1282–1288. [Google Scholar] [CrossRef]
- D’Attoma, G.; Morelli, M.; Saldarelli, P.; Saponari, M.; Giampetruzzi, A.; Boscia, D.; Savino, V.N.; De La Fuente, L.; Cobine, P.A. Ionomic differences between susceptible and resistant olive cultivars infected by Xylella fastidiosa in the outbreak area of Salento, Italy. Pathogens 2019, 8, 272. [Google Scholar] [CrossRef]
- Spadoni, A.; Sion, S.; Gadaleta, S.; Savoia, M.; Piarulli, L.; Fanelli, V.; di Rienzo, V.; Taranto, F.; Miazzi, M.M.; Montemurro, C.; et al. A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. J. Agric. Sci. Technol. 2019, 21, 1215–1226. Available online: http://jast.modares.ac.ir/article-23-13213-en.html (accessed on 14 November 2024).
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef]
- De La Rosa, R.; James, C.M.; Tobutt, K.R. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol. Ecol. Notes 2002, 2, 265–267. [Google Scholar] [CrossRef]
- Miazzi, M.M.; Pasqualone, A.; Zammit-Mangion, M.; Savoia, M.A.; Fanelli, V.; Procino, S.; Gadaleta, S.; Aurelio, F.L.; Montemurro, C. A Glimpse into the Genetic Heritage of the Olive Tree in Malta. Agriculture 2024, 14, 495. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Morelli, M.; García-Madero, J.M.; Jos, Á.; Saldarelli, P.; Dongiovanni, C.; Kovacova, M.; Saponari, M.; Arjona, B.A.; Hackl, E.; Webb, S.; et al. Xylella fastidiosa in olive: A review of control attempts and current management. Microorganisms 2021, 9, 1771. [Google Scholar] [CrossRef] [PubMed]
- Murray-Watson, R.; Cunniffe, N. How the epidemiology of disease-resistant and disease-tolerant varieties affects grower behaviour. J. R. Soc. Interface 2022, 19, 195. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, V.; Mascio, I.; Falek, W.; Miazzi, M.M.; Montemurro, C. Current Status of Biodiversity Assessment and Conservation of Wild Olive (Olea europaea L. subsp. europaea var. sylvestris). Plants 2022, 11, 480. [Google Scholar] [CrossRef]
- Pavan, S.; Vergine, M.; Nicolì, F.; Sabella, E.; Aprile, A.; Negro, C.; Fanelli, V.; Savoia, M.A.; Montilon, V.; Susca, L.; et al. Screening of olive biodiversity defines genotypes potentially resistant to Xylella fastidiosa. Front. Plant Sci. 2021, 12, 723879. [Google Scholar] [CrossRef]
- Roux, F.; Gao, L.; Bergelson, J. Impact of initial pathogen density on resistance and tolerance in a polymorphic disease resistance gene system in Arabidopsis thaliana. Genetics 2010, 185, 283–291. [Google Scholar] [CrossRef][Green Version]
- Medina, I.; Langmore, N. The evolution of acceptance and tolerance in hosts of avian brood parasites. Biol. Rev. 2015, 91, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Pearse, I.; Aguilar, J.; Schroder, J.; Strauss, S. Macroevolutionary constraints to tolerance: Trade-offs with drought tolerance and phenology, but not resistance. Ecology 2017, 98, 2758–2772. [Google Scholar] [CrossRef] [PubMed]
- Walker, N.C.; Ruiz, S.A.; Ferreira, T.R.; Coletta-Filho, H.D.; Le Houx, J.; McKay Fletcher, D.; White, S.M.; Roose, T. A high-throughput analysis of high-resolution X-ray CT images of stems of olive and citrus plants resistant and susceptible to Xylella fastidiosa. Plant Pathol. 2024, 73, 630–643. [Google Scholar] [CrossRef]
- Vergine, M.; Meyer, J.B.; Cardinale, M.; Sabella, E.; Hartmann, M.; Cherubini, P.; De Bellis, L.; Luvisi, A. The Xylella fastidiosa-resistant olive cultivar “Leccino” has stable endophytic microbiota during the olive quick decline syndrome (OQDS). Pathogens 2019, 9, 35. [Google Scholar] [CrossRef]
- Mourou, M.; Hanani, A.; D’Onghia, A.M.; Davino, S.W.; Balestra, G.M.; Valentini, F. Antagonism and antimicrobial capacity of epiphytic and endophytic bacteria against the phytopathogen Xylella fastidiosa. Agronomy 2022, 12, 1266. [Google Scholar] [CrossRef]
- D’Agostino, N.; Taranto, F.; Camposeo, S.; Mangini, G.; Fanelli, V.; Gadaleta, S.; Miazzi, M.M.; Pavan, S.; di Rienzo, V.; Sabetta, W.; et al. GBS-derived SNP catalogue unveiled wide genetic variability and geographical relationships of Italian olive cultivars. Sci. Rep. 2018, 8, 15877. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C. Plant–vertebrate seed dispersal systems in the Mediterranean: Ecological, evolutionary, and historical determinants, Annu. Rev. Ecol. Evol. Syst. 1995, 26, 705–727. Available online: https://www.jstor.org/stable/2097225 (accessed on 10 October 2024). [CrossRef]
- Alcantara, J.M.; Rey, P.J. Conflicting selection pressures on seed size: Evolutionary ecology of fruit size in a bird-dispersed tree, Olea europaea. J. Evol. Biol. 2003, 16, 1168–1176. [Google Scholar] [CrossRef] [PubMed]
- Tarifa, R.; González-Varo, J.P.; Camacho, F.M.; Pérez, A.J.; Salido, T.; Rey, P.J. Woodland loss differently affects seed dispersal by resident and migratory avian frugivores in olive grove-dominated landscapes. Agric. Ecosyst. Environ. 2024, 359, 108752. [Google Scholar] [CrossRef]
- Pasqualone, A.; Di Rienzo, V.; Blanco, A.; Summo, C.; Caponio, F.; Montemurro, C. Characterization of virgin olive oil from Leucocarpa cultivar by chemical and DNA analysis. Food Res. Int. 2012, 47, 188–193. [Google Scholar] [CrossRef]
- Fontanazza, G.; Baldoni, L.; Corona, C. Osservazioni sull’impiego di portainnesti clonali negli olivi “Ascolana tenera” e “Giarraffa”. Riv. Frutticol. 1992, 54, 65–69. [Google Scholar]
- Ranalli, A.; Lucera, L.; Contento, S.; Fontanazza, G.; Patumi, M. Assessment of physico-chemical, sensory and nutritional parameters in virgin olive oil from the new genotype Favolosa (FS17). Acta Hortic. 2008, 791, 697–704. [Google Scholar] [CrossRef]
- Sefc, K.M.; Lopes, M.S.; Mendonça, D.; Santos, M.R.D.; Machado, M.L.D.C.; Machado, A.D.C. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 2000, 9, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
| Genotypes | Symptom Severity | |||||
|---|---|---|---|---|---|---|
| 2019 | 2020 | 2021 | 2022 | 2023 | 2024 | |
| P1_A | 1 | 1 | 0 | 0 | 0 | 1 |
| P1_B | 0 | 0 | 1,4 | 0 | 1 | 0 |
| P2 | 0 | 0 | 1 | 0 | 1 | 1,3 |
| P3_Sud | 0 | 0 | 1 | 0 | 1,5 | 1 |
| P3_Nord | 1 | 1 | 1 | 0 | 1 | 1 |
| P6 | - | - | - | 1 | 0 | 1 |
| P7 | 1 | 0 | 0 | 0 | 0 | 0 |
| P8 | 1 | 0 | 0 | 0 | 0 | 0 |
| P10 | 1 | 1 | 1 | 1 | 1 | 1 |
| P11 | 1 | 1 | 0 | 0 | 0 | 1 |
| P12 | 0 | 0 | 0 | 0 | 0 | 0 |
| P13 | 1 | 1 | 1 | 0 | 1 | 1 |
| P14 | 0 | 1 | 0 | 1 | 1 | 1 |
| P15 | - | - | - | - | - | 1 |
| P16 | - | - | - | - | - | 1 |
| P17 | - | - | - | - | - | 0 |
| Genotypes | 2019 | 2020 | 2021 | 2022 | 2023 | 2024 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cq | Cfu/mL | Cq | Cfu/mL | Cq | Cfu/mL | Cq | Cfu/mL | Cq | Cfu/mL | Cq | Cfu/mL | |
| P1_A | 35.87 | 4.5 × 103 | 34.66 | 3.2 × 103 | U | U | 35.67 | 4.24 × 103 | U | U | 34.22 | 3.52 × 103 |
| P1_B | U | U | U | U | 24.78 | 3.12 × 106 | U | U | U | U | 37.24 | 8.58 × 102 |
| P2 | U | U | U | U | 31.77 | 4.35 × 104 | U | U | 36.63 | 1.55 × 103 | 27.53 | 2.43 × 105 |
| P3_Sud | U | U | U | U | 30.58 | 5.71 × 104 | U | U | 28.40 | 2.77 × 105 | 32.68 | 9.61 × 103 |
| P3_Nord | 33.40 | 6.8 × 104 | U | U | 34.87 | 2.97 × 103 | 35.67 | 1.27 × 103 | 30.33 | 8.10 × 104 | U | U |
| P6 | NA | NA | NA | NA | NA | NA | 35.77 | 1.72 × 103 | 37.24 | 2.43 × 102 | U | U |
| P7 | U | U | U | U | U | U | U | U | U | U | U | U |
| P8 | U | U | U | U | U | U | U | U | U | U | U | U |
| P10 | 32.13 | 7.5 × 104 | 32.24 | 7.1 × 104 | 31.46 | 1.24 × 104 | 29.45 | 3.65 × 105 | 31.39 | 9.4 × 104 | U | U |
| P11 | 34.70 | 4.6 × 103 | 35.67 | 1.4 × 103 | U | U | U | U | U | U | NA | NA |
| P12 | U | U | U | U | U | U | U | U | U | U | NA | NA |
| P13 | 33.12 | 5.2 × 104 | 33.61 | 2.7 × 104 | 32.19 | 8.67 × 103 | 35.62 | 6.48 × 103 | 33.44 | 8.6 × 103 | 23.78 | 2.94 × 106 |
| P14 | U | U | 31.73 | 8.3 × 104 | U | U | 33.65 | 2.72 × 104 | 32.87 | 2.6 × 104 | 34.14 | 4.30 × 103 |
| P15 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 20.46 | 2.35 × 107 |
| P16 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 21.15 | 1.53 × 107 |
| P17 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 35.19 | 2.38 × 103 |
| Genotype 1 | Genotype 2 | LRM Value | Putative Parents of Genotype 1 | |
|---|---|---|---|---|
| First Candidate | Second Candidate | |||
| P1_A | P1_B | 0.50 | - | - |
| P3_Sud | 0.36 | - | - | |
| P11 | 0.25 | - | - | |
| P1_B | P3_Sud | 0.36 | - | - |
| P11 | 0.25 | - | - | |
| P2 | - | - | Alb_Kalinjot_Oval | - |
| P6 | Ciciariello | 0.25 | IT_Bianca | GRE_Leucocarpa |
| P7 | Pendolino | 0.30 | IT_Pendolino | - |
| P8 | - | - | IT_Framichele | - |
| P11 | P3_Sud | 0.34 | - | - |
| P12 | P2 | 0.29 | - | - |
| P3_Nord | 0.28 | - | - | |
| Leccino_REF | 0.27 | - | - | |
| P13 | P14 | 0.48 | IT_Leccino_REF | Alb_Kalinjot_Oval |
| P3_Sud | 0.30 | |||
| Alb Kalinjot oval | 0.27 | |||
| P14 | P3_Sud | 0.30 | IT_Leccino_Lazio | - |
| P3_Nord | 0.27 | |||
| P7 | 0.27 | |||
| P15 | Taggiasca | 0.28 | - | - |
| P16 | P3_Nord | 0.29 | ALG_Aayrouni | - |
| Ascolana Tenera | 0.26 | |||
| P17 | Cipressino | 0.29 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carlucci, M.; Savoia, M.A.; Lucchese, P.G.; Fanelli, V.; Mascio, I.; Aurelio, F.L.; Miazzi, M.M.; Pacifico, A.; Montemurro, C.; Nigro, F. Behavior of Olive Genotypes Against Quick Decline Syndrome (QDS) Caused by Xylella fastidiosa subsp. pauca in Apulia. Plants 2025, 14, 157. https://doi.org/10.3390/plants14020157
Carlucci M, Savoia MA, Lucchese PG, Fanelli V, Mascio I, Aurelio FL, Miazzi MM, Pacifico A, Montemurro C, Nigro F. Behavior of Olive Genotypes Against Quick Decline Syndrome (QDS) Caused by Xylella fastidiosa subsp. pauca in Apulia. Plants. 2025; 14(2):157. https://doi.org/10.3390/plants14020157
Chicago/Turabian StyleCarlucci, Mariangela, Michele Antonio Savoia, Pompea Gabriella Lucchese, Valentina Fanelli, Isabella Mascio, Francesco Luigi Aurelio, Monica Marilena Miazzi, Andrea Pacifico, Cinzia Montemurro, and Franco Nigro. 2025. "Behavior of Olive Genotypes Against Quick Decline Syndrome (QDS) Caused by Xylella fastidiosa subsp. pauca in Apulia" Plants 14, no. 2: 157. https://doi.org/10.3390/plants14020157
APA StyleCarlucci, M., Savoia, M. A., Lucchese, P. G., Fanelli, V., Mascio, I., Aurelio, F. L., Miazzi, M. M., Pacifico, A., Montemurro, C., & Nigro, F. (2025). Behavior of Olive Genotypes Against Quick Decline Syndrome (QDS) Caused by Xylella fastidiosa subsp. pauca in Apulia. Plants, 14(2), 157. https://doi.org/10.3390/plants14020157












