Classification of Calcium-Dependent Protein Kinases and Their Transcriptional Response to Abiotic Stresses in Halophyte Nitraria sibirica
Abstract
1. Introduction
2. Results
2.1. Identification and Identity Analysis of N. sibirica CDPK Family Members
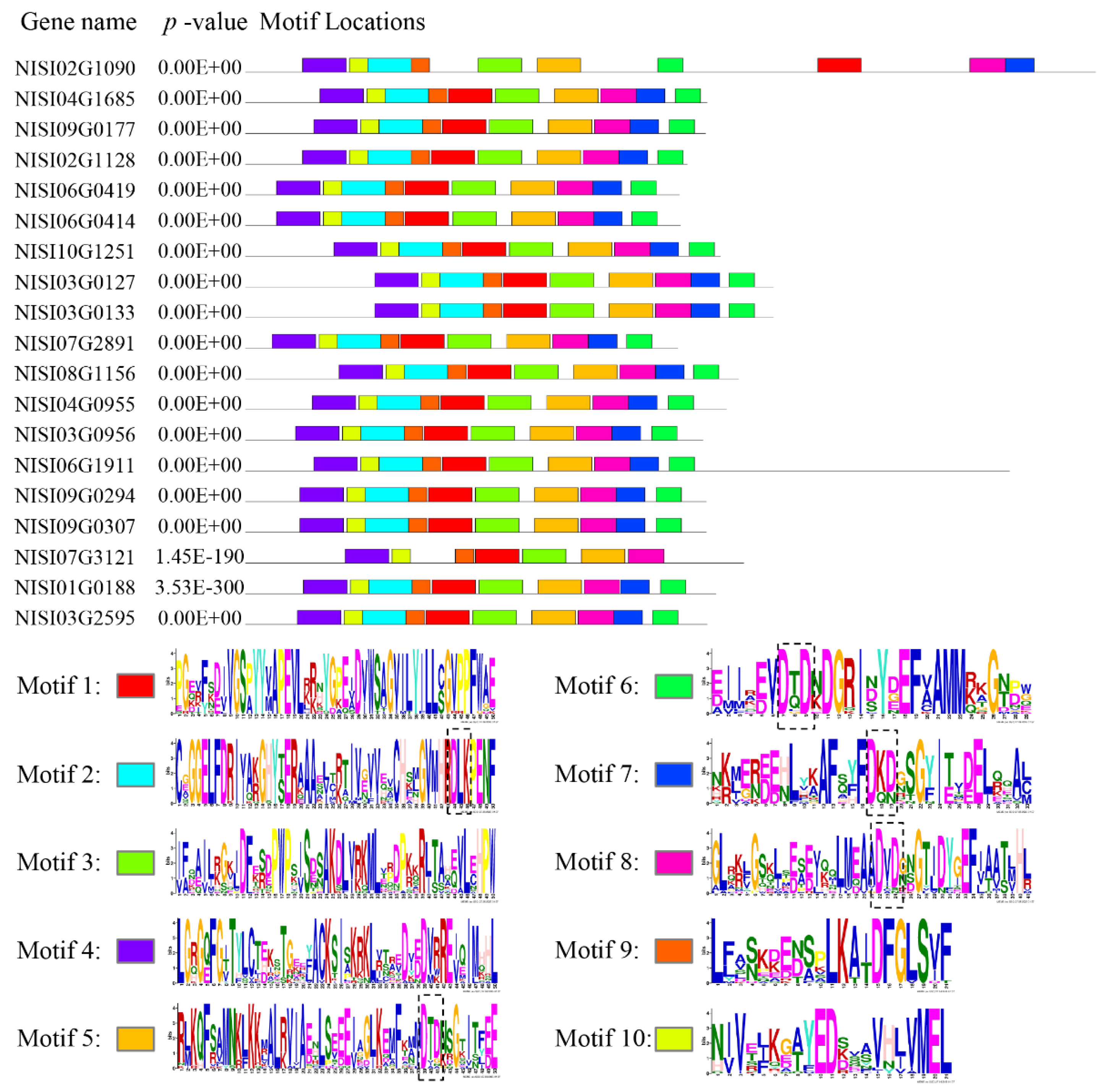
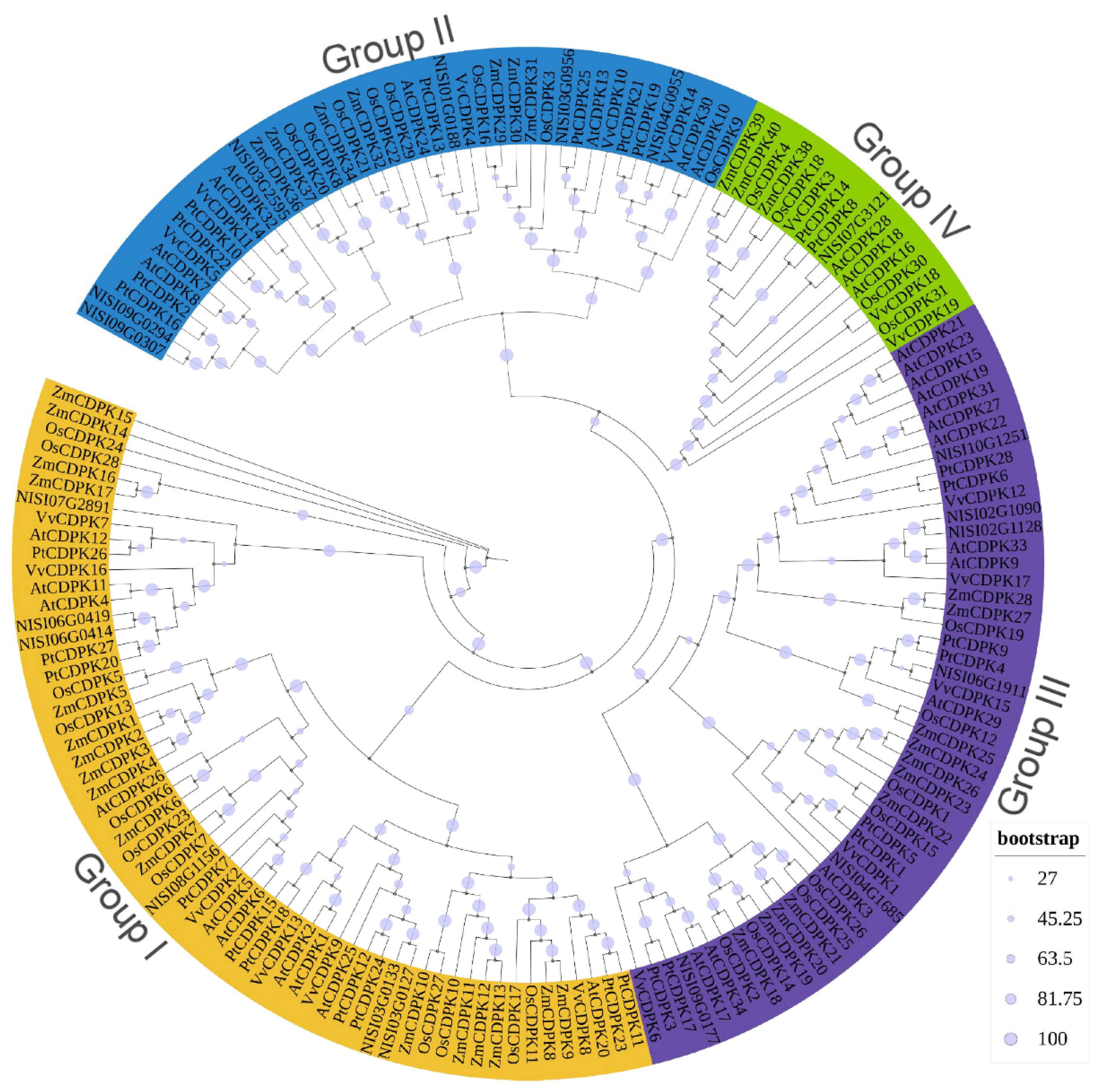
2.2. Conserved Motifs Analysis and Phylogenetic Study for NsCDPK Proteins
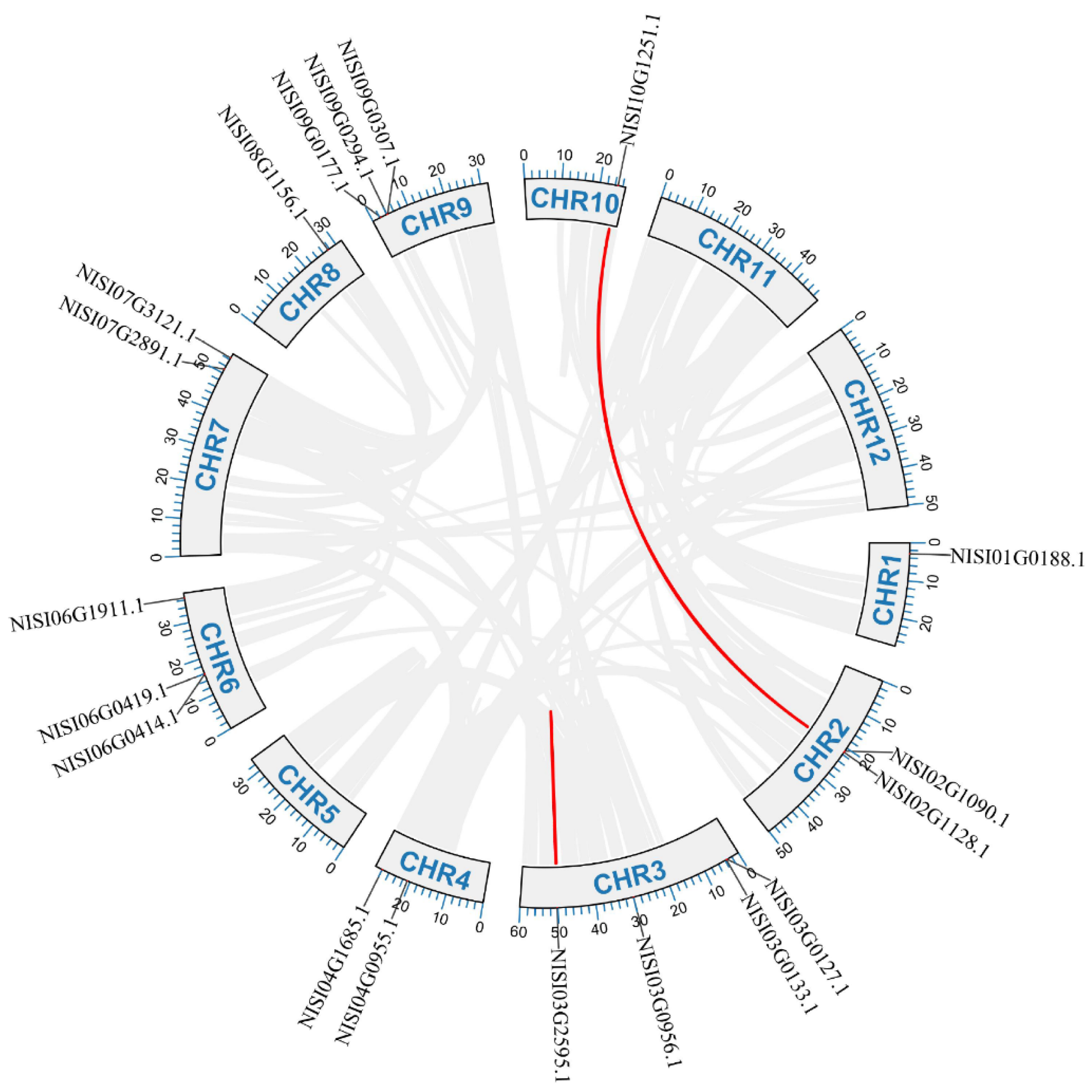
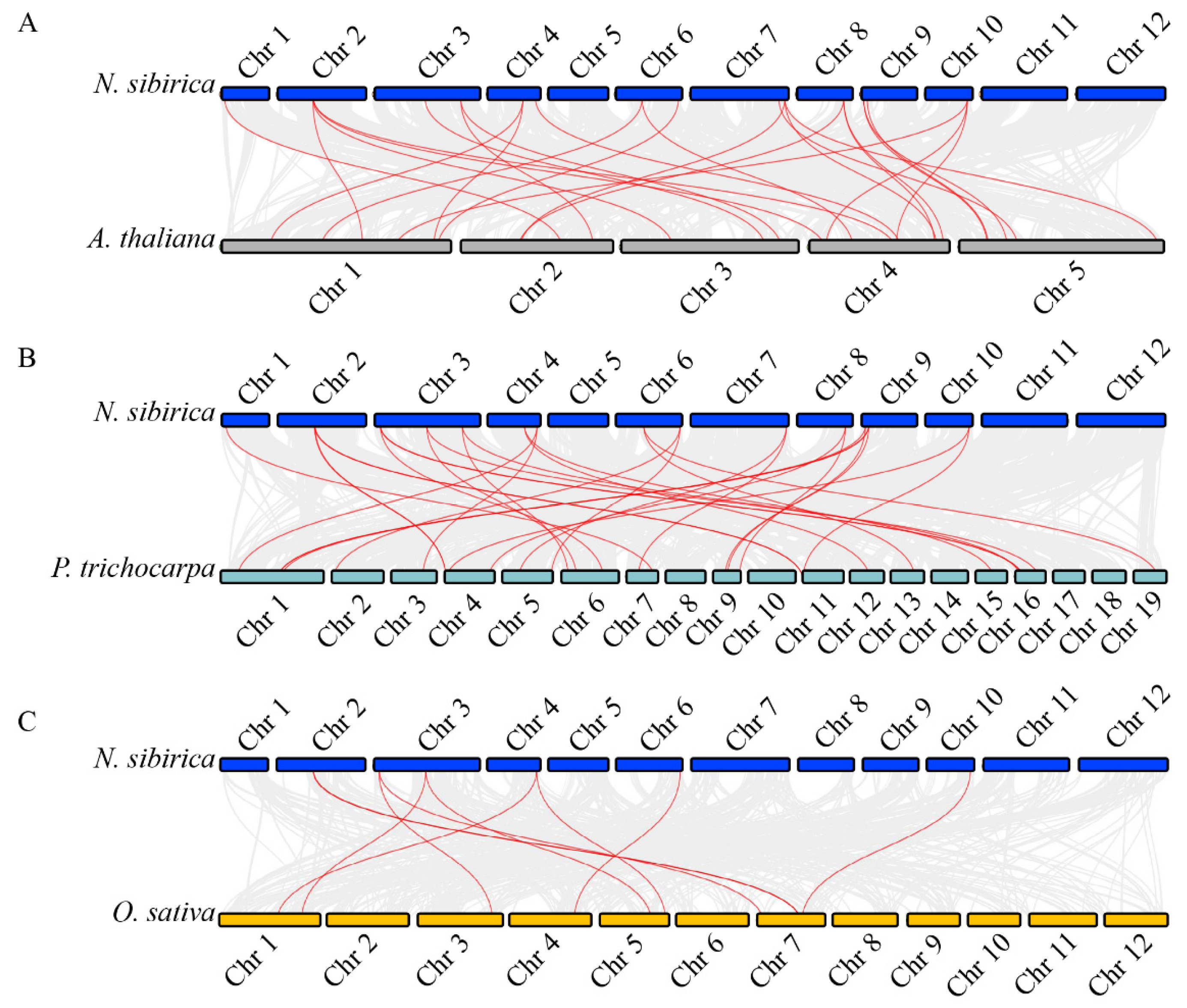
2.3. Chromosomal Location and Synteny Analysis of NsCDPKs
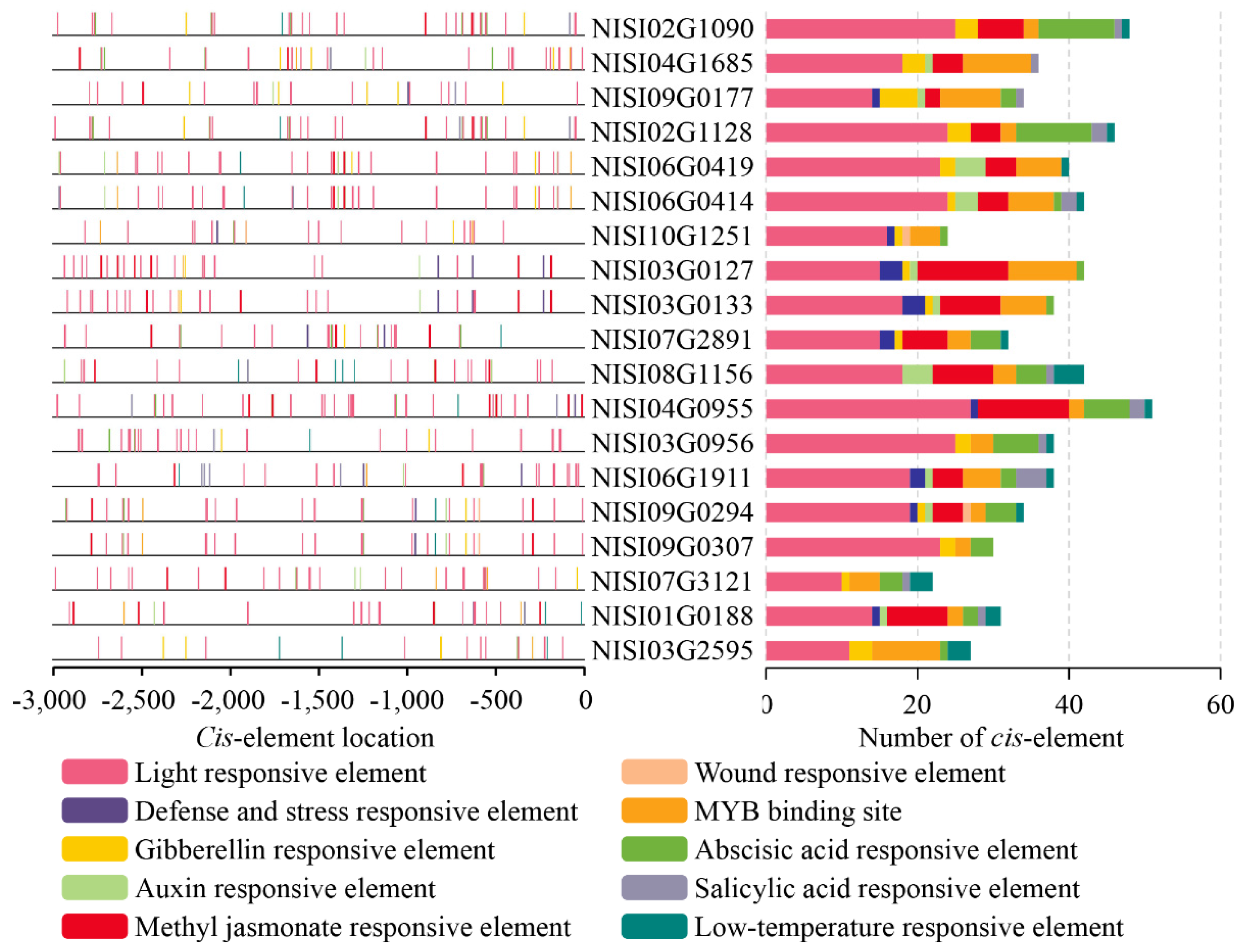
2.4. Cis-Acting Element Analysis of NsCDPKs Promoters
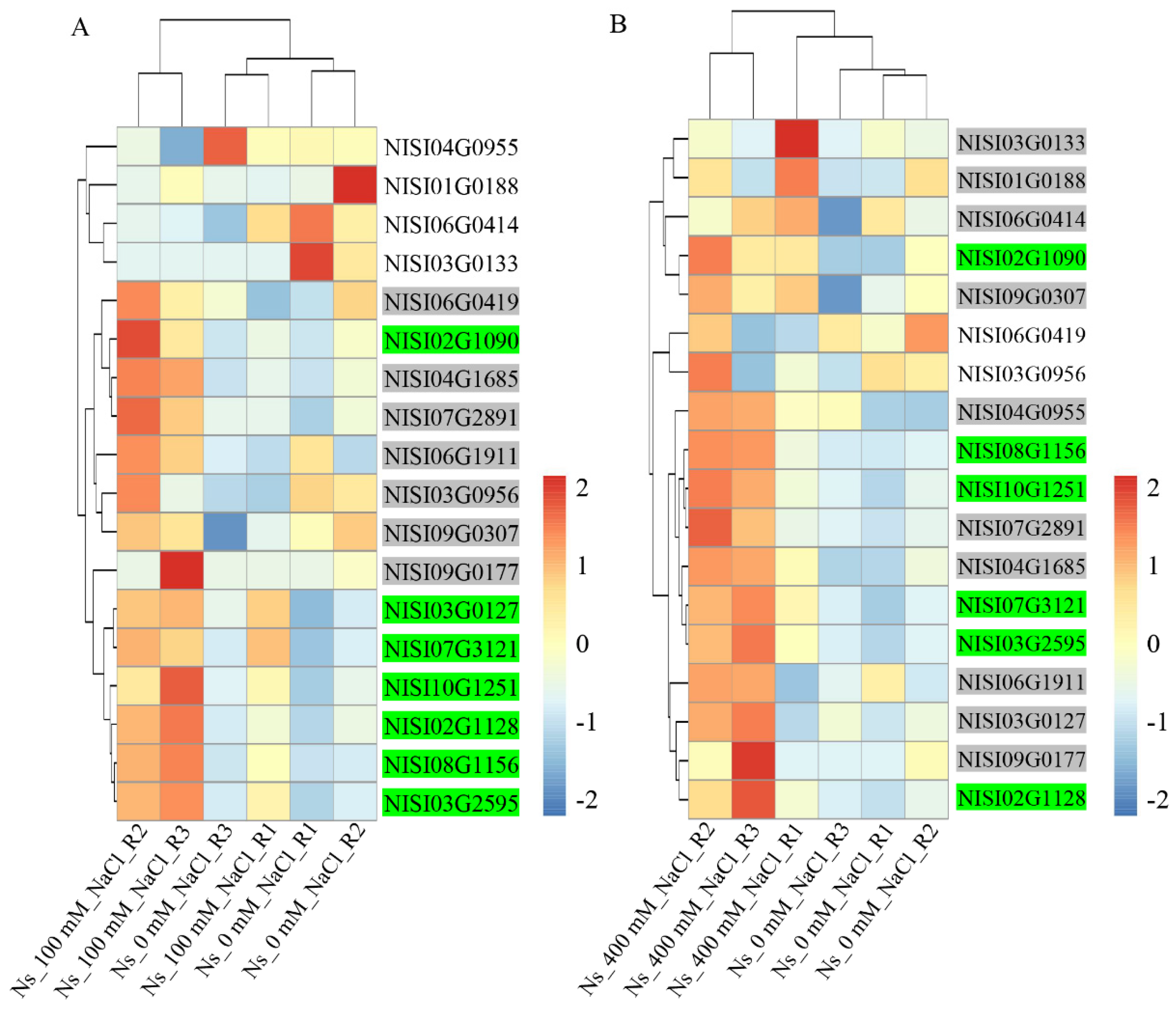
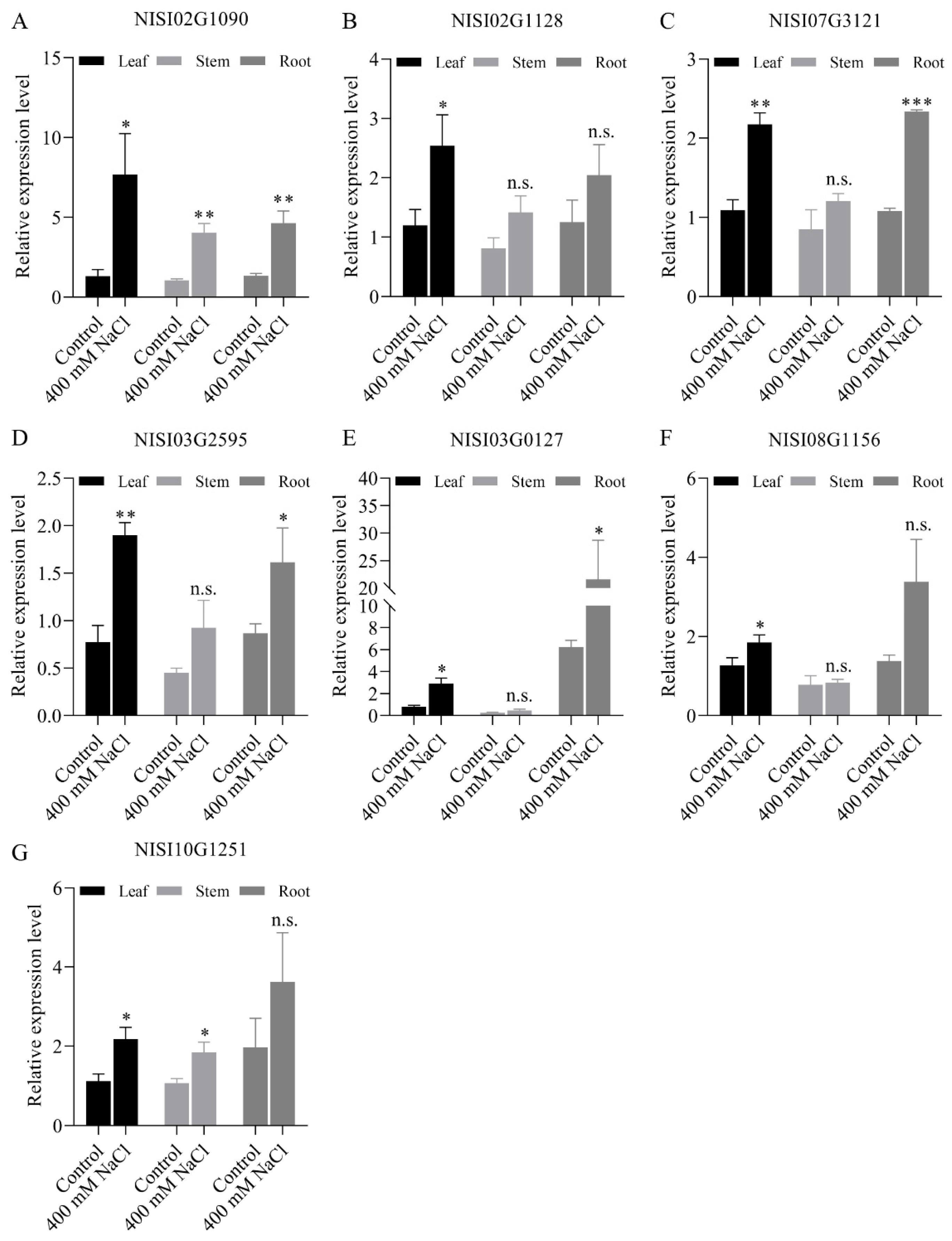
2.5. NsCDPKs Positively Respond to Salt Treatment
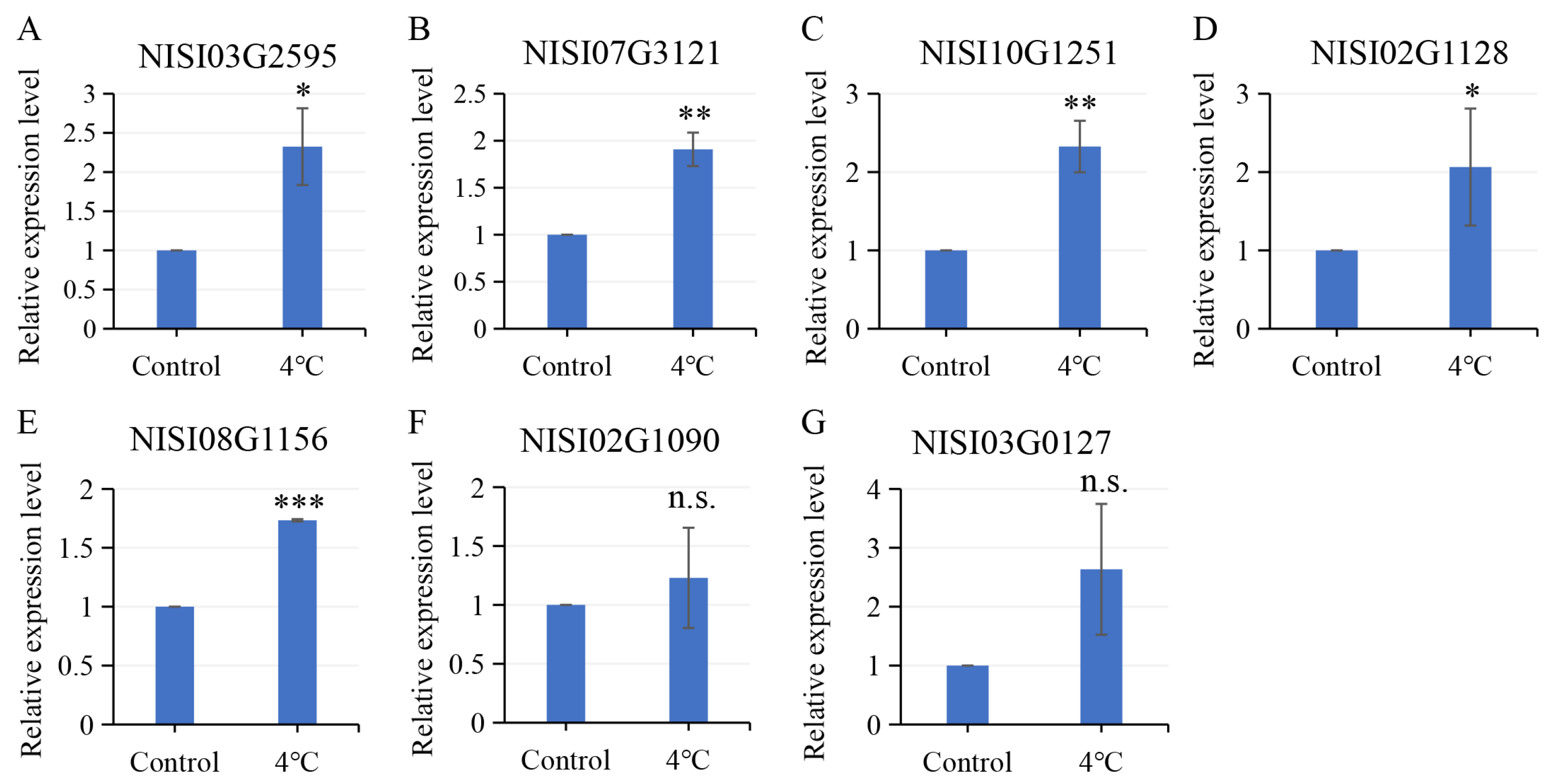
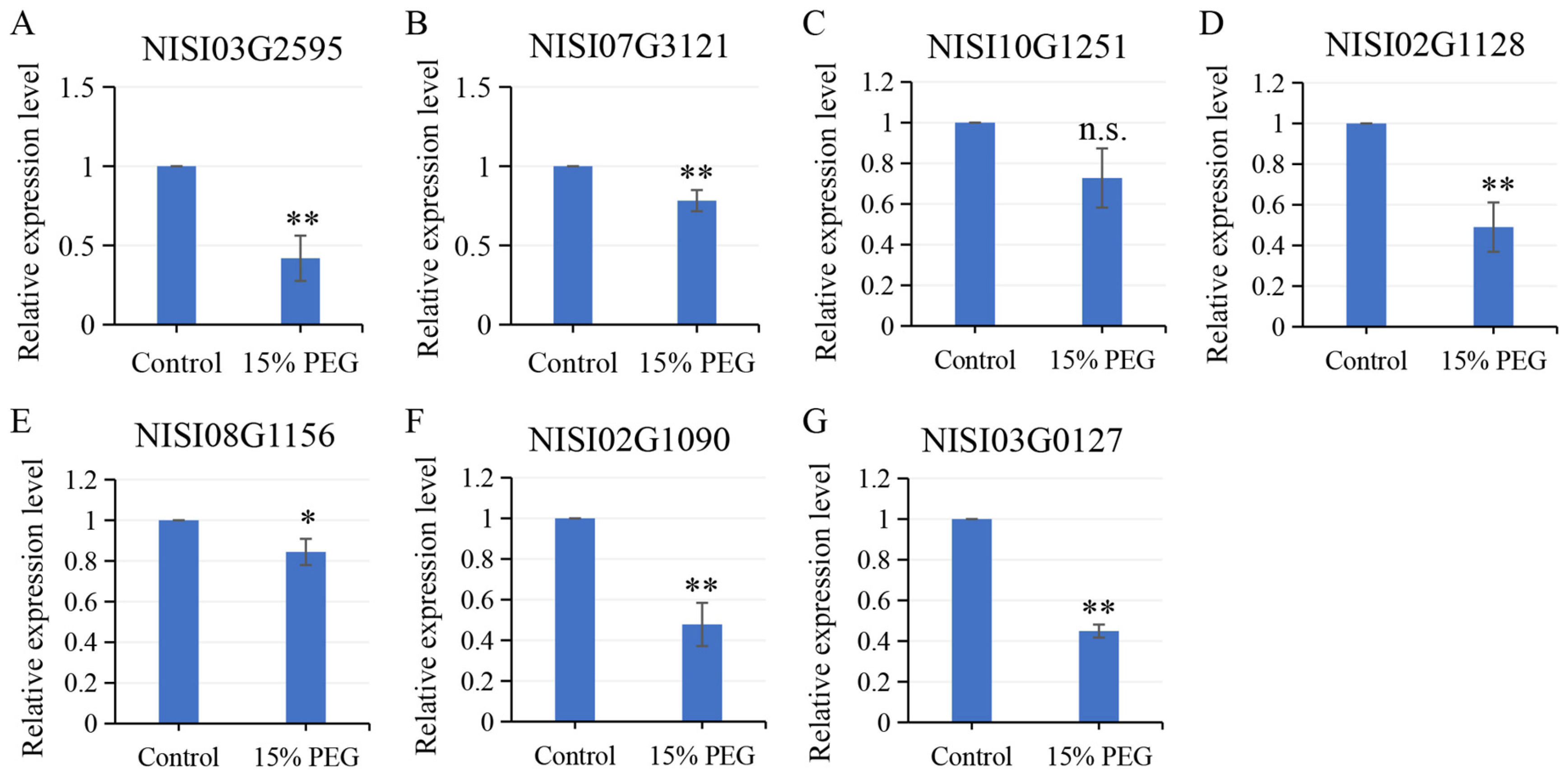
2.6. Salt-Responsive NsCDPKs Altered Expression in Response to Cold and Drought Treatment
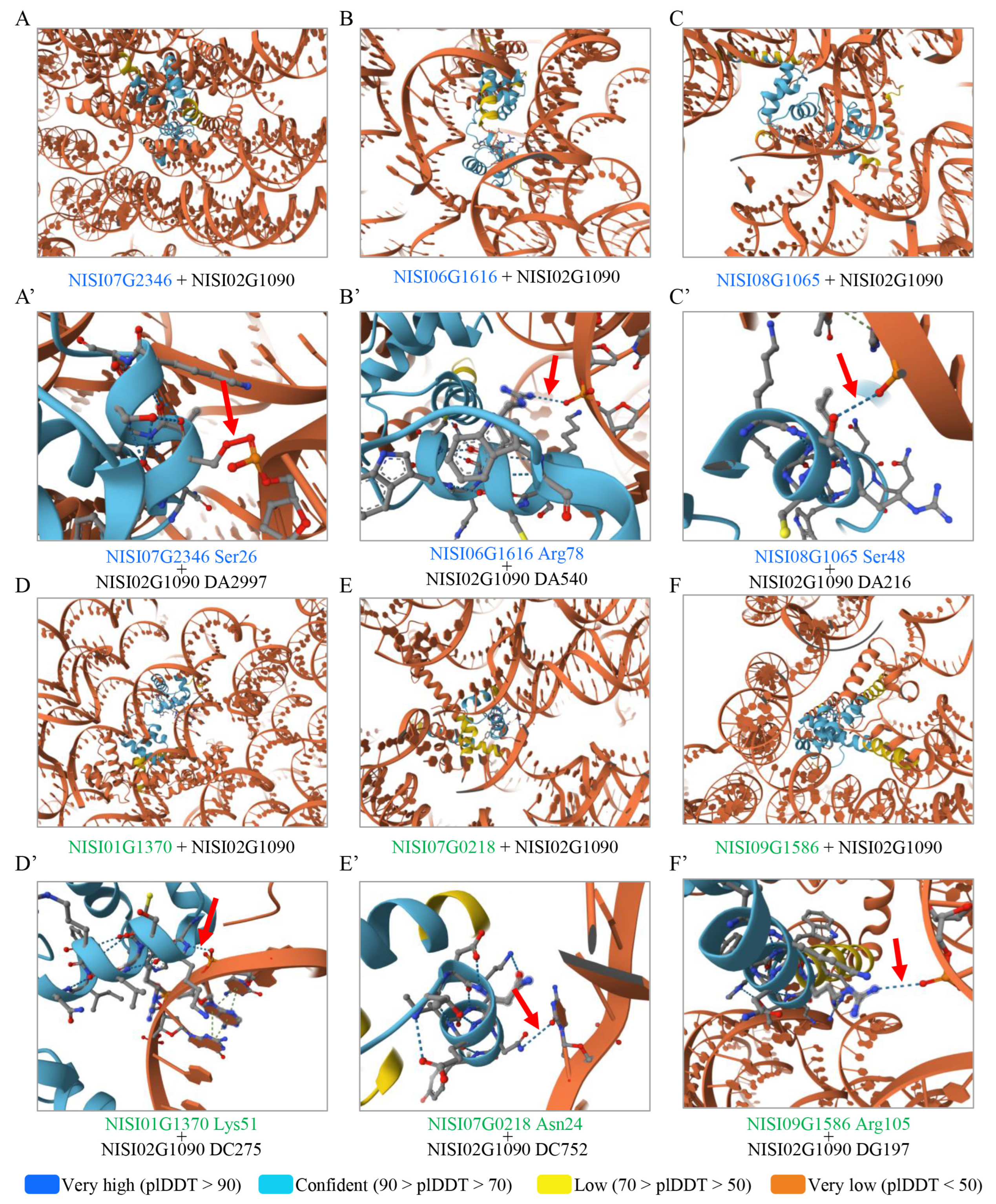
2.7. Identification of Candidate Transcription Factors Regulating NISI02G1090 Transcription
3. Discussion
3.1. NsCDPKs Responded to Various Abiotic Stresses in N. sibirica
3.2. The Prediction of Potential Upstream TF Regulating NsCDPKs
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Identification of N. sibirica CDPK Family Members
4.3. Basic Information of NsCDPK Proteins
4.4. Phylogenetic Study for NsCDPK Proteins
4.5. Collinearity Analysis of CDPKs Between N. sibirica and Other Plants
4.6. Cis-Acting Element Analysis of NsCDPK Promoters
4.7. Transcriptome Data Analysis
4.8. Quantitative Real-Time PCR Analyses
4.9. The Prediction for the Binding of NsMYB to NsCDPK Promoter
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef]
- Hepler, P.K. Calcium: A central regulator of plant growth and development. Plant Cell 2005, 17, 2142–2155. [Google Scholar] [CrossRef]
- Sanders, D.; Brownlee, C.; Harper, J.F. Communicating with calcium. Plant Cell 1999, 11, 691–706. [Google Scholar] [CrossRef]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-H.; Willmann, M.R.; Chen, H.-C.; Sheen, J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef] [PubMed]
- Trewavas, A. Le calcium, c’est la vie: Calcium makes waves. Plant Physiol. 1999, 120, 1–6. [Google Scholar] [CrossRef]
- Cui, M.; Gupta, S.K.; Bauer, P. Role of the plant-specific calcium-binding C2-DOMAIN ABSCISIC ACID-RELATED (CAR) protein family in environmental signaling. Eur. J. Cell Biol. 2023, 102, 151322. [Google Scholar] [CrossRef]
- Zhang, H.; Zeng, Y.; Seo, J.; Kim, Y.J.; Kim, S.T.; Kwon, S.W. Global Identification and Characterization of C2 Domain-Containing Proteins Associated with Abiotic Stress Response in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 2221. [Google Scholar] [CrossRef]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Bouché, N.; Yellin, A.; Snedden, W.A.; Fromm, H. Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 2005, 56, 435–466. [Google Scholar] [CrossRef]
- Hoeflich, K.P.; Ikura, M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 2002, 108, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Luan, S. The CBL–CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Wang, C. Calcium signaling mechanisms across kingdoms. Annu. Rev. Cell Dev. Biol. 2021, 37, 311–340. [Google Scholar] [CrossRef] [PubMed]
- McCormack, E.; Tsai, Y.-C.; Braam, J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005, 10, 383–389. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, H.; Wu, S.; Fu, D.; Li, M.; Gong, Z.; Yang, S. CPK28-NLP7 module integrates cold-induced Ca2+ signal and transcriptional reprogramming in Arabidopsis. Sci. Adv. 2022, 8, eabn7901. [Google Scholar] [CrossRef]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 2013, 1833, 1582–1589. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhao, J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 2647–2668. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Agarwal, P.; Arora, R.; Kapoor, S.; Tyagi, A.K. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol. Genet. Genom. 2007, 278, 493–505. [Google Scholar] [CrossRef]
- Li, A.L.; Zhu, Y.F.; Tan, X.M.; Wang, X.; Wei, B.; Guo, H.Z.; Zhang, Z.L.; Chen, X.B.; Zhao, G.Y.; Kong, X.Y.; et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 66, 429–443. [Google Scholar] [CrossRef]
- Liu, W.; Li, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS ONE 2014, 9, e98189. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.; Lu, L.; He, M.; Qu, W.; Xu, Q.; Qi, X.; Chen, X. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 2015, 290, 1403–1414. [Google Scholar] [CrossRef]
- Hu, Z.; Lv, X.; Xia, X.; Zhou, J.; Shi, K.; Yu, J.; Zhou, Y. Genome-Wide Identification and Expression Analysis of Calcium-dependent Protein Kinase in Tomato. Front. Plant Sci. 2016, 7, 469. [Google Scholar] [CrossRef]
- Alves, H.L.; Matiolli, C.C.; Soares, R.C.; Almadanim, M.C.; Oliveira, M.M.; Abreu, I.A. Carbon/nitrogen metabolism and stress response networks–calcium-dependent protein kinases as the missing link? J. Exp. Bot. 2021, 72, 4190–4201. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef]
- Goher, F.; Bai, X.; Liu, S.; Pu, L.; Xi, J.; Lei, J.; Kang, Z.; Jin, Q.; Guo, J. The calcium-dependent protein kinase TaCDPK7 positively regulates wheat resistance to Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2024, 25, 1048. [Google Scholar] [CrossRef]
- Sun, L.; Qin, J.; Wu, X.; Zhang, J.; Zhang, J. TOUCH 3 and CALMODULIN 1/4/6 cooperate with calcium-dependent protein kinases to trigger calcium-dependent activation of CAM-BINDING PROTEIN 60-LIKE G and regulate fungal resistance in plants. Plant Cell 2022, 34, 4088–4104. [Google Scholar] [CrossRef]
- Zhao, L.; Cassan-Wang, H.; Zhao, Y.; Bao, Y.; Hou, Y.; Liu, Y.; Wu, Z.; Bouzayen, M.; Zheng, Y.; Jin, P. Calcium-dependent protein kinase PpCDPK29-mediated Ca2+-ROS signal and PpHSFA2a phosphorylation regulate postharvest chilling tolerance of peach fruit. Plant Biotechnol. J. 2025, 23, 1938–1953. [Google Scholar] [CrossRef]
- Huang, J.; Yang, X.; Wang, M.M.; Tang, H.J.; Ding, L.Y.; Shen, Y.; Zhang, H.S. A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim. Biophys. Acta 2007, 1769, 220–227. [Google Scholar] [CrossRef]
- Yu, W.-W.; Chen, Q.-F.; Liao, K.; Zhou, D.-M.; Yang, Y.-C.; He, M.; Yu, L.-J.; Guo, D.-Y.; Xiao, S.; Xie, R.-H. The calcium-dependent protein kinase CPK16 regulates hypoxia-induced ROS production by phosphorylating the NADPH oxidase RBOHD in Arabidopsis. Plant Cell 2024, 36, 3451–3466. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Chai, M.; Li, L.; Dong, Z.; Jin, H.; Tan, M.; Ye, Z.; Yu, S.; Feng, Z. Calcium-Dependent Protein Kinase GhCDPK16 Exerts a Positive Regulatory Role in Enhancing Drought Tolerance in Cotton. Int. J. Mol. Sci. 2024, 25, 8308. [Google Scholar] [CrossRef] [PubMed]
- Bundó, M.; Coca, M. Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J. Exp. Bot. 2017, 68, 2963–2975. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Tanaka, N.; Yang, G.; Hayashi, N.; Komatsu, S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 2005, 46, 356–366. [Google Scholar] [CrossRef]
- Zhu, H.; He, M.; Jahan, M.S.; Wu, J.; Gu, Q.; Shu, S.; Sun, J.; Guo, S. CsCDPK6, a CsSAMS1-interacting protein, affects polyamine/ethylene biosynthesis in cucumber and enhances salt tolerance by overexpression in tobacco. Int. J. Mol. Sci. 2021, 22, 11133. [Google Scholar] [CrossRef]
- Ma, W.; Berkowitz, G.A. The grateful dead: Calcium and cell death in plant innate immunity. Cell. Microbiol. 2007, 9, 2571–2585. [Google Scholar] [CrossRef] [PubMed]
- Giammaria, V.; Grandellis, C.; Bachmann, S.; Gargantini, P.R.; Feingold, S.E.; Bryan, G.; Ulloa, R.M. StCDPK2 expression and activity reveal a highly responsive potato calcium-dependent protein kinase involved in light signalling. Planta 2011, 233, 593–609. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Yu, T.F.; Zhao, W.Y.; Fu, J.D.; Liu, Y.W.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; Xi, Y.J. Genome-Wide Analysis of CDPK Family in Foxtail Millet and Determination of SiCDPK24 Functions in Drought Stress. Front. Plant Sci. 2018, 9, 651. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity Tolerance in Halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Yuan, F.; Leng, B.; Wang, B. Progress in Studying Salt Secretion from the Salt Glands in Recretohalophytes: How Do Plants Secrete Salt? Front. Plant Sci. 2016, 7, 977. [Google Scholar] [CrossRef]
- Lu, L.; Wang, Y.; Chen, Y.; Zhu, L.; Wu, X.; Shi, J.; Chen, J.; Cheng, T. Salt stimulates carbon fixation in the halophyte Nitraria sibirica to enhance growth. For. Res. 2025, 5, e004. [Google Scholar] [CrossRef]
- Li, H.; Tang, X.; Zhu, J.; Yang, X.; Zhang, H. De Novo Transcriptome Characterization, Gene Expression Profiling and Ionic Responses of Nitraria sibirica Pall. under Salt Stress. Forests 2017, 8, 211. [Google Scholar] [CrossRef]
- Wu, X.; Zhu, J.; Zhu, L.; Tang, Y.; Hao, Z.; Zhang, J.; Shi, J.; Cheng, T.; Lu, L. Genome-wide analyses of calmodulin and calmodulin-like proteins in the halophyte Nitraria sibirica reveal their involvement in response to salinity, drought and cold stress. Int. J. Biol. Macromol. 2023, 253, 127442. [Google Scholar] [CrossRef] [PubMed]
- Gamage, D.G.; Gunaratne, A.; Periyannan, G.R.; Russell, T.G. Applicability of Instability Index for In vitro Protein Stability Prediction. Protein Pept. Lett. 2019, 26, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Pati, P.K.; Pati, A.M.; Nagpal, A.K. Physico-chemical characterization and topological analysis of pathogenesis-related proteins from Arabidopsis thaliana and Oryza sativa using in-silico approaches. PLoS ONE 2020, 15, e0239836. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, H.; Tang, X.; Yang, X.; Zhang, H. Comprehensive transcriptome and metabolome profiling reveal metabolic mechanisms of Nitraria sibirica Pall. to salt stress. Sci. Rep. 2021, 11, 12878. [Google Scholar] [CrossRef]
- Yue, J.Y.; Jiao, J.L.; Wang, W.W.; Wang, H.Z. The Calcium-Dependent Protein Kinase TaCDPK27 Positively Regulates Salt Tolerance in Wheat. Int. J. Mol. Sci. 2022, 23, 7341. [Google Scholar] [CrossRef]
- Li, Y.N.; Lei, C.; Yang, Q.; Yu, X.; Li, S.; Sun, Y.; Ji, C.; Zhang, C.; Xue, J.A.; Cui, H.; et al. Identification and expression analysis of calcium-dependent protein kinase family in oat (Avena sativa L.) and their functions in response to saline-alkali stresses. Front. Plant Sci. 2024, 15, 1395696. [Google Scholar] [CrossRef]
- Su, S.; Jiang, Y.; Zhu, X.; Yu, S.; Wang, F.; Xue, L.; Cui, H. Calcium-dependent protein kinases 5 and 13 enhance salt tolerance in rice by directly activating OsMPK3/6 kinases. Plant Physiol. 2024, 196, 3033–3047. [Google Scholar] [CrossRef]
- Li, J.; Ishii, T.; Yoshioka, M.; Hino, Y.; Nomoto, M.; Tada, Y.; Yoshioka, H.; Takahashi, H.; Yamauchi, T.; Nakazono, M. CDPK5 and CDPK13 play key roles in acclimation to low oxygen through the control of RBOH-mediated ROS production in rice. Plant Physiol. 2024, 197, kiae293. [Google Scholar] [CrossRef]
- Marques, J.; Matiolli, C.C.; Abreu, I.A. Visualization of a curated Oryza sativa L. CDPKs Protein-Protein Interaction Network (CDPK-OsPPIN). Micropublication Biol. 2022, 2022, 10-17912. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Liang, D.; Wang, P.; Liu, J.; Ma, F. Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol. Genet. Genom. 2012, 287, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Wang, L.; Li, F.; Zhang, W.; Chen, G.; Lin, X. Isolation and characterization of Nitraria sibirica actin gene. Acta Prataculturae Sin. 2012, 21, 151–158. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Yuan, Q.; Chen, S.; Rao, J.; Zheng, S.; Zhao, H.; Yang, Y. AlphaFold2-aware protein–DNA binding site prediction using graph transformer. Brief. Bioinform. 2022, 23, bbab564. [Google Scholar] [CrossRef]



| Gene ID | AA | MW (kDa) | pI | II | AI | GRAVY |
|---|---|---|---|---|---|---|
| NISI02G1090 | 979 | 110.78 | 5.90 | 32.41 | 83.09 | −0.356 |
| NISI04G1685 | 532 | 59.77 | 6.02 | 37.06 | 77.74 | −0.474 |
| NISI09G0177 | 530 | 58.77 | 5.72 | 37.15 | 76.75 | −0.493 |
| NISI02G1128 | 509 | 57.04 | 6.18 | 31.45 | 81.45 | −0.412 |
| NISI06G0419 | 500 | 56.20 | 5.51 | 39.71 | 88.14 | −0.296 |
| NISI06G0414 | 501 | 56.23 | 5.51 | 39.80 | 88.34 | −0.296 |
| NISI10G1251 | 547 | 61.59 | 6.35 | 42.80 | 74.41 | −0.559 |
| NISI03G0127 | 608 | 68.41 | 5.68 | 44.52 | 78.87 | −0.460 |
| NISI03G0133 | 608 | 68.41 | 5.68 | 44.52 | 78.87 | −0.460 |
| NISI07G2891 | 498 | 56.00 | 5.35 | 39.43 | 83.39 | −0.374 |
| NISI08G1156 | 568 | 63.71 | 5.72 | 41.85 | 80.18 | −0.448 |
| NISI04G0955 | 554 | 63.00 | 6.99 | 39.95 | 85.00 | −0.459 |
| NISI03G0956 | 527 | 59.53 | 6.03 | 37.86 | 83.23 | −0.449 |
| NISI06G1911 | 880 | 98.07 | 6.86 | 42.00 | 89.07 | −0.311 |
| NISI09G0294 | 531 | 59.66 | 6.40 | 32.95 | 81.90 | −0.470 |
| NISI09G0307 | 531 | 59.66 | 6.40 | 32.95 | 81.90 | −0.470 |
| NISI07G3121 | 574 | 65.28 | 9.25 | 42.30 | 76.11 | −0.695 |
| NISI01G0188 | 542 | 61.95 | 5.89 | 38.26 | 79.67 | −0.491 |
| NISI03G2595 | 532 | 60.62 | 6.78 | 36.20 | 81.35 | −0.536 |
| Gene ID | Alpha Helix% | Extended Strand% | Beta Turn% | Random Coil% |
|---|---|---|---|---|
| NISI02G1090 | 48.31 | 11.85 | 9.70 | 30.13 |
| NISI04G1685 | 41.35 | 9.40 | 7.89 | 41.35 |
| NISI09G0177 | 43.02 | 9.43 | 8.30 | 39.25 |
| NISI02G1128 | 44.60 | 10.02 | 9.23 | 36.15 |
| NISI06G0419 | 47.00 | 10.60 | 9.40 | 33.00 |
| NISI06G0414 | 46.91 | 10.18 | 8.78 | 34.13 |
| NISI10G1251 | 40.77 | 8.96 | 8.59 | 41.68 |
| NISI03G0127 | 40.30 | 9.54 | 7.57 | 42.60 |
| NISI03G0133 | 40.30 | 9.54 | 7.57 | 42.60 |
| NISI07G2891 | 45.58 | 13.25 | 9.44 | 31.73 |
| NISI08G1156 | 40.14 | 10.56 | 8.10 | 41.20 |
| NISI04G0955 | 43.50 | 10.83 | 8.66 | 37.00 |
| NISI03G0956 | 47.63 | 10.63 | 8.35 | 33.40 |
| NISI06G1911 | 36.25 | 15.45 | 8.98 | 39.32 |
| NISI09G0294 | 45.76 | 10.17 | 8.85 | 35.22 |
| NISI09G0307 | 45.76 | 10.17 | 8.85 | 35.22 |
| NISI07G3121 | 40.42 | 12.20 | 7.14 | 40.24 |
| NISI01G0188 | 43.54 | 10.89 | 8.86 | 36.72 |
| NISI03G2595 | 47.74 | 10.53 | 8.83 | 32.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Chen, T.; Yang, T.; Han, C.; Zhang, J.; Chen, J.; Cheng, T. Classification of Calcium-Dependent Protein Kinases and Their Transcriptional Response to Abiotic Stresses in Halophyte Nitraria sibirica. Plants 2025, 14, 3091. https://doi.org/10.3390/plants14193091
Lu L, Chen T, Yang T, Han C, Zhang J, Chen J, Cheng T. Classification of Calcium-Dependent Protein Kinases and Their Transcriptional Response to Abiotic Stresses in Halophyte Nitraria sibirica. Plants. 2025; 14(19):3091. https://doi.org/10.3390/plants14193091
Chicago/Turabian StyleLu, Lu, Ting Chen, Tiangui Yang, Chunxia Han, Jingbo Zhang, Jinhui Chen, and Tielong Cheng. 2025. "Classification of Calcium-Dependent Protein Kinases and Their Transcriptional Response to Abiotic Stresses in Halophyte Nitraria sibirica" Plants 14, no. 19: 3091. https://doi.org/10.3390/plants14193091
APA StyleLu, L., Chen, T., Yang, T., Han, C., Zhang, J., Chen, J., & Cheng, T. (2025). Classification of Calcium-Dependent Protein Kinases and Their Transcriptional Response to Abiotic Stresses in Halophyte Nitraria sibirica. Plants, 14(19), 3091. https://doi.org/10.3390/plants14193091






