Genome-Wide Identification and Expression Analysis of the SPL Gene Family in Phalaenopsis equestris
Abstract
1. Introduction
2. Results
2.1. Identification of PeqSPL Family Members
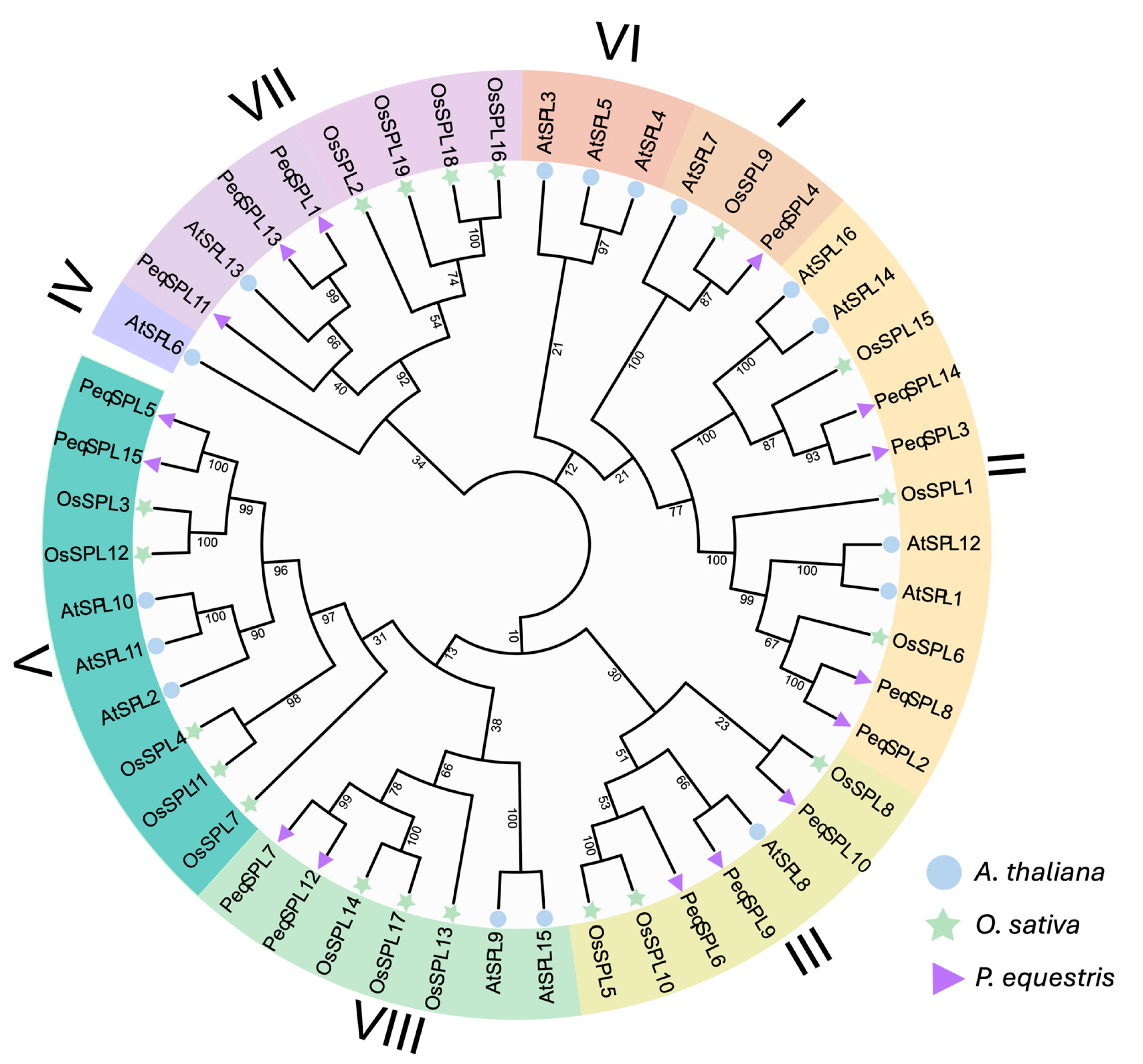
2.2. Phylogenetic Relationships and Evolutionary Divergence of PeqSPL Genes
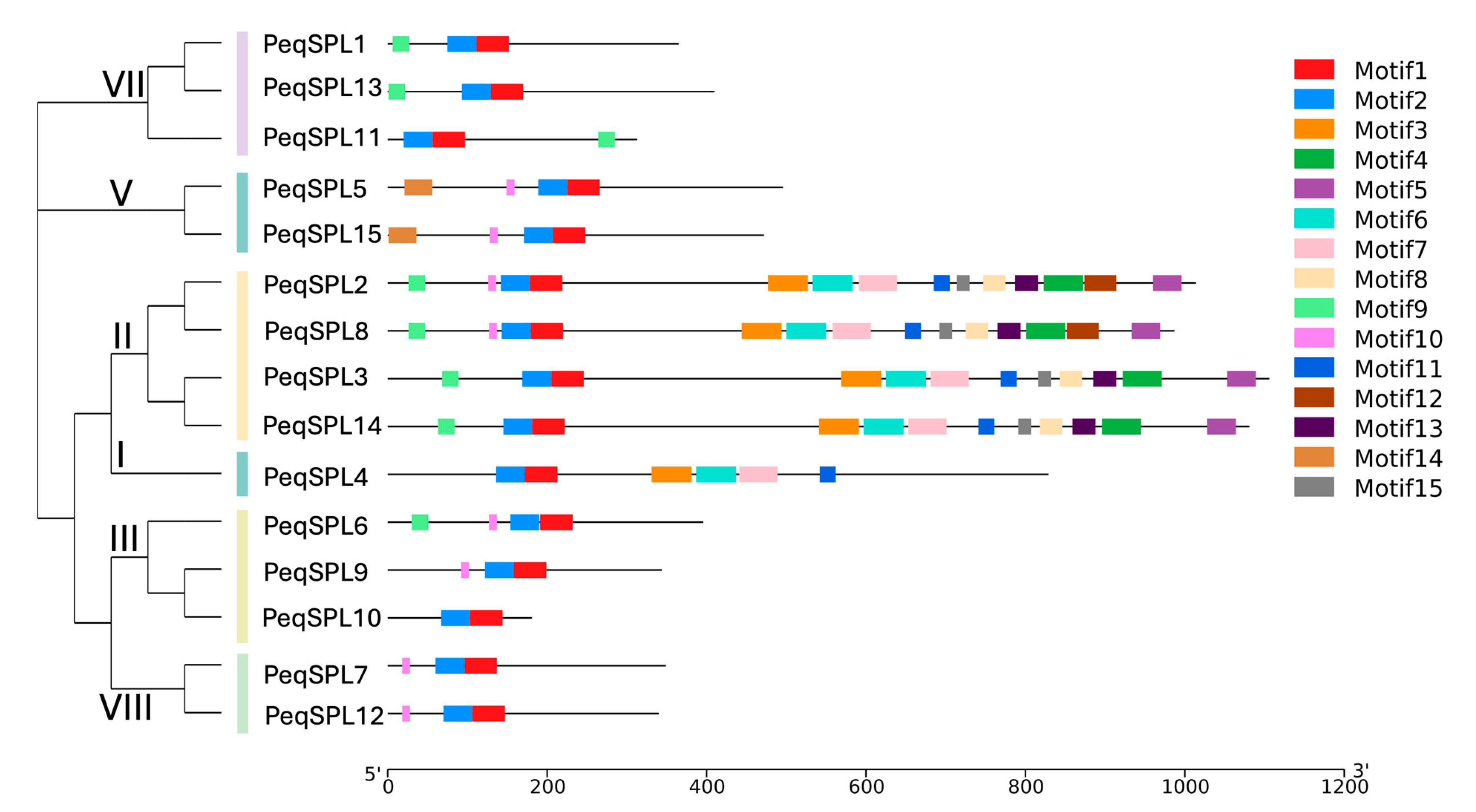
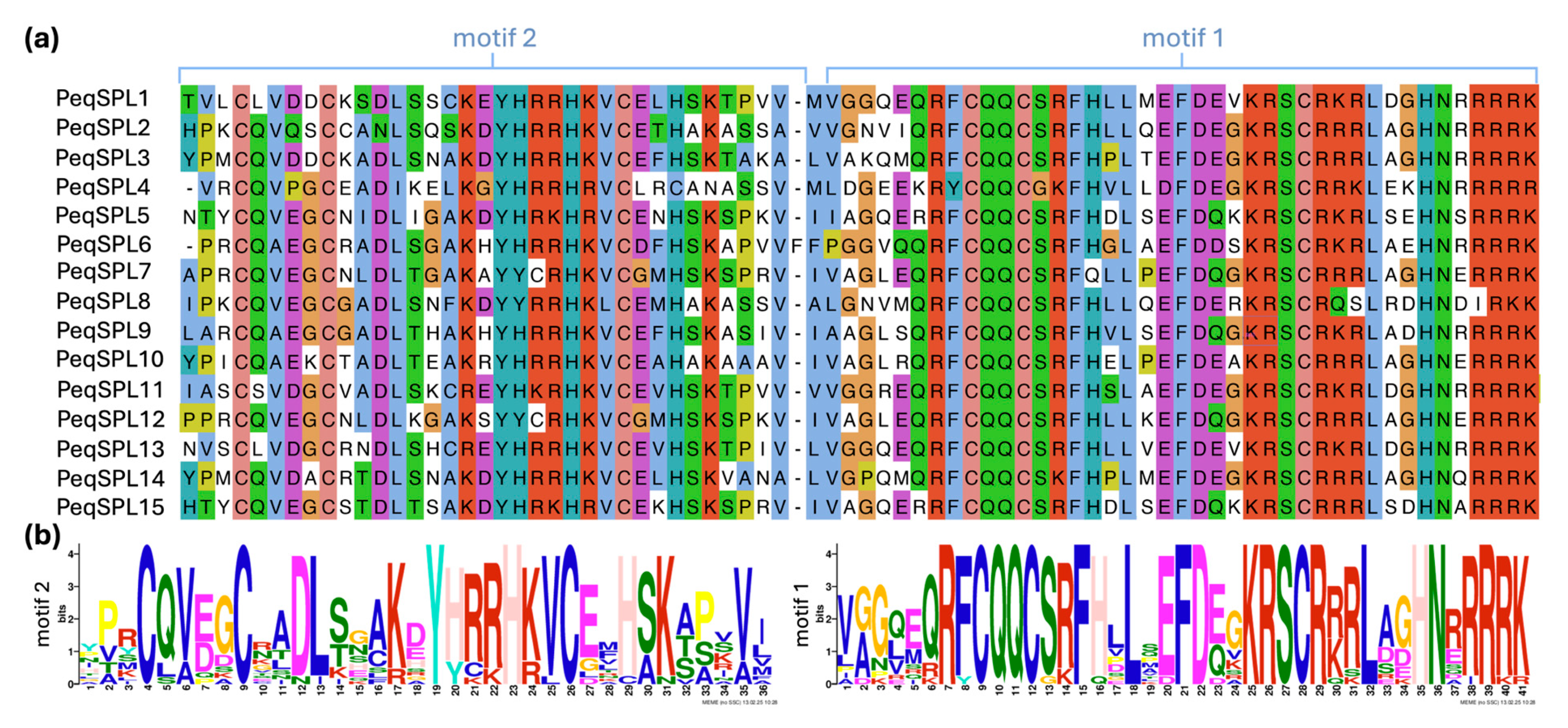
2.3. Protein Conservative Domain and Functional Motifs Analysis
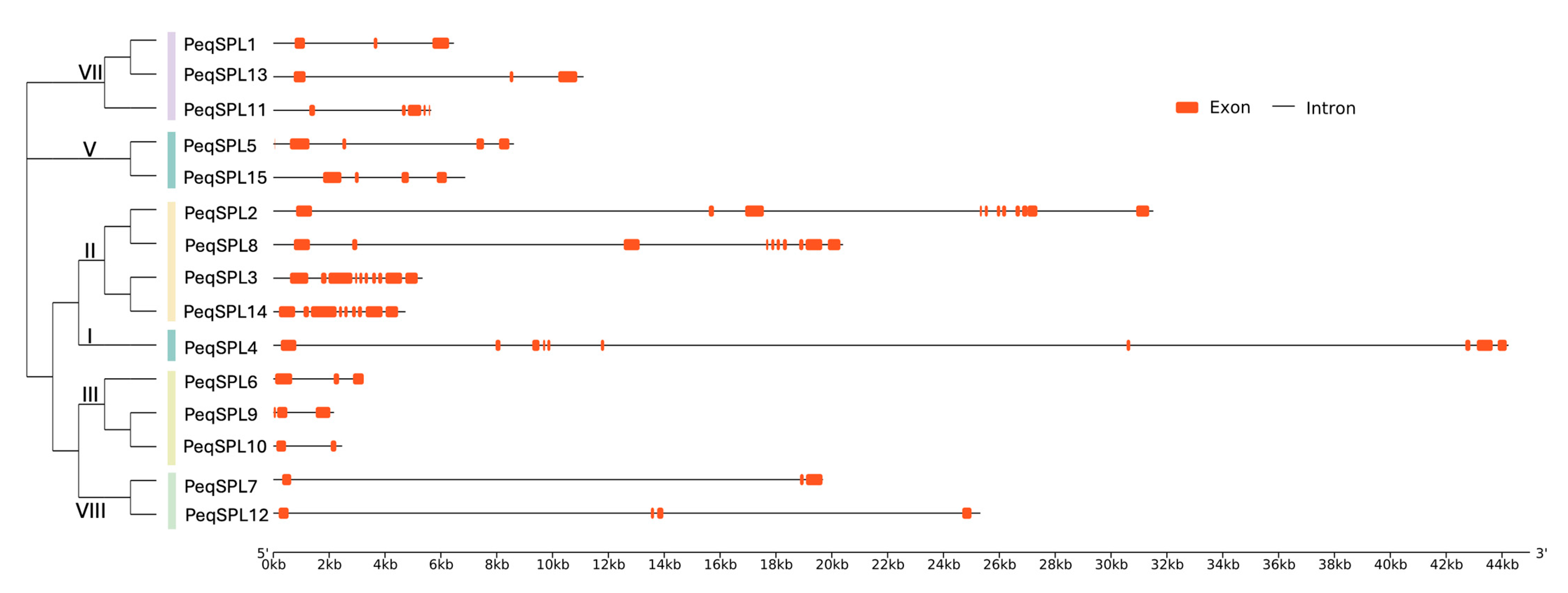
2.4. Gene Structure Variation Among PeqSPL Family Members
2.5. Collinearity and Synteny Analysis of PeqSPL Genes
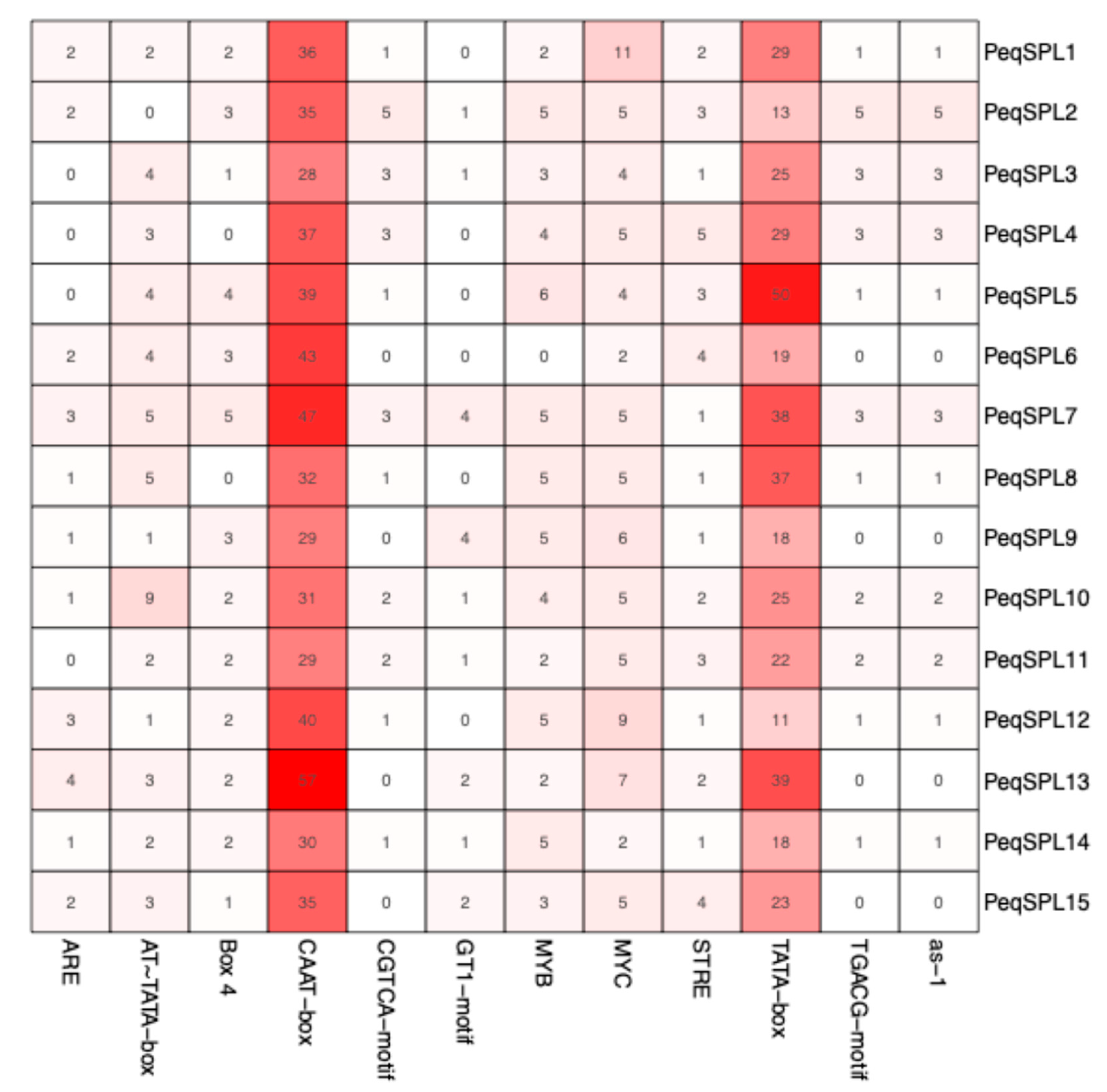
2.6. Cis-Element Analysis Reveals Regulatory Potential of PeqSPL Promoters
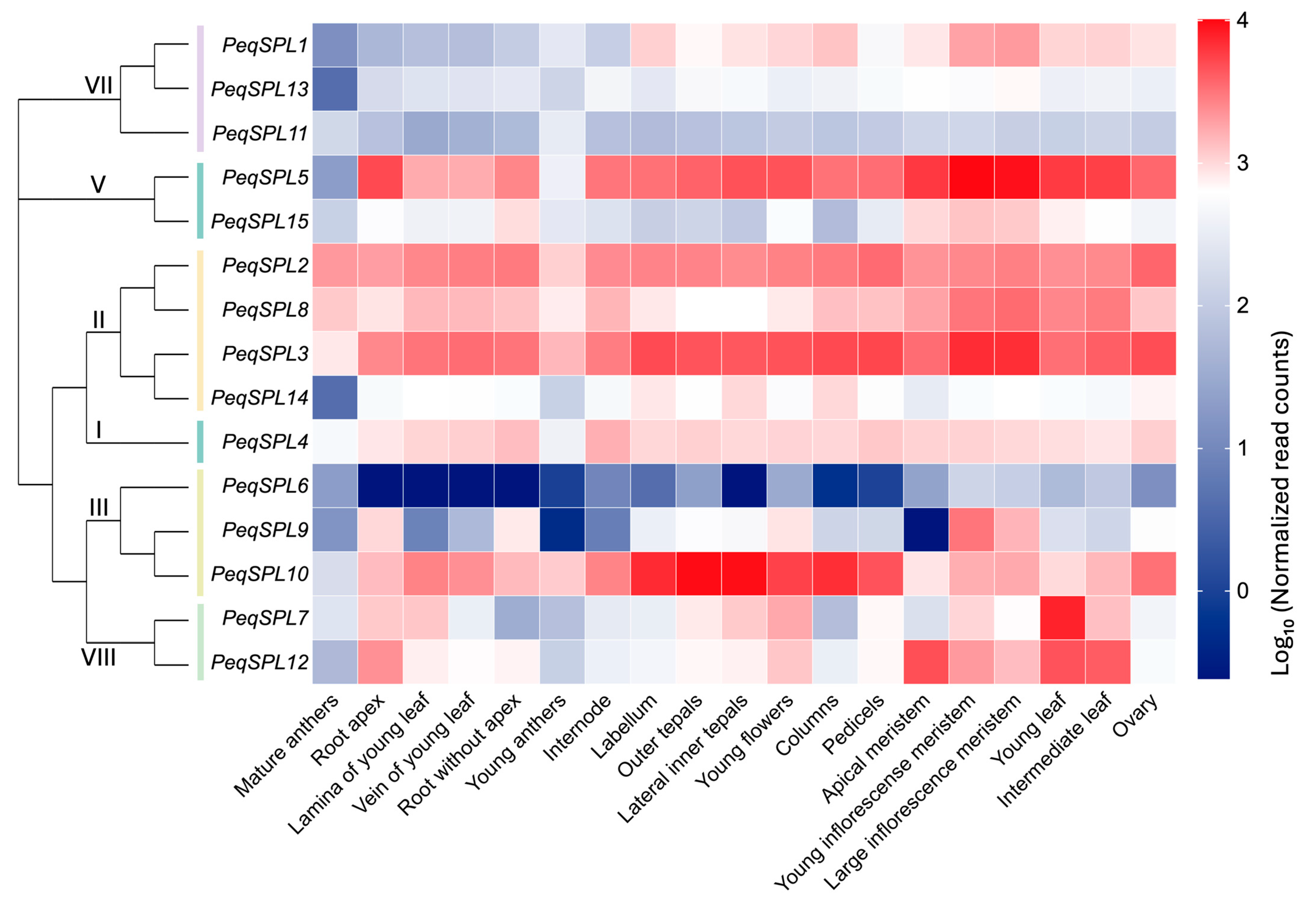
2.7. Tissue-Specific Expression Patterns of PeqSPLs
3. Discussion
4. Materials and Methods
4.1. Whole-Genome Identification of SPL Gene in P. equestris
4.2. Physicochemical Properties of the SPLs
4.3. Phylogenetic Analysis
4.4. Protein Domain and Gene Structure Analysis
4.5. Collinearity and Location Analysis on Chromosome
4.6. Cis-Acting Regulatory Elements Analysis
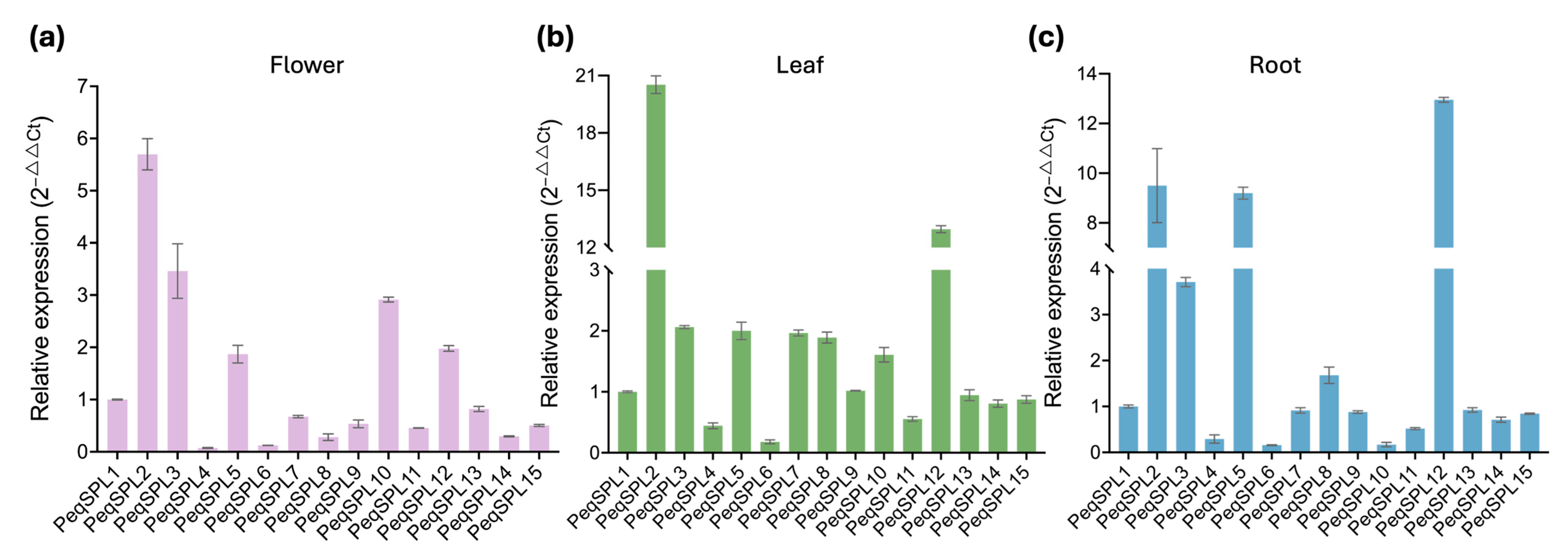
4.7. Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, A.Y.; Zhu, Q.H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-Wide Identification and Evolutionary Analysis of the Plant Specific Sbp-Box Transcription Factor Family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional Dissection of the Plant-Specific Sbp-Domain: Overlap of the DNA-Binding and Nuclear Localization Domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Ming, M.; Yi, M.; Qin, C.; Yan, L.; Sun, Y.; Zhang, J.; Cao, F.; Fu, F. Genome-Wide Identification of Ginkgo biloba SPL Gene Family and Expression Analysis in Flavonoid Biosynthesis and Water Stress. Int. J. Mol. Sci. 2025, 26, 4932. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Saedler, H.; Huijser, P. A New Family of DNA Binding Proteins Includes Putative Transcriptional Regulators of The Antirrhinum majus Floral Meristem Identity Gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [PubMed]
- Li, Y.; Deng, Y.; Qin, D.; An, X. Study of the SPL Gene Family and Mir156-SPL Module in Populus Tomentosa: Potential Roles in Juvenile-to-Adult Phase Transition and Reproductive Phase. Int. J. Biol. Macromol. 2025, 296, 139547. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The Mir156/SPL Module Regulates Apple Salt Stress Tolerance by Activating Mdwrky100 Expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef]
- Chao, L.M.; Liu, Y.Q.; Chen, D.Y.; Xue, X.Y.; Mao, Y.B.; Chen, X.Y. Arabidopsis Transcription Factors SPL1 and SPL12 Confer Plant Thermotolerance at Reproductive Stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef]
- Fan, E.; Liu, C.; Wang, Z.; Wang, S.; Ma, W.; Lu, N.; Liu, Y.; Fu, P.; Wang, R.; Lv, S.; et al. Genome-Wide Identification and Expression Analysis of the Squamosa Promoter-Binding Protein-Like (SPL) Transcription Factor Family in Catalpa bungei. Int. J. Mol. Sci. 2023, 25, 97. [Google Scholar] [CrossRef]
- He, A.; Zhou, H.; Ma, C.; Bai, Q.; Yang, H.; Yao, X.; Wu, W.; Xue, G.; Ruan, J. Genome-Wide Identification and Expression Analysis of the SPL Gene Family and Its Response to Abiotic Stress in Barley (Hordeum vulgare L.). BMC Genom. 2024, 25, 846. [Google Scholar] [CrossRef]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL 14, a Plant-Specific Sbp-Domain Transcription Factor, Participates in Plant Development and Sensitivity to Fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-Box Gene That Affects Pollen Sac Development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef]
- Wang, J.; Ye, Y.; Xu, M.; Feng, L.; Xu, L.A. Roles of the SPL Gene Family and Mir156 in the Salt Stress Responses of Tamarisk (Tamarix chinensis). BMC Plant Biol. 2019, 19, 370. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of Mir156-Regulated Squamosa Promoter Binding Protein-Like (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. Spl8, a Local Regulator in a Subset of Gibberellin-Mediated Developmental Processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Wu, C.; Xiong, L. Genomic Organization, Differential Expression, and Interaction of Squamosa Promoter-Binding-Like Transcription Factors and Microrna156 in Rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Li, Y.; Han, S.; Sun, X.; Khan, N.U.; Zhong, Q.; Zhang, Z.; Zhang, H.; Ming, F.; Li, Z.; Li, J. Variations in Osspl10 Confer Drought Tolerance by Directly Regulating Osnac2 Expression and Ros Production in Rice. J. Integr. Plant Biol. 2023, 65, 918–933. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Q. Boosting Rice Yield by Fine-Tuning SPL Gene Expression. Trends Plant Sci. 2017, 22, 643–646. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of Osspl14 by Osmir156 Defines Ideal Plant Architecture in Rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The Osspl16-Gw7 Regulatory Module Determines Grain Shape and Simultaneously Improves Rice Yield and Grain Quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The Microrna-Regulated SBP-Box Transcription Factor SPL3 Is a Direct Upstream Activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef]
- Sun, Y.; Fu, M.; Wang, L.; Bai, Y.; Fang, X.; Wang, Q.; He, Y.; Zeng, H. Osspls Regulate Male Fertility in Response to Different Temperatures by Flavonoid Biosynthesis and Tapetum PCD in PTGMS Rice. Int. J. Mol. Sci. 2022, 23, 3744. [Google Scholar] [CrossRef]
- Yang, J.; Wei, H.; Hou, M.; Chen, L.; Zou, T.; Ding, H.; Jing, Y.; Zhang, X.; Zhao, Y.; Liu, Q.; et al. Zmspl13 and Zmspl29 Act Together to Promote Vegetative and Reproductive Transition in Maize. New Phytol. 2023, 239, 1505–1520. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Pan, X.; Jing, Y.; Zhao, Y.; Duan, Y.; Yang, J.; Wang, B.; Liu, Y.; Shen, R.; Cao, Y.; et al. Zmspl10/14/26 Are Required for Epidermal Hair Cell Fate Specification on Maize Leaf. New Phytol. 2021, 230, 1533–1549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, X.W.; Li, Y.Y.; Ke, S.J.; Yin, W.L.; Lan, S.; Liu, Z.J. Advances and Prospects of Orchid Research and Industrialization. Hortic. Res. 2022, 9, uhac220. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, M.; He, X.; Zheng, Q.; Huang, Y.; Li, Y.; Ahmad, S.; Liu, D.; Lan, S.; Liu, Z. Genome-Wide Identification and Expression Analysis of the SPL Gene Family in Three Orchids. Int. J. Mol. Sci. 2023, 24, 10039. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sun, Y.; Minne, M.; Ge, Y.; Yue, Q.; Goossens, V.; Mor, E.; Callebaut, B.; Bevernaege, K.; Winne, J.M.; et al. SPL13 Controls a Root Apical Meristem Phase Change by Triggering Oriented Cell Divisions. Science 2024, 386, eado4298. [Google Scholar] [CrossRef]
- Zhao, Y.; He, J.; Liu, M.; Miao, J.; Ma, C.; Feng, Y.; Qian, J.; Li, H.; Bi, H.; Liu, W. The SPL Transcription Factor TaSPL6 Negatively Regulates Drought Stress Response in Wheat. Plant Physiol. Biochem. 2024, 206, 108264. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Qian, Q.; Liu, Q.; Li, Q.; Pan, Y.; Ye, Y.; Liu, X.; Wang, J.; Zhang, J.; et al. Non-Canonical Regulation of SPL Transcription Factors by a Human Otub1-Like Deubiquitinase Defines a New Plant Type Rice Associated with Higher Grain Yield. Cell Res. 2017, 27, 1142–1156. [Google Scholar] [CrossRef]
- Wang, W.; Luo, L.; Shi, H.; Song, Y.; Wang, J.; Chen, C.; Shen, Z.; Rouached, H.; Zheng, L. The Transcription Factor OsSPL9 Endows Rice with Copper Deficiency Resilience. J. Exp. Bot. 2024, 75, 5909–5922. [Google Scholar] [CrossRef]
- Jiang, M.; He, Y.; Chen, X.; Zhang, X.; Guo, Y.; Yang, S.; Huang, J.; Traw, M.B. Crispr-Based Assessment of Genomic Structure in the Conserved Squamosa Promoter-Binding-Like Gene Clusters in Rice. Plant J. 2020, 104, 1301–1314. [Google Scholar] [CrossRef]
- Li, F.; Tan, Q.; Gan, Z.; Han, D.; Yang, W.; Luan, X.; Liu, J.; Zhao, H.; Fu, Y.; Wang, S.; et al. OsSPL5 Promotes Rice Outcrossing Efficiency by G-Protein Pathway. Plant Biotechnol. J. 2025, 23, 509–511. [Google Scholar] [CrossRef]
- Xing, S.; Quodt, V.; Chandler, J.; Höhmann, S.; Berndtgen, R.; Huijser, P. SPL8 Acts Together with the Brassinosteroid-Signaling Component BIM1 in Controlling Arabidopsis thaliana Male Fertility. Plants 2013, 2, 416–428. [Google Scholar] [CrossRef] [PubMed]
- He, D.Y.; Liang, Q.Y.; Xiang, C.B.; Xia, J.Q. Loss of OsSPL8 Function Confers Improved Resistance to Glufosinate and Abiotic Stresses in Rice. Plant Cell Environ. 2025, 48, 682–698. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ma, Y.; Zhang, M.; Lyu, M.; Yuan, Y.; Wu, B. Expression Pattern of FT/TFL1 and Mir156-Targeted SPL Genes Associated with Developmental Stages in Dendrobium catenatum. Int. J. Mol. Sci. 2019, 20, 2725. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Ma, R.; Fan, Y.; Zhao, B.; Cheng, P.; Fan, Y.; Wang, B. Genome-Wide Identification and Expression Analysis of the SPL Transcription Factor Family and Its Response to Abiotic Stress in Quinoa (Chenopodium quinoa). BMC Genom. 2022, 23, 773. [Google Scholar] [CrossRef]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel Positive Regulatory Role for the SPL6 Transcription Factor in the N Tir-Nb-Lrr Receptor-Mediated Plant Innate Immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef]
- Sommer, F.; Kropat, J.; Malasarn, D.; Grossoehme, N.E.; Chen, X.; Giedroc, D.P.; Merchant, S.S. The Crr1 Nutritional Copper Sensor in Chlamydomonas Contains Two Distinct Metal-Responsive Domains. Plant Cell 2010, 22, 4098–4113. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. Wolf Psort: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. Mafft Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (Itol) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The Meme Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. Tbtools-Ii: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep Neural Networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. Gsds 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Ezhova, M.A.; Penin, A.A.; Logacheva, M.D. Transcriptome Atlas of Phalaenopsis equestris. PeerJ 2021, 9, e12600. [Google Scholar] [CrossRef]
- Lin, Y.F.; Chen, Y.Y.; Hsiao, Y.Y.; Shen, C.Y.; Hsu, J.L.; Yeh, C.M.; Mitsuda, N.; Ohme-Takagi, M.; Liu, Z.J.; Tsai, W.C. Genome-Wide Identification and Characterization of TCP Genes Involved in Ovule Development of Phalaenopsis equestris. J. Exp. Bot. 2016, 67, 5051–5066. [Google Scholar] [CrossRef]

| Name | Gene ID | CDS 1 (bp) | Amino Acid 2 (No.) | MW 3 (Da) | pI 4 | II 5 | AI 6 | GRAVY 7 | Homolog 8 |
|---|---|---|---|---|---|---|---|---|---|
| PeqSPL1 | LOC110018951 | 1098 | 365 | 40729.45 | 5.88 | 57.13 | 62.27 | −0.592 | AtSPL13 [26] |
| PeqSPL2 | LOC110021553 | 3045 | 1014 | 112500.42 | 6.28 | 58.01 | 80.5 | −0.345 | OsSPL6 [27] |
| PeqSPL3 | LOC110021701 | 3321 | 1106 | 121984.69 | 7.11 | 59.63 | 74.98 | −0.438 | OsSPL15 [28] |
| PeqSPL4 | LOC110022067 | 2490 | 829 | 93005.98 | 5.8 | 48.94 | 84.09 | −0.251 | OsSPL9 [29] |
| PeqSPL5 | LOC110023454 | 1491 | 496 | 54162.83 | 8.63 | 46.46 | 57.08 | −0.661 | OsSPL3, OsSPL12 [30] |
| PeqSPL6 | LOC110024613 | 1191 | 396 | 43610.53 | 9.07 | 59.48 | 49.07 | −0.645 | OsSPL5, OsSPL10 [31] |
| PeqSPL7 | LOC110026415 | 1050 | 349 | 38391.89 | 8.99 | 50.13 | 55.7 | −0.658 | OsSPL14, OsSPL17 [18] |
| PeqSPL8 | LOC110027702 | 2964 | 987 | 110148.37 | 7.3 | 46.62 | 82.88 | −0.319 | OsSPL6 [27] |
| PeqSPL9 | LOC110030741 | 1035 | 344 | 37084.14 | 8.89 | 55.37 | 57.73 | −0.538 | AtSPL8 [32] |
| PeqSPL10 | LOC110030902 | 546 | 181 | 19991.32 | 9.28 | 66.96 | 48.73 | −0.959 | OsSPL8 [33] |
| PeqSPL11 | LOC110031164 | 942 | 313 | 34984.28 | 8.49 | 51.34 | 66.61 | −0.485 | AtSPL13 [26] |
| PeqSPL12 | LOC110033253 | 1023 | 340 | 36708.28 | 8.57 | 57.42 | 60.88 | −0.554 | OsSPL14, OsSPL17 [18] |
| PeqSPL13 | LOC110033310 | 1233 | 410 | 45150.41 | 6.13 | 57.77 | 67.29 | −0.496 | AtSPL13 [26] |
| PeqSPL14 | LOC110033499 | 3246 | 1081 | 119828.82 | 8.33 | 50.77 | 75.76 | −0.389 | OsSPL15 [28] |
| PeqSPL15 | LOC110039299 | 1419 | 472 | 52527.42 | 8.82 | 48.64 | 59.34 | −0.719 | OsSPL3, OsSPL12 [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Feng, L.; Hu, Q.; Hu, Y.; Ma, X.; Zheng, J. Genome-Wide Identification and Expression Analysis of the SPL Gene Family in Phalaenopsis equestris. Plants 2025, 14, 3090. https://doi.org/10.3390/plants14193090
Zhang X, Feng L, Hu Q, Hu Y, Ma X, Zheng J. Genome-Wide Identification and Expression Analysis of the SPL Gene Family in Phalaenopsis equestris. Plants. 2025; 14(19):3090. https://doi.org/10.3390/plants14193090
Chicago/Turabian StyleZhang, Xule, Lei Feng, Qingdi Hu, Yaping Hu, Xiaohua Ma, and Jian Zheng. 2025. "Genome-Wide Identification and Expression Analysis of the SPL Gene Family in Phalaenopsis equestris" Plants 14, no. 19: 3090. https://doi.org/10.3390/plants14193090
APA StyleZhang, X., Feng, L., Hu, Q., Hu, Y., Ma, X., & Zheng, J. (2025). Genome-Wide Identification and Expression Analysis of the SPL Gene Family in Phalaenopsis equestris. Plants, 14(19), 3090. https://doi.org/10.3390/plants14193090






