Genotyping of Commercial European Cannabis Seeds Based on Multiple Mapped Marker Loci: A Comparative Study of Drug and Hemp Varieties
Abstract
1. Introduction
2. Materials and Methods
2.1. Cannabis Plant Cultivation
2.2. Cannabis Sampling and DNA Extraction
2.3. Analysis of SSR Marker Loci
2.4. Molecular Data Analysis
3. Results and Discussion
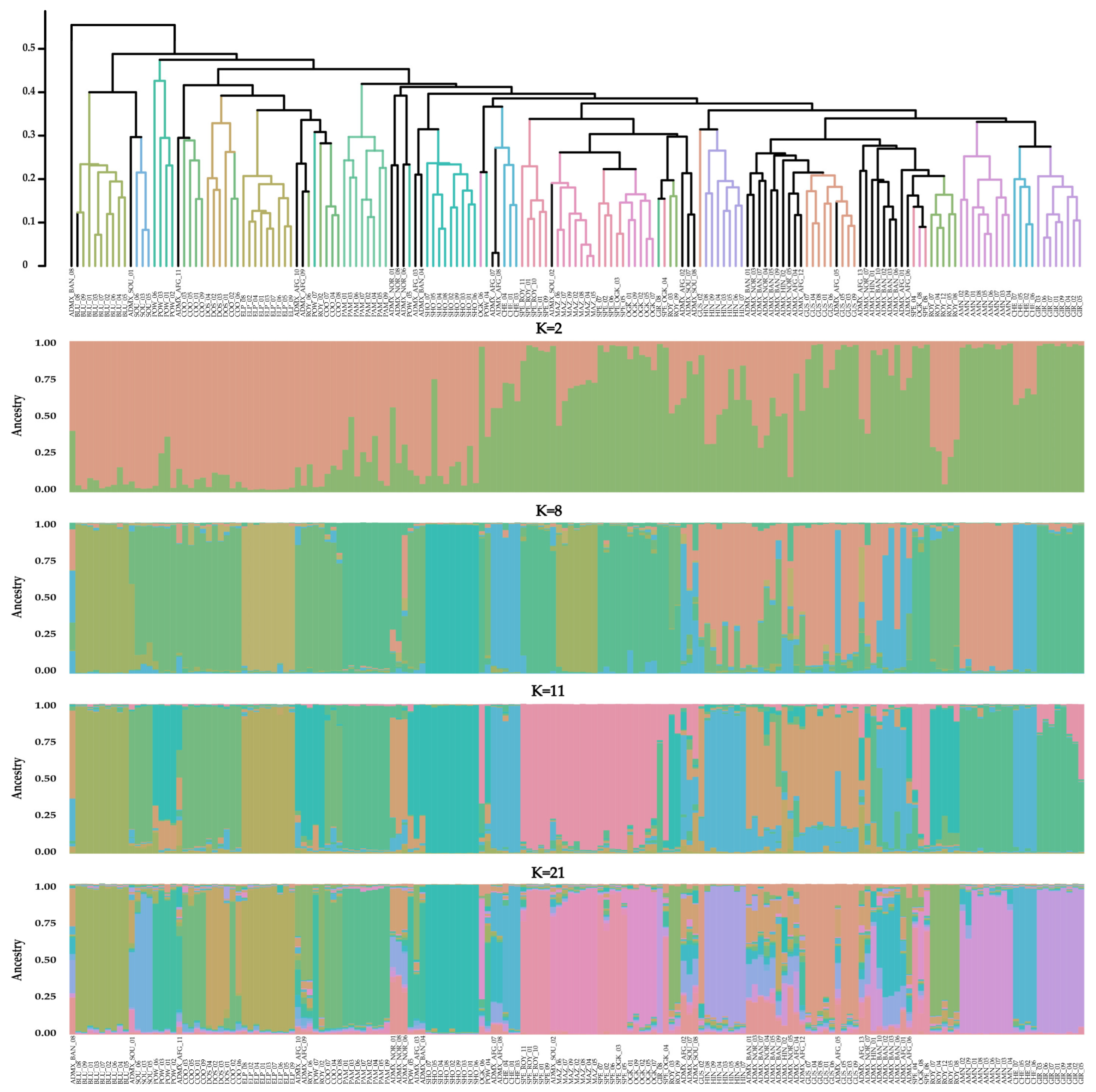
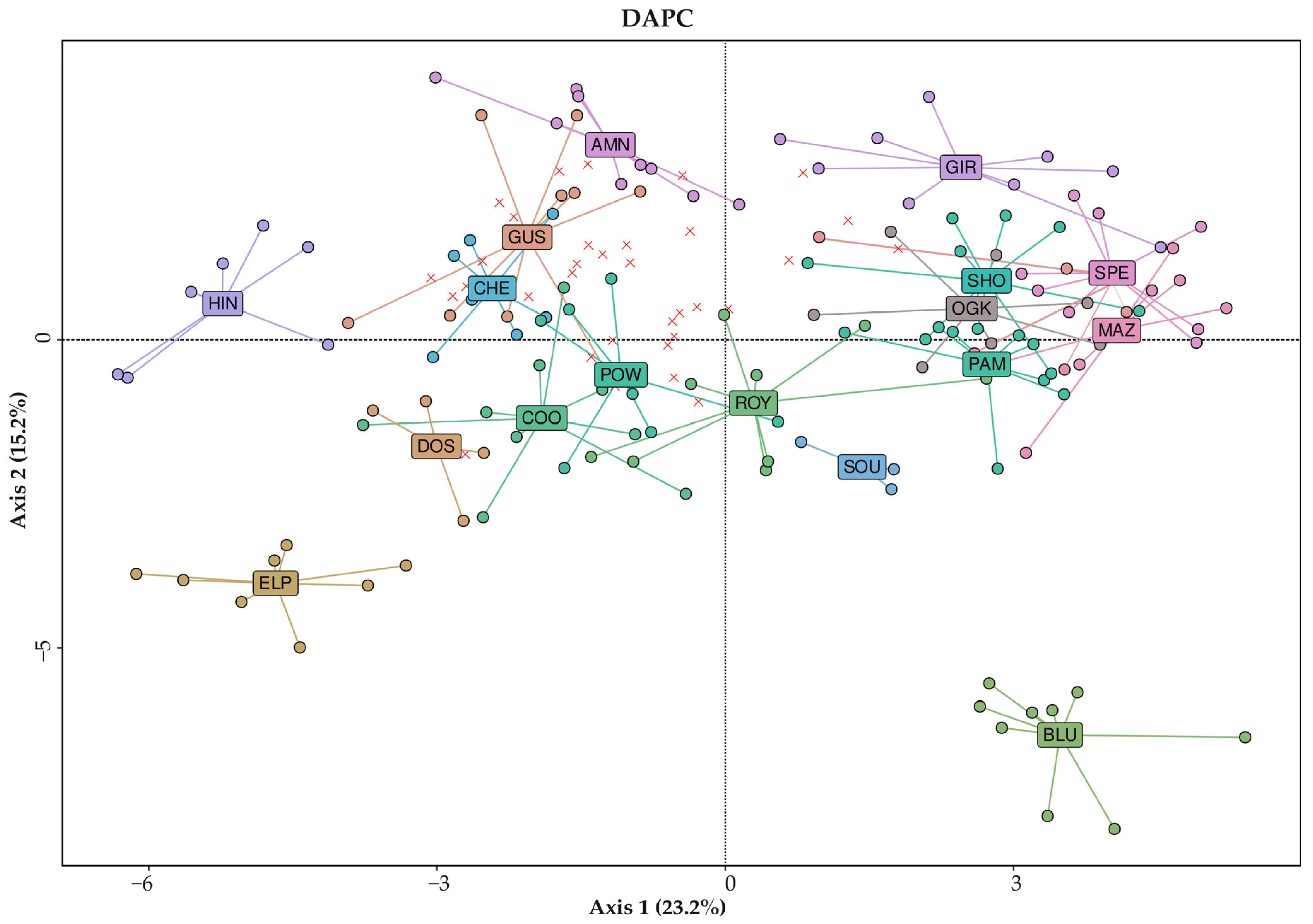
3.1. Genetic Analysis of 171 Drug-Type Cannabis Samples from European Seeds
3.2. Labeling Concerns with the Acquired European Seeds
- (1)
- Numerous samples, especially those from the AFG, BAN, and NOR varieties, had no clear population identity;
- (2)
- Seeds labeled as ROY and OGK were found to be part of the SPE population rather than their respective populations (like the majority of their respective samples);
- (3)
- Samples from the HIN varieties were found to be not only genetically but also phenotypically distinct, despite belonging to the same batch of acquired seeds.
3.3. Discriminating Between Drug- and Hemp-Type Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kovalchuk, I.; Pellino, M.; Rigault, P.; van Velzen, R.; Ebersbach, J.; Ashnest, J.R.; Mau, M.; Schranz, M.E.; Alcorn, J.; Laprairie, R.B.; et al. The Genomics of Cannabis and Its Close Relatives. Annu. Rev. Plant Biol. 2020, 71, 713–739. [Google Scholar] [CrossRef]
- Yazici, L. Optimizing plant density for fiber and seed production in industrial hemp (Cannabis sativa L.). J. King Saud Univ. Sci. 2023, 35, 102419. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.; Tang, Q.; Dai, Z.G.; Deng, C.H.; Chen, Y.; Cheng, C.H.; Yang, Z.M.; Zhang, X.Y.; Chen, J.Q.; et al. Integrated metabolomic and transcriptomic analysis revealed the regulation of yields, cannabinoid, and terpene biosynthesis in Cannabis sativa L. under different photoperiods. S. Afr. J. Bot. 2024, 174, 735–746. [Google Scholar] [CrossRef]
- Kalousek, P.; Holátko, J.; Schreiber, P.; Pluhácek, T.; Lónová, K.S.; Radziemska, M.; Tarkowski, P.; Vyhnánek, T.; Hammerschmiedt, T.; Brtnicky, M. The effect of chelating agents on the Zn-phytoextraction potential of hemp and soil microbial activity. Chem. Biol. Technol. Agric. 2024, 11, 23. [Google Scholar] [CrossRef]
- Malík, M.; Praus, L.; Kuklina, A.; Velechovský, J.; Janatová, A.K.; Klouček, P.; Mládek, V.; Tlustoš, P. Cannabis yield and cannabinoid profile affected by plant nutrition and planting density. Ind. Crops Prod. 2025, 223, 120293. [Google Scholar] [CrossRef]
- Jin, D.; Henry, P.; Shan, J.; Chen, J. Identification of Phenotypic Characteristics in Three Chemotype Categories in the Genus Cannabis. HortScience 2021, 56, 481–490. [Google Scholar] [CrossRef]
- Schultes, R.E. Random Thoughts and Queries on the Botany of Cannabis; J.A.Churchill: London, UK, 1970; pp. 11–33. [Google Scholar]
- Small, E.; Cronquist, A. A Practical and Natural Taxonomy for Cannabis. Taxon 1976, 25, 405–435. [Google Scholar] [CrossRef]
- Schultes, R.E.; Hofmann, A. The Botany and Chemistry of Hallucinogens, 2nd ed.; Charles C Thomas Pub Ltd.: Springfield, IL, USA, 1980. [Google Scholar]
- Gerarde, J. The Herball or General Historie of Plants; John Norton: London, UK, 1597. [Google Scholar]
- Clarke, C. Marijuana Botany. An Advanced Study: The Propagation and Breeding of Distinctive Cannabis; Ronin Publishing: Berkeley, CA, USA, 1981; Volume 13, p. 395. [Google Scholar]
- Ming, R.; Bendahmane, A.; Renner, S.S. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 2011, 62, 485–514. [Google Scholar] [CrossRef]
- Schultes, R.E. Marihuana. The first twelve thousand years. J. Ethnopharmacol. 1982, 5, 115–116. [Google Scholar] [CrossRef]
- Small, E. Evolution and Classification of (Marijuana, Hemp) in Relation to Human Utilization. Bot. Rev. 2015, 81, 189–294. [Google Scholar] [CrossRef]
- Lapierre, E.; de Ronne, M.; Boulanger, R.; Torkamaneh, D. Comprehensive Phenotypic Characterization of Diverse Drug-Type Cannabis Varieties from the Canadian Legal Market. Plants 2023, 12, 3756. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Pepe, M.; Alizadeh, M.; Rakei, A.; Baiton, A.; Jones, A.M.P. Recent advances in Cannabis biotechnology. Ind. Crops Prod. 2020, 158, 113026. [Google Scholar] [CrossRef]
- Barcaccia, G.; Palumbo, F.; Scariolo, F.; Vannozzi, A.; Borin, M.; Bona, S. Potentials and Challenges of Genomics for Breeding Cannabis Cultivars. Front. Plant Sci. 2020, 11, 573299. [Google Scholar] [CrossRef]
- Jin, D.; Henry, P.; Shan, J.; Chen, J. Identification of Chemotypic Markers in Three Chemotype Categories of Cannabis Using Secondary Metabolites Profiled in Inflorescences, Leaves, Stem Bark, and Roots. Front. Plant Sci. 2021, 12, 699530. [Google Scholar] [CrossRef]
- Grassa, C.J.; Weiblen, G.D.; Wenger, J.P.; Dabney, C.; Poplawski, S.G.; Timothy Motley, S.; Michael, T.P.; Schwartz, C.J. A new Cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytol. 2021, 230, 1665–1679. [Google Scholar] [CrossRef]
- Lapierre, E.; Monthony, A.S.; Torkamaneh, D. Genomics-based taxonomy to clarify Cannabis classification. Genome 2023, 66, 202–211. [Google Scholar] [CrossRef]
- Pancaldi, F.; Salentijn, E.M.J.; Trindade, L.M. From fibers to flowering to metabolites: Unlocking hemp (Cannabis sativa) potential with the guidance of novel discoveries and tools. J. Exp. Bot. 2025, 76, 109–123. [Google Scholar] [CrossRef]
- Cull, A.; Joly, D.L. Development and validation of a minimal SNP genotyping panel for the differentiation of Cannabis sativa cultivars. BMC Genom. 2025, 26, 83. [Google Scholar] [CrossRef]
- Zhang, L.; Yuan, M.; Tao, A.; Xu, J.; Lin, L.; Fang, P.; Qi, J. Genetic Structure and Relationship Analysis of an Association Population in Jute (Corchorus spp.) Evaluated by SSR Markers. PLoS ONE 2015, 10, e0128195. [Google Scholar] [CrossRef]
- Gonzaga, Z.J.; Aslam, K.; Septiningsih, E.M.; Collard, B.C.Y. Evaluation of SSR and SNP Markers for Molecular Breeding in Rice. Plant Breed. Biotech. 2015, 3, 139–152. [Google Scholar] [CrossRef]
- Ghedina, A.; Galla, G.; Cadalen, T.; Hilbert, J.L.; Caenazzo, S.T.; Barcaccia, G. A method for genotyping elite breeding stocks of leaf chicory (Cichorium intybus L.) by assaying mapped microsatellite marker loci. BMC Res. Notes 2015, 8, 831. [Google Scholar] [CrossRef]
- Adedze, Y.M.N.; Lu, X.; Fan, W.Y.; Zhang, W.T.; Yang, X.; Deng, Z.J.; Alam, M.A.; Xu, G.L.; Zhang, L.H.; Li, W.H. Development of PCR-based markers associated with powdery mildew resistance using bulked segregant analysis (BSA-seq) in melon. Czech J. Genet. Plant 2024, 60, 25–33. [Google Scholar] [CrossRef]
- Tian, F.; Tian, Y.; Yu, F.; Qian, J.; Wang, F.; Li, X.; Li, T.; Zhang, X.; Huang, D.; Zhao, X. Association analysis of the molecular characteristics and floral traits of Iris × germanica. Czech J. Genet. Plant 2025, 61, 55–66. [Google Scholar] [CrossRef]
- Soler, S.; Gramazio, P.; Figàs, M.R.; Vilanova, S.; Rosa, E.; Llosa, E.R.; Borràs, D.; Plazas, M.; Prohens, J. Genetic structure of var. cultivars based on genomic SSR (gSSR) markers: Implications for breeding and germplasm management. Ind. Crops Prod. 2017, 104, 171–178. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, J.; Huang, S.; Pan, G.; Chang, L.; Li, J.; Zhang, C.; Tang, H.; Chen, A.; Peng, D.; et al. Genetic Diversity and Population Structure of Cannabis Based on the Genome-Wide Development of Simple Sequence Repeat Markers. Front. Genet. 2020, 11, 958. [Google Scholar] [CrossRef] [PubMed]
- Borin, M.; Palumbo, F.; Vannozzi, A.; Scariolo, F.; Sacilotto, G.B.; Gazzola, M.; Barcaccia, G. Developing and Testing Molecular Markers in Cannabis sativa (Hemp) for Their Use in Variety and Dioecy Assessments. Plants 2021, 10, 2174. [Google Scholar] [CrossRef]
- Alghanim, H.J.; Almirall, J.R. Development of microsatellite markers in Cannabis sativa for DNA typing and genetic relatedness analyses. Anal. Bioanal. Chem. 2003, 376, 1225–1233. [Google Scholar] [CrossRef]
- Gilmore, S.; Peakall, R.; Robertson, J. Short tandem repeat (STR) DNA markers are hypervariable and informative in Cannabis sativa: Implications for forensic investigations. Forensic Sci. Int. 2003, 131, 65–74. [Google Scholar] [CrossRef]
- Hsieh, H.M.; Hou, R.J.; Tsai, L.C.; Wei, C.S.; Liu, S.W.; Huang, L.H.; Kuo, Y.C.; Linacre, A.; Lee, J.C. A highly polymorphic STR locus in Cannabis sativa. Forensic Sci. Int. 2003, 131, 53–58. [Google Scholar] [CrossRef]
- Dufresnes, C.; Jan, C.; Bienert, F.; Goudet, J.; Fumagalli, L. Broad-Scale Genetic Diversity of Cannabis for Forensic Applications. PLoS ONE 2017, 12, e0170522. [Google Scholar] [CrossRef]
- Gao, C.; Xin, P.; Cheng, C.; Tang, Q.; Chen, P.; Wang, C.; Zang, G.; Zhao, L. Diversity analysis in Cannabis sativa based on large-scale development of expressed sequence tag-derived simple sequence repeat markers. PLoS ONE 2014, 9, e110638. [Google Scholar] [CrossRef] [PubMed]
- Grassa, C.J.; Wenger, J.P.; Dabney, C.; Poplawski, S.G.; Motley, S.T.; Michael, T.P.; Schwartz, C.J.; Weiblen, G.D. A complete Cannabis chromosome assembly and adaptive admixture for elevated cannabidiol (CBD) content. BioRxiv 2018. [Google Scholar] [CrossRef]
- Hillig, K.W. Genetic evidence for speciation in Cannabis (Cannabaceae). Genet. Resour. Crop Evol. 2005, 52, 161–180. [Google Scholar] [CrossRef]
- Lynch, R.C.; Vergara, D.; Tittes, S.; White, K.; Schwartz, C.J.; Gibbs, M.J.; Ruthenburg, T.C.; deCesare, K.; Land, D.P.; Kane, N.C. Genomic and Chemical Diversity in Cannabis. Crit. Rev. Plant Sci. 2017, 35, 349–363. [Google Scholar] [CrossRef]
- Sawler, J.; Stout, J.M.; Gardner, K.M.; Hudson, D.; Vidmar, J.; Butler, L.; Page, J.E.; Myles, S. The Genetic Structure of Marijuana and Hemp. PLoS ONE 2015, 10, e0133292. [Google Scholar] [CrossRef]
- Soorni, A.; Fatahi, R.; Haak, D.C.; Salami, S.A.; Bombarely, A. Assessment of Genetic Diversity and Population Structure in Iranian Cannabis Germplasm. Sci. Rep. 2017, 7, 15668. [Google Scholar] [CrossRef]
- Fisarova, L.; Surinova, M.; Jarosova, A.; Krejcik, J.; Vosatka, M. Evidence of the Ability of Microsatellite Method to Distinguish Cannabis Strains with High Cannabinoid Content. Cannabis Cannabinoid Res. 2024, 9, 513–522. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Houston, R. The development of two fast genotyping assays for the differentiation of hemp from marijuana. J. Forensic Sci. 2025, 70, 49–60. [Google Scholar] [CrossRef]
- Vyhnanek, T.; Nevrtalova, E.; Bjelkova, M.; Balgova, B. SSR loci survey of technical hemp cultivars: The optimization of a cost-effective analyses to study genetic variability. Plant Sci. 2020, 298, 110551. [Google Scholar] [CrossRef]
- Hu, Z.G.; Guo, H.Y.; Hu, X.L.; Chen, X.; Liu, X.Y.; Guo, M.B.; Zhang, Q.Y.; Xu, Y.P.; Guo, L.F.; Yang, M. Genetic diversity research of hemp (Cannabis sativa L) cultivar based on AFLP analysis. Plant Gene. Res. 2012, 13, 555–561. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, X.; Guo, H.; Trindade, L.M.; Salentijn, E.M.J.; Guo, R.; Guo, M.; Xu, Y.; Yang, M. Latitudinal Adaptation and Genetic Insights Into the Origins of Cannabis sativa L. Front. Plant Sci. 2018, 9, 1876. [Google Scholar] [CrossRef]
- Benkirane, C.; Charif, M.; Müller, C.M.; Taaifi, Y.; Mansouri, F.; Addi, M.; Bellaoui, M.; Serghini-Caid, H.; Elamrani, A.; Abid, M. Population structure and genetic diversity of Moroccan Cannabis (Cannabis sativa L.) germplasm through simple sequence repeat (SSR) analysis. Genet. Resour. Crop Evol. 2023, 71, 2037–2051. [Google Scholar] [CrossRef]
- Rull, V. Origin, early expansion, domestication and anthropogenic diffusion of Cannabis, with emphasis on Europe and the Iberian Peninsula. Perspect. Plant Ecol. 2022, 55, 125670. [Google Scholar] [CrossRef]
- Groom, Q. R.C. Clarke & M.D. Merlin (2013)–Cannabis: Evolution and Ethnobotany. Plant Ecol. Evol. 2014, 147, 149. [Google Scholar] [CrossRef]
- Clarke, R.C.; Merlin, M.D. Domestication, Breeding History, Present-day Genetic Diversity, and Future Prospects. Crit. Rev. Plant Sci. 2016, 35, 293–327. [Google Scholar] [CrossRef]
- Borin, M. Overcoming Challenges in Cannabis sativa Breeding Research with Conventional and Biotechnological Tools. Ph.D. Thesis, University of Verona, Verona, Italy, 2024. [Google Scholar]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T.J.B. Population Genetic Analysis of Codominant and Dominant Markers and Quantitative Traits. Belg. J. Bot. 1997, 129, 157–163. [Google Scholar]
- Levene, H. On a matching problem arising in genetics. Ann. Math. Stat. 1949, 20, 91–94. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System Ver. 2.2; Exeter Publishing: New York, NY, USA, 2009; pp. 1–50. [Google Scholar]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Wickham, M.H. ggplot2. Use R! Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Brooks, J.C.; Grunwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Scariolo, F.; Palumbo, F.; Farinati, S.; Barcaccia, G. Pipeline to Design Inbred Lines and F1 Hybrids of Leaf Chicory (Radicchio) Using Male Sterility and Genotyping-by-Sequencing. Plants 2023, 12, 1242. [Google Scholar] [CrossRef]
- Mufti, S.; Afroza, B.; Bhat, R.; Mushtaq, S.; Nabi, A. DUS testing for plant variety protection. Ann. Agri Bio Res. 2017, 22, 43–48. [Google Scholar]
- Hall, J.; Bhattarai, S.P.; Midmore, D.J. Review of Flowering Control in Industrial Hemp. J. Nat. Fibers 2012, 9, 23–36. [Google Scholar] [CrossRef]
- Vergara, D.; Huscher, E.L.; Keepers, K.G.; Pisupati, R.; Schwabe, A.L.; McGlaughlin, M.E.; Kane, N.C. Genomic Evidence That Governmentally Produced Cannabis sativa Poorly Represents Genetic Variation Available in State Markets. Front. Plant Sci. 2021, 12, 668315. [Google Scholar] [CrossRef]
- de Oliveira Pereira Ribeiro, L.; Avila, E.; Mariot, R.F.; Fett, M.S.; de Oliveira Camargo, F.A.; Alho, C.S. Evaluation of two 13-loci STR multiplex system regarding identification and origin discrimination of Brazilian Cannabis sativa samples. Int. J. Leg. Med. 2020, 134, 1603–1612. [Google Scholar] [CrossRef]
- Palumbo, F.; Galla, G.; Barcaccia, G. Developing a Molecular Identification Assay of Old Landraces for the Genetic Authentication of Typical Agro-Food Products: The Case Study of the Barley ‘Agordino’. Food Technol. Biotechnol. 2017, 55, 29–39. [Google Scholar] [CrossRef]

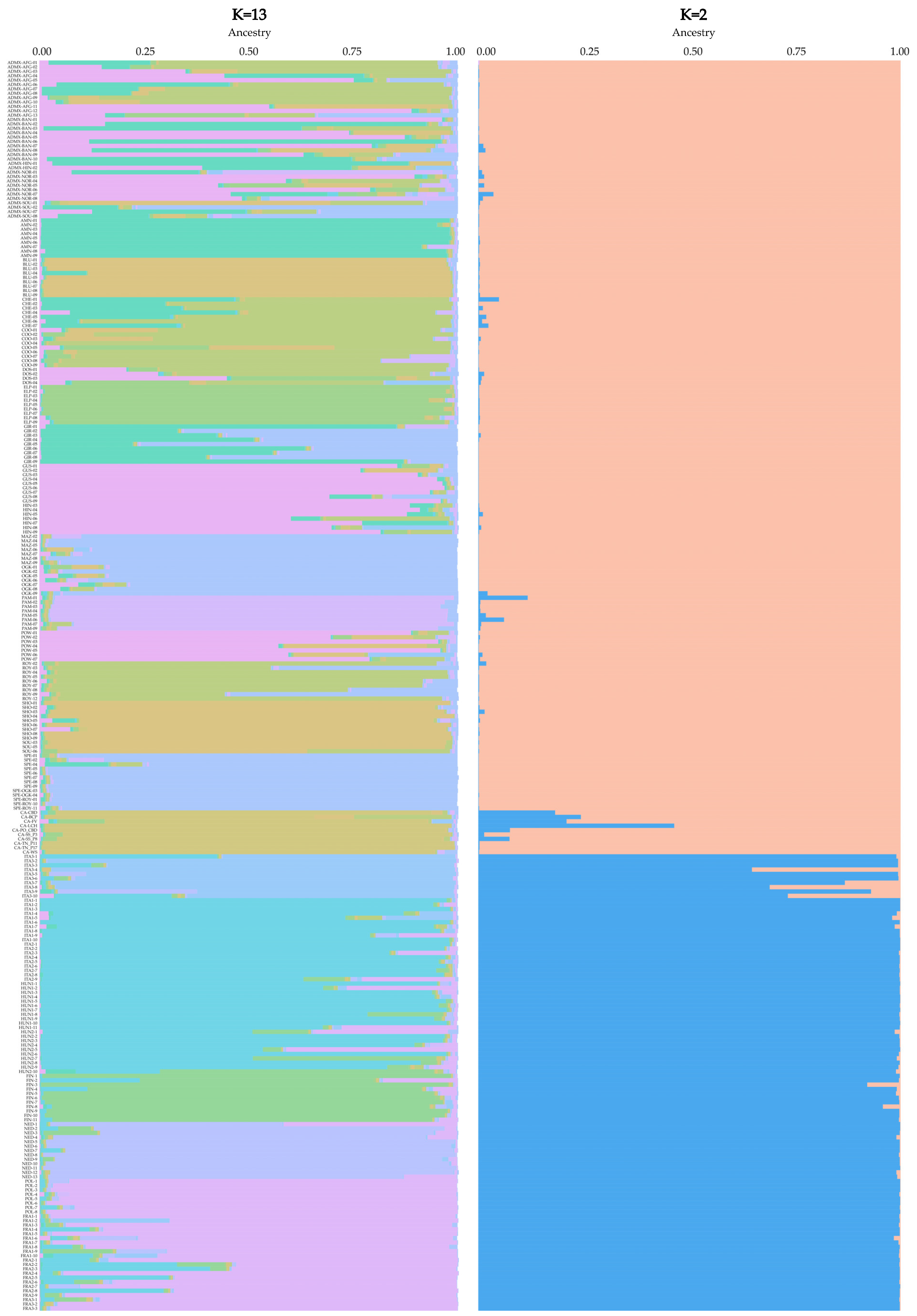

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borin, M.; Scariolo, F.; Cappello Fusaro, M.; Lucchetta, I.; Sacilotto, G.B.; Gazzola, M.; Bona, S.; Barcaccia, G. Genotyping of Commercial European Cannabis Seeds Based on Multiple Mapped Marker Loci: A Comparative Study of Drug and Hemp Varieties. Plants 2025, 14, 3050. https://doi.org/10.3390/plants14193050
Borin M, Scariolo F, Cappello Fusaro M, Lucchetta I, Sacilotto GB, Gazzola M, Bona S, Barcaccia G. Genotyping of Commercial European Cannabis Seeds Based on Multiple Mapped Marker Loci: A Comparative Study of Drug and Hemp Varieties. Plants. 2025; 14(19):3050. https://doi.org/10.3390/plants14193050
Chicago/Turabian StyleBorin, Marcello, Francesco Scariolo, Maddalena Cappello Fusaro, Irene Lucchetta, Gio Batta Sacilotto, Marco Gazzola, Stefano Bona, and Gianni Barcaccia. 2025. "Genotyping of Commercial European Cannabis Seeds Based on Multiple Mapped Marker Loci: A Comparative Study of Drug and Hemp Varieties" Plants 14, no. 19: 3050. https://doi.org/10.3390/plants14193050
APA StyleBorin, M., Scariolo, F., Cappello Fusaro, M., Lucchetta, I., Sacilotto, G. B., Gazzola, M., Bona, S., & Barcaccia, G. (2025). Genotyping of Commercial European Cannabis Seeds Based on Multiple Mapped Marker Loci: A Comparative Study of Drug and Hemp Varieties. Plants, 14(19), 3050. https://doi.org/10.3390/plants14193050






