Genome-Wide Identification of the TCP Gene Family in Chimonanthus praecox and Functional Analysis of CpTCP2 Regulating Leaf Development and Flowering in Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Properties of CpTCP Genes
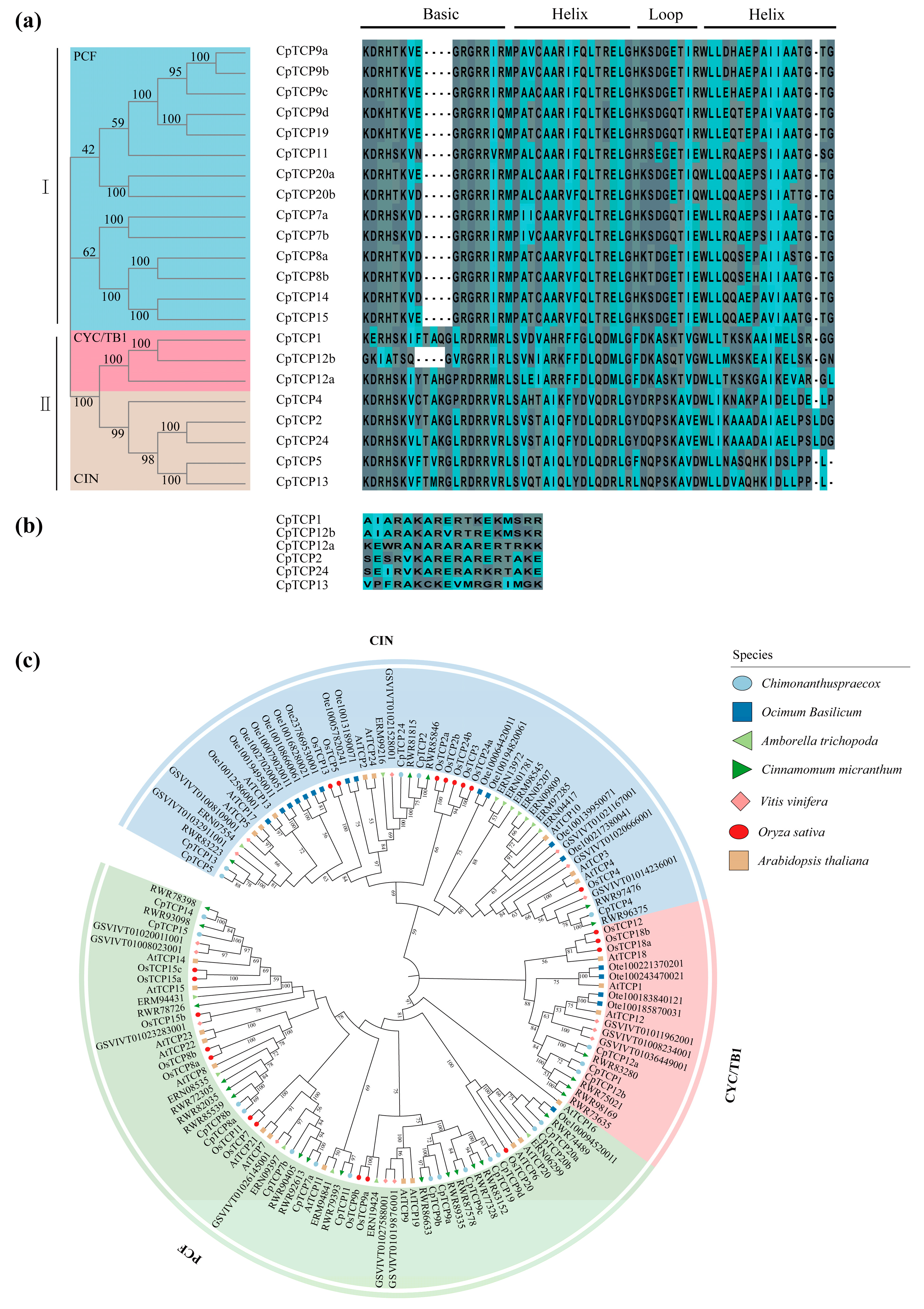
2.2. Multiple Sequence Alignment and Phylogenetic Tree Analysis of CpTCP Proteins
2.3. Conserved Motif Analysis and Promoter Cis-Element Analysis of CpTCPs
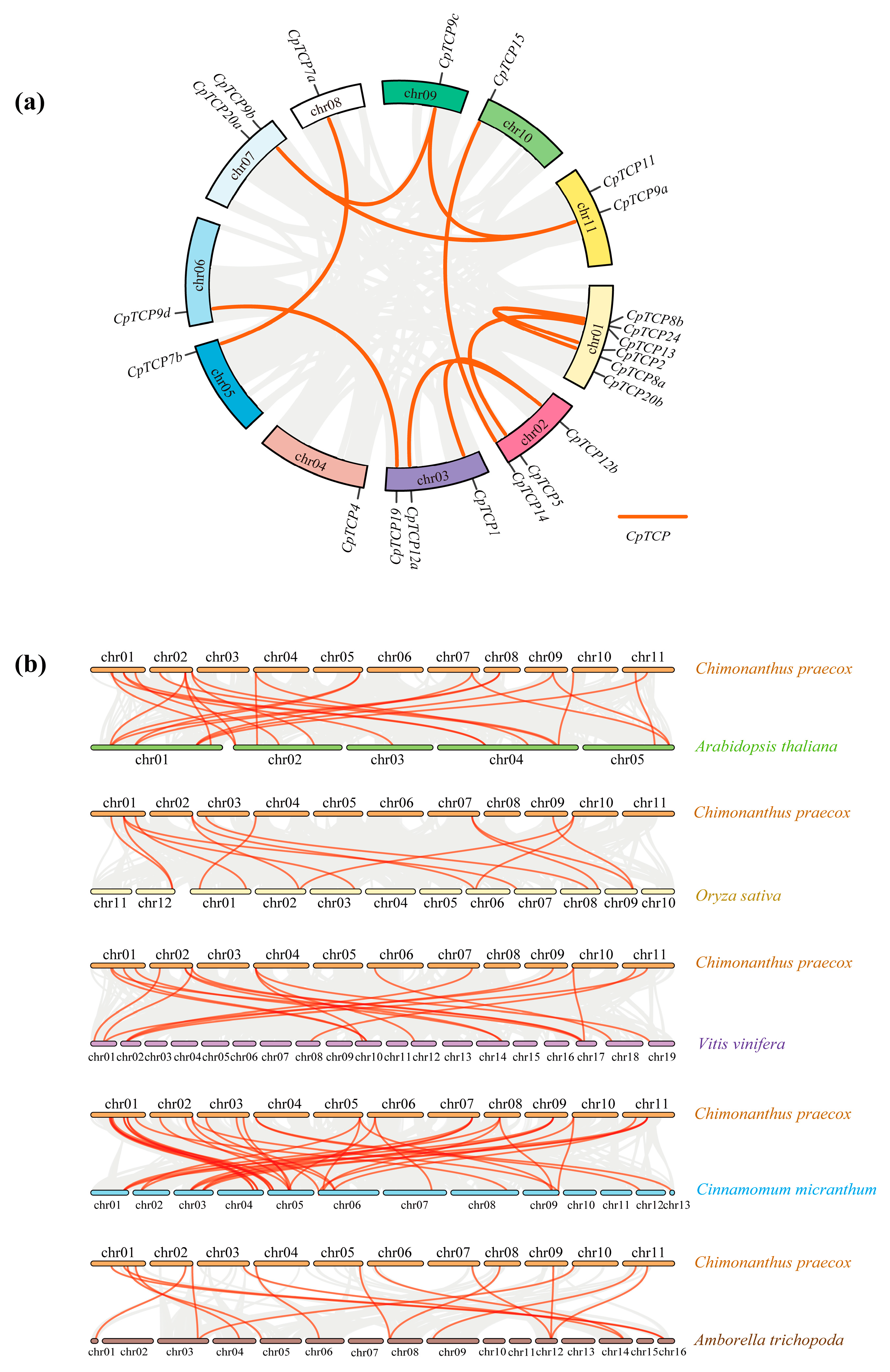
2.4. Chromosomal Localization and Synteny Analysis of CpTCP Genes
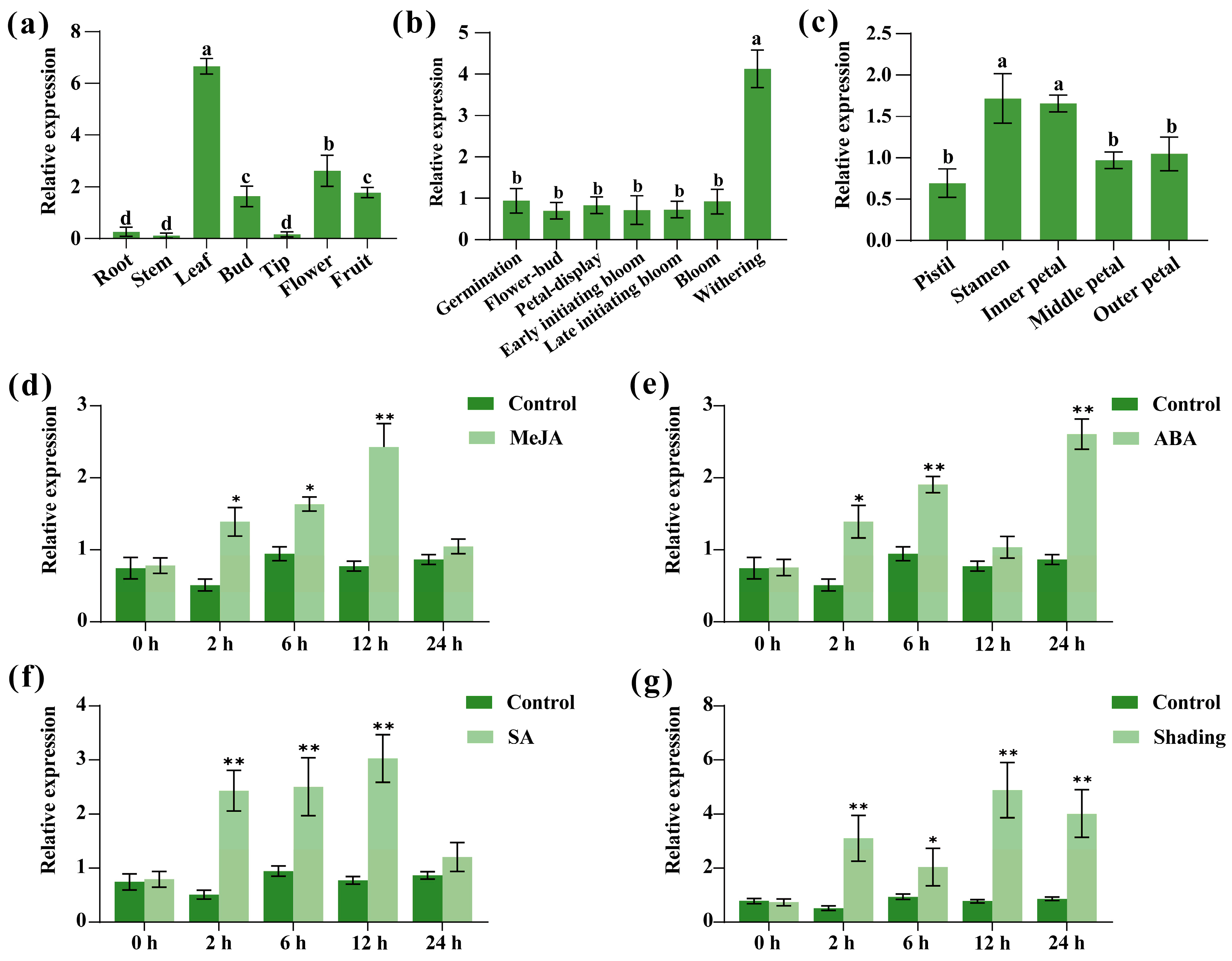
2.5. Analysis of the Sequence Characteristics of CpTCP2
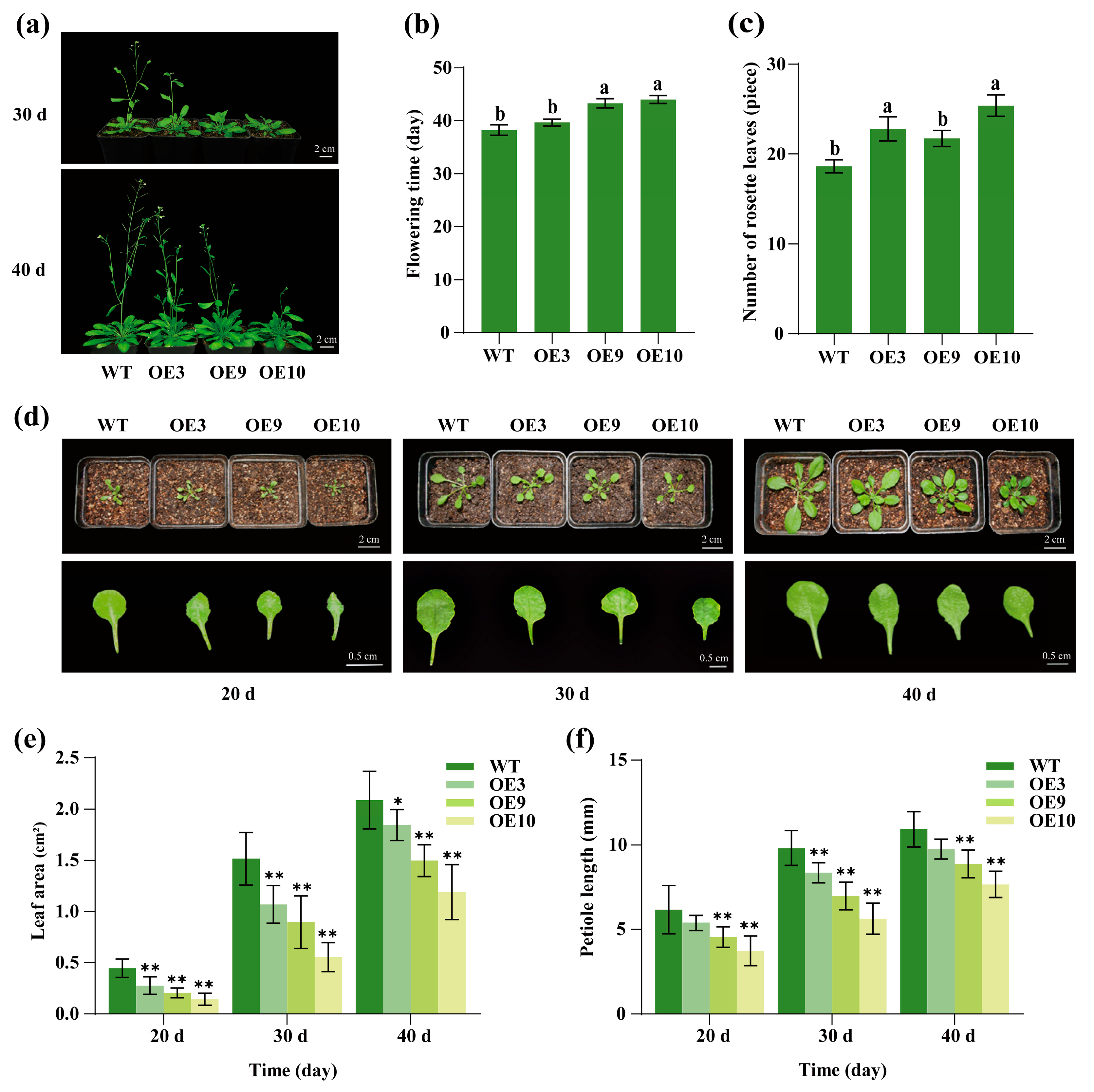
2.6. Regulation of Flowering Time and Leaf Development by CpTCP2 in Arabidopsis
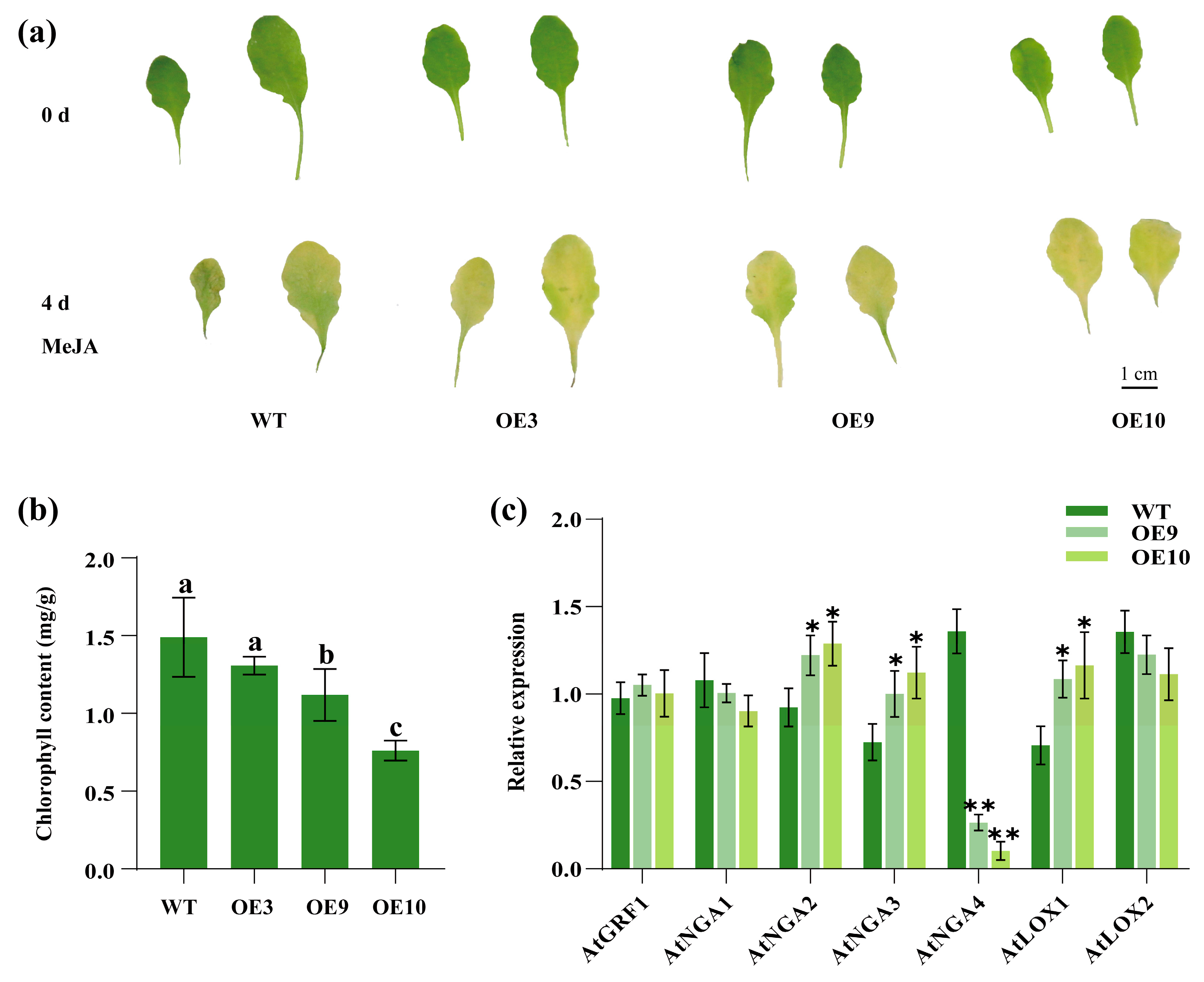
2.7. Regulation of Leaf Senescence by CpTCP2 in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Identification, Multiple Sequence Alignment, and Evolutionary Analysis of the CpTCP Gene Family
4.2. Gene Structure, Conserved Motif, and Promoter Element Analysis
4.3. Chromosome Localization and Synteny Analysis
4.4. Cloning of CpTCP2 Gene and Arabidopsis Transformation
4.5. Phenotypic Observation and Measurements of Transgenic Plants
4.6. Gene Expression Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Liu, N.; Zhang, W.; Wu, C.; Jiang, Y.; Ma, J.; Li, M.; Sui, S. Integrated transcriptome and proteome analysis provides insight into chilling-induced dormancy breaking in Chimonanthus praecox. Hortic. Res. 2020, 7, 198. [Google Scholar] [CrossRef]
- Shang, J.; Tian, J.; Cheng, H.; Yan, Q.; Li, L.; Jamal, A.; Xu, Z.; Xiang, L.; Saski, C.A.; Jin, S.; et al. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 2020, 21, 200. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, F.; Song, A.; Zhao, Y.; Chen, X.; Gao, Y.; Wei, G.; Zhang, W.; Guan, Y.; Fu, J.; et al. The genome assembly of Chimonanthus praecox var. concolor and comparative genomic analysis highlight the genetic basis underlying conserved and variable floral traits of wintersweet. Ind. Crops Prod. 2023, 206, 117603. [Google Scholar] [CrossRef]
- Shen, Z.; Li, W.; Li, Y.; Liu, M.; Cao, H.; Provart, N.; Ding, X.; Sun, M.; Tang, Z.; Yue, C.; et al. The red flower wintersweet genome provides insights into the evolution of magnoliids and the molecular mechanism for tepal color development. Plant J. 2021, 108, 1662–1678. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, R.; Ma, J.; Sui, S.; Guo, Y.; Liu, D.; Li, Z.; Lin, Y.; Li, M. Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis. Genes 2017, 8, 199. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, X.; Cao, Y.; Zhang, H.; Hua, R.; Liu, H.; Sui, S. CpBBX19, a B-Box Transcription Factor Gene of Chimonanthus praecox, Improves Salt and Drought Tolerance in Arabidopsis. Genes 2021, 12, 1456. [Google Scholar] [CrossRef]
- Lin, J.; Liu, D.; Wang, X.; Ahmed, S.; Li, M.; Kovinich, N.; Sui, S. Transgene CpNAC68 from Wintersweet (Chimonanthus praecox) Improves Arabidopsis Survival of Multiple Abiotic Stresses. Plants 2021, 10, 1403. [Google Scholar] [CrossRef]
- Wu, H.; Liu, B.; Cao, Y.; Ma, G.; Zheng, X.; Zhu, H.; Sui, S. Genome-Wide Identification of WOX Gene Family in Chimonanthus praecox and a Functional Analysis of CpWUS. Plants 2025, 14, 1144. [Google Scholar] [CrossRef]
- Wang, X.; Liu, D.; Lin, J.; Zhu, T.; Liu, N.; Yang, X.; Ma, J.; Sui, S. Carotenoid Cleavage Dioxygenase Genes of Chimonanthus praecox, CpCCD7 and CpCCD8, Regulate Shoot Branching in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 8750. [Google Scholar] [CrossRef]
- Tian, M.; Li, Q.; Liu, N.; Li, J.; Huo, J.; Sui, S.; Li, Z. Ca2+-induced CpCBL8-CpCIPK9 module negatively regulates dormancy breaking and cold tolerance in winter-flowering wintersweet. Hortic. Plant J. 2025, 11, 877–890. [Google Scholar] [CrossRef]
- Zhang, H.; Hua, R.; Wang, X.; Wu, H.; Ou, H.; Lu, X.; Huang, Y.; Liu, D.; Sui, S. CpMAX1a, a Cytochrome P450 Monooxygenase Gene of Chimonanthus praecox Regulates Shoot Branching in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 10888. [Google Scholar] [CrossRef]
- Li, R.; Ma, J.; Liu, H.; Wang, X.; Li, J.; Li, Z.; Li, M.; Sui, S.; Liu, D. Overexpression of CpSIZ1, a SIZ/PIAS-Type SUMO E3 Ligase from Wintersweet (Chimonanthus praecox), Delays Flowering, Accelerates Leaf Senescence and Enhances Cold Tolerance in Arabidopsis. Plant Mol. Biol. Report. 2021, 39, 301–316. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar] [CrossRef]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef]
- Manassero, N.G.; Viola, I.L.; Welchen, E.; Gonzalez, D.H. TCP transcription factors: Architectures of plant form. Biomol. Concepts 2013, 4, 111–127. [Google Scholar] [CrossRef]
- Das Gupta, M.; Aggarwal, P.; Nath, U. CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 2014, 204, 901–912. [Google Scholar] [CrossRef]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial Control of Gene Expression by miR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol. 2018, 176, 1694–1708. [Google Scholar] [CrossRef]
- Schommer, C.; Debernardi, J.M.; Bresso, E.G.; Rodriguez, R.E.; Palatnik, J.F. Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 2014, 7, 1533–1544. [Google Scholar] [CrossRef]
- He, Z.M.; Zhou, X.M.; Chen, J.M.; Yin, L.T.; Zeng, Z.H.; Xiang, J.; Liu, S.C. Identification of a consensus DNA-binding site for the TCP domain transcription factor TCP2 and its important roles in the growth and development of Arabidopsis. Mol. Biol. Rep. 2021, 48, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Hu, S.; Yu, Q.; Wang, C.; Yang, Y.; Sun, H.; Yang, Y.; Sun, X. Genome-Wide Identification and Characterization of BrrTCP Transcription Factors in Brassica rapa ssp. rapa. Front. Plant Sci. 2017, 8, 1588. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Sinha, N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 2013, 4, 406. [Google Scholar] [CrossRef] [PubMed]
- Cruz, R.; Melo-de-Pinna, G.F.A.; Vasco, A.; Prado, J.; Ambrose, B.A. Class I KNOX Is Related to Determinacy during the Leaf Development of the Fern Mickelia scandens (Dryopteridaceae). Int. J. Mol. Sci. 2020, 21, 4295. [Google Scholar] [CrossRef]
- Mizukami, Y. A matter of size: Developmental control of organ size in plants. Curr. Opin. Plant Biol. 2001, 4, 533–539. [Google Scholar] [CrossRef]
- Challa, K.R.; Rath, M.; Nath, U. The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet. 2019, 15, e1007988. [Google Scholar] [CrossRef]
- Nag, A.; King, S.; Jack, T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 22534–22539. [Google Scholar] [CrossRef]
- Chen, D.; Yan, W.; Fu, L.-Y.; Kaufmann, K. Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat. Commun. 2018, 9, 4534. [Google Scholar] [CrossRef]
- Lin, Y.F.; Chen, Y.Y.; Hsiao, Y.Y.; Shen, C.Y.; Hsu, J.L.; Yeh, C.M.; Mitsuda, N.; Ohme-Takagi, M.; Liu, Z.J.; Tsai, W.C. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 2016, 67, 5051–5066. [Google Scholar] [CrossRef]
- Liu, H.; Sun, M.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q. Two Cyc2CL transcripts (Cyc2CL-1 and Cyc2CL-2) may play key roles in the petal and stamen development of ray florets in chrysanthemum. BMC Plant Biol. 2021, 21, 105. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Huang, D.; Yang, Y.; Sun, M.; Cheng, T.; Wang, J.; Pan, H.; Zhang, Q. CmCYC2-like transcription factors may interact with each other or bind to the promoter to regulate floral symmetry development in Chrysanthemum morifolium. Plant Mol. Biol. 2020, 103, 159–171. [Google Scholar] [CrossRef] [PubMed]
- He, J.; He, X.; Chang, P.; Jiang, H.; Gong, D.; Sun, Q. Genome-wide identification and characterization of TCP family genes in Brassica juncea var. tumida. PeerJ 2020, 8, e9130. [Google Scholar] [CrossRef] [PubMed]
- Li, S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 2015, 10, e1044192. [Google Scholar] [CrossRef]
- Leng, X.; Wei, H.; Xu, X.; Ghuge, S.A.; Jia, D.; Liu, G.; Wang, Y.; Yuan, Y. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom. 2019, 20, 786. [Google Scholar] [CrossRef]
- Chai, W.; Jiang, P.; Huang, G.; Jiang, H.; Li, X. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants 2017, 23, 779–791. [Google Scholar] [CrossRef]
- Dong, Z.; Hao, Y.; Zhao, Y.; Tang, W.; Wang, X.; Li, J.; Wang, L.; Hu, Y.; Guan, X.; Gu, F.; et al. Genome-Wide Analysis of the TCP Transcription Factor Gene Family in Pepper (Capsicum annuum L.). Plants 2024, 13, 641. [Google Scholar] [CrossRef]
- Huang, F.; Shi, C.; Zhang, Y.; Hou, X. Genome-Wide Identification and Characterization of TCP Family Genes in Pak-Choi [Brassica campestris (syn. Brassica rapa) ssp. chinensis var. communis]. Front. Plant Sci. 2022, 13, 854171. [Google Scholar] [CrossRef]
- Jiang, M.Q.; Xue, X.; Su, L.; Chen, Y.; Zhang, S.; Li, X.; Wang, P.; Zhang, Z.; Lai, Z.; Lin, Y. Genome-wide Identification and Expression Analysis of TCP Family in Dimocarpus longan. Acta Hortic. Sin. 2021, 48, 2481–2496. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Z.; Zhao, K.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Genome-Wide Identification, Characterization and Expression Analysis of the TCP Gene Family in Prunus mume. Front. Plant Sci. 2016, 7, 1301. [Google Scholar] [CrossRef]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011, 67, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S.; van der Wal, F.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Ballester, P.; Navarrete-Gomez, M.; Carbonero, P.; Onate-Sanchez, L.; Ferrandiz, C. Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol. Plant 2015, 155, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wang, Z.; Jia, C.H.; Miao, H.X.; Zhang, J.N.; Liu, J.H.; Xu, B.Y.; Jin, Z.Q. Genome-Wide Identification and Transcript Analysis of TCP Gene Family in Banana (Musa acuminata L.). Biochem. Genet. 2022, 60, 204–222. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Name | Gene ID | Homolog | AA | MW | pI | II | GRAVY | AI | SP | TH | SL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpTCP1 | Ws006179 | AtTCP1 | 365 | 40,312.96 | 6.57 | 51.63 | −0.707 | 59.42 | NO | 0 | Nucleus |
| CpTCP2 | Ws001834 | AtTCP2 | 436 | 47,788.71 | 6.77 | 45.49 | −0.782 | 58.51 | NO | 0 | Nucleus |
| CpTCP4 | Ws008336 | AtTCP4 | 508 | 55,927.39 | 6.38 | 68.71 | −0.884 | 60.39 | NO | 0 | Nucleus |
| CpTCP5 | Ws004919 | AtTCP5 | 345 | 38,523.92 | 8.43 | 62.36 | −0.743 | 64.99 | NO | 0 | Nucleus |
| CpTCP7a | Ws020128 | AtTCP7 | 249 | 25,889.27 | 9.90 | 54.46 | −0.244 | 76.59 | NO | 0 | Nucleus |
| CpTCP7b | Ws013215 | AtTCP7 | 209 | 22,059.89 | 9.29 | 57.09 | −0.340 | 67.42 | NO | 0 | Nucleus |
| CpTCP8a | Ws002199 | AtTCP8 | 452 | 47,290.06 | 7.07 | 53.85 | −0.563 | 65.95 | NO | 0 | Nucleus |
| CpTCP8b | Ws000567 | AtTCP8 | 442 | 46,544.66 | 7.94 | 63.48 | −0.486 | 66.33 | NO | 0 | Nucleus |
| CpTCP9a | Ws026740 | AtTCP9 | 338 | 35,322.83 | 9.55 | 59.55 | −0.315 | 73.11 | NO | 0 | Nucleus |
| CpTCP9b | Ws018174 | AtTCP9 | 353 | 36,308.07 | 10.20 | 56.89 | −0.209 | 77.76 | NO | 0 | Nucleus |
| CpTCP9c | Ws021384 | AtTCP9 | 306 | 32,376.73 | 9.13 | 69.60 | −0.279 | 79.15 | NO | 0 | Nucleus |
| CpTCP9d | Ws013991 | AtTCP9 | 603 | 65,091.48 | 5.90 | 55.38 | −0.502 | 71.38 | NO | 0 | Nucleus |
| CpTCP11 | Ws025658 | AtTCP11 | 234 | 24,461.38 | 6.65 | 56.45 | −0.355 | 63.55 | NO | 0 | Nucleus |
| CpTCP12a | Ws007064 | AtTCP12 | 389 | 43,437.09 | 6.79 | 45.16 | −0.801 | 59.51 | NO | 0 | Nucleus |
| CpTCP12b | Ws004363 | AtTCP12 | 288 | 32,374.64 | 9.91 | 52.48 | −0.755 | 65.69 | NO | 0 | Nucleus |
| CpTCP13 | Ws000794 | AtTCP13 | 376 | 42,297.49 | 8.99 | 57.15 | −0.700 | 65.35 | NO | 0 | Nucleus |
| CpTCP14 | Ws005371 | AtTCP14 | 373 | 40,123.80 | 7.24 | 60.09 | −0.591 | 60.72 | NO | 0 | Nucleus |
| CpTCP15 | Ws022851 | AtTCP15 | 350 | 37,916.24 | 7.22 | 51.49 | −0.610 | 63.94 | NO | 0 | Nucleus |
| CpTCP19 | Ws007603 | AtTCP19 | 569 | 61,907.04 | 6.35 | 53.18 | −0.543 | 70.47 | NO | 0 | Nucleus |
| CpTCP20a | Ws017440 | AtTCP20 | 270 | 28,772.23 | 9.13 | 58.33 | −0.554 | 65.81 | NO | 0 | Nucleus |
| CpTCP20b | Ws002910 | AtTCP20 | 340 | 36,678.30 | 6.83 | 58.50 | −0.426 | 71.41 | NO | 0 | Nucleus |
| CpTCP24 | Ws000671 | AtTCP24 | 427 | 46,947.91 | 8.27 | 54.99 | −0.791 | 59.72 | NO | 0 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Guo, G.; Wu, H.; Wang, X.; Liu, B.; Yang, X.; Dai, Q.; Zhu, H.; Lu, M.; Zhu, H.; et al. Genome-Wide Identification of the TCP Gene Family in Chimonanthus praecox and Functional Analysis of CpTCP2 Regulating Leaf Development and Flowering in Transgenic Arabidopsis. Plants 2025, 14, 3039. https://doi.org/10.3390/plants14193039
Cao Y, Guo G, Wu H, Wang X, Liu B, Yang X, Dai Q, Zhu H, Lu M, Zhu H, et al. Genome-Wide Identification of the TCP Gene Family in Chimonanthus praecox and Functional Analysis of CpTCP2 Regulating Leaf Development and Flowering in Transgenic Arabidopsis. Plants. 2025; 14(19):3039. https://doi.org/10.3390/plants14193039
Chicago/Turabian StyleCao, Yinzhu, Gangyu Guo, Huafeng Wu, Xia Wang, Bin Liu, Ximeng Yang, Qianli Dai, Hengxing Zhu, Min Lu, Haoxiang Zhu, and et al. 2025. "Genome-Wide Identification of the TCP Gene Family in Chimonanthus praecox and Functional Analysis of CpTCP2 Regulating Leaf Development and Flowering in Transgenic Arabidopsis" Plants 14, no. 19: 3039. https://doi.org/10.3390/plants14193039
APA StyleCao, Y., Guo, G., Wu, H., Wang, X., Liu, B., Yang, X., Dai, Q., Zhu, H., Lu, M., Zhu, H., Li, Z., Jin, C., Li, S., & Sui, S. (2025). Genome-Wide Identification of the TCP Gene Family in Chimonanthus praecox and Functional Analysis of CpTCP2 Regulating Leaf Development and Flowering in Transgenic Arabidopsis. Plants, 14(19), 3039. https://doi.org/10.3390/plants14193039






