Genome-Wide Identification and Expression Profiling of Sugar Transport Protein Response to Fusarium Head Blight in Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Results
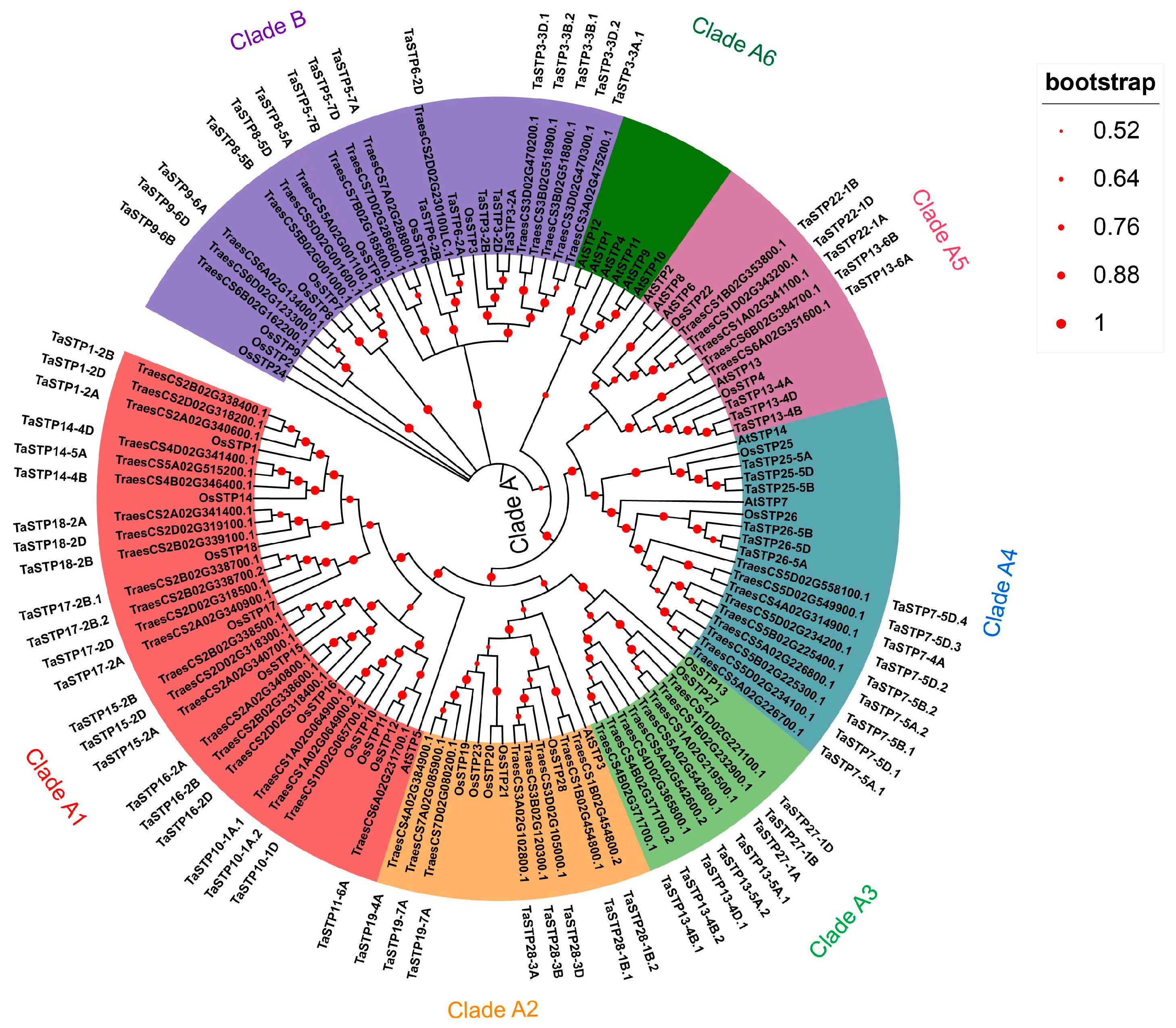
2.1. Identification and Phylogenetic Analysis of TaSTP Genes in Wheat
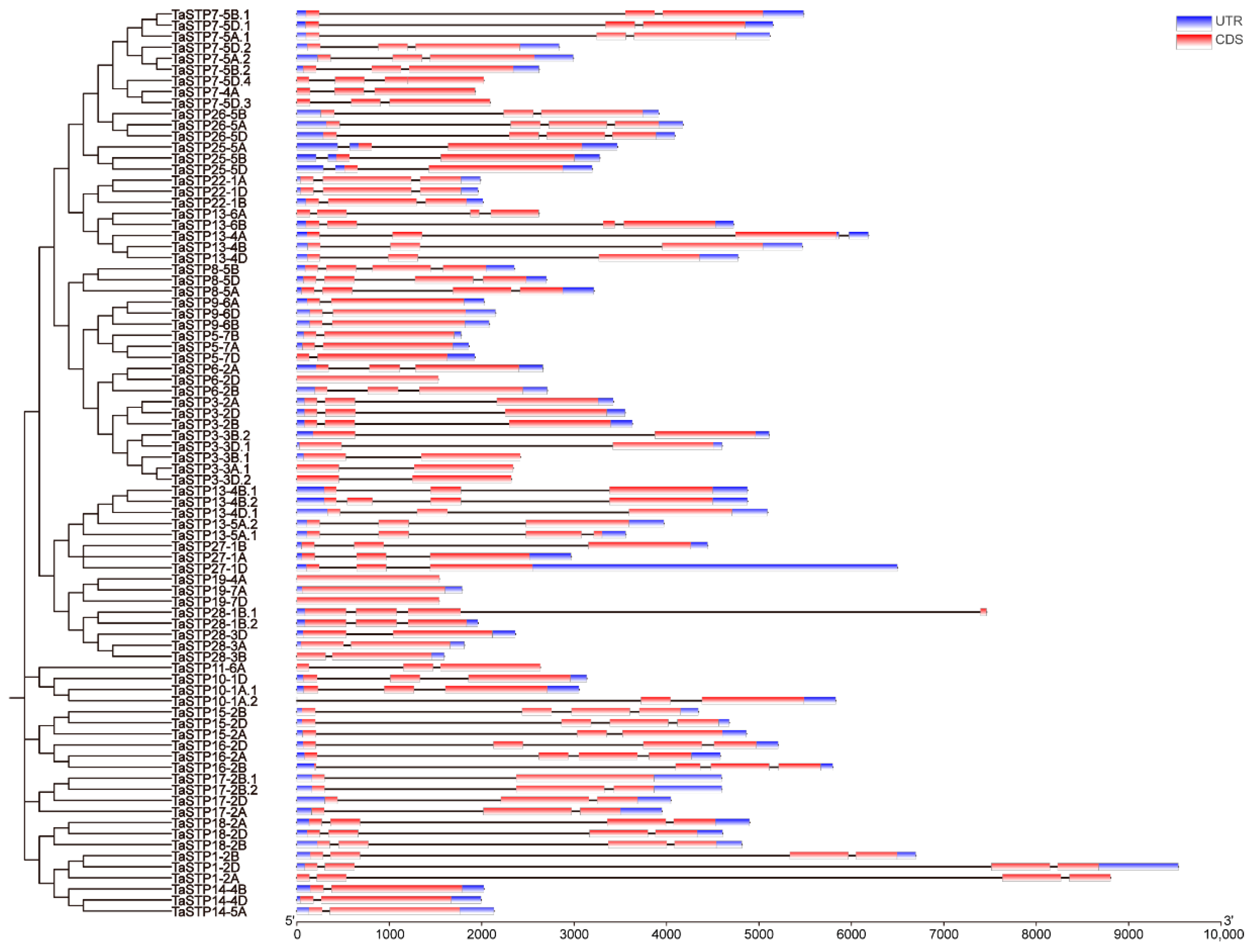
2.2. Genome Distribution of Wheat TaSTP Genes
2.3. Expression Profiles of TaSTP Genes
2.3.1. Gene Expression in Wheat Tissues
2.3.2. Gene Expression in Wheat Developmental Stages
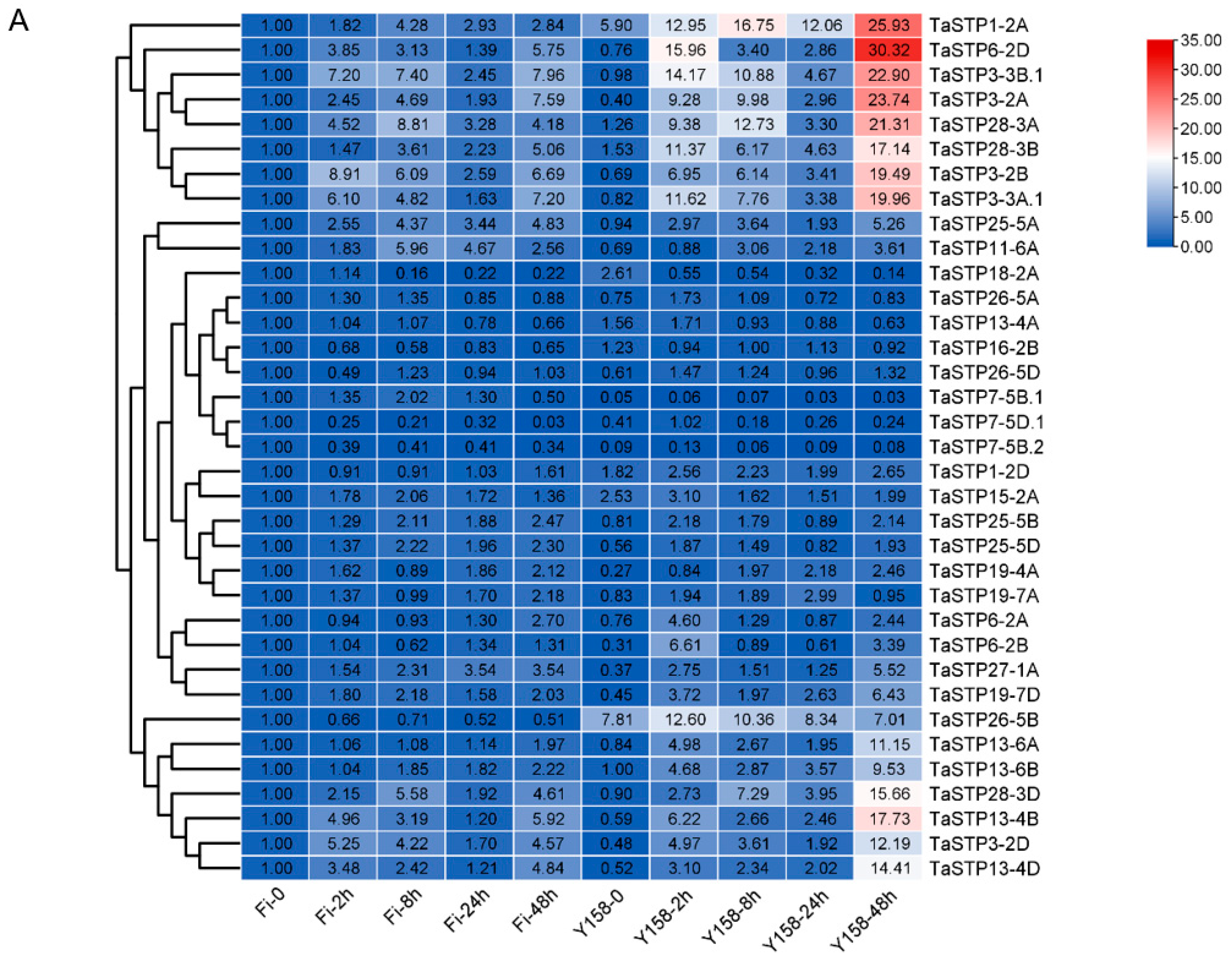
2.3.3. Gene Expression Under Abiotic, Biotic, and Hormonal Stress Conditions
2.4. FHB Resistance of the Fielder and Yangmai 158
2.5. Validation of the Expression of Wheat TaSTP Genes by RT-qPCR
2.5.1. Chitin Treatment
2.5.2. F. graminearum Infection
2.5.3. DON Treatment
3. Discussion
3.1. Expansion and Conservation of the TaSTP Gene Family in Wheat
3.2. Expression Patterns of TaSTP Genes Hint at Functional Diversification
3.3. TaSTP Genes Are Potentially Pivotal in Wheat’s Response to FHB
3.4. Limitations and Future Perspectives
4. Materials and Methods
4.1. Database Mining and Identification of TaSTP Genes in Wheat
4.2. Phylogenetic Analysis, Motif Prediction, and Conserved Domain Analysis
4.3. Naming of TaSTP Genes
4.4. Plant Materials, Growth Conditions, and Stress Treatments
4.5. F. graminearum, Chitin, and DON Configuration
4.6. RNA Extraction, Reverse Transcriptase, and RT-qPCR
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di, P.A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Mesterhazy, A. What Is Fusarium Head Blight (FHB) Resistance and What Are Its Food Safety Risks in Wheat? Problems and Solutions—A Review. Toxins 2024, 16, 31. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhang, X.; Yao, J.; Cheng, S. Breeding for the resistance to Fusarium head blight of wheat in China. Front. Agric. Sci. Eng. 2019, 6, 251–264. [Google Scholar] [CrossRef]
- Cainong, J.C.; Bockus, W.W.; Feng, Y.; Chen, P.; Qi, L.; Sehgal, S.K.; Danilova, T.V.; Koo, D.H.; Friebe, B.; Gill, B.S. Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor. Appl. Genet. 2015, 128, 1019–1027. [Google Scholar] [CrossRef]
- Zhou, M.P.; Hayden, M.J.; Zhang, Z.Y.; Lu, W.Z.; Ma, H.X. Saturation and mapping of a major Fusarium head blight resistance QTL on chromosome 3BS of Sumai 3 wheat. J. Appl. Genet. 2010, 51, 19–25. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selected for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Ollier, M.; Talle, V.; Brisset, A.L.; Le, B.Z.; Duerr, S.; Lemmens, M.; Goudemand, E.; Robert, O.; Hilbert, J.L.; Buerstmayr, H. QTL mapping and successful introgression of the spring wheat-derived QTL Fhb1 for Fusarium head blight resistance in three European triticale populations. Theor. Appl. Genet. 2020, 133, 457–477. [Google Scholar] [CrossRef]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef]
- Li, G.; Zhou, J.; Jia, H.; Gao, Z.; Fan, M.; Luo, Y.; Zhao, P.; Xue, S.; Li, N.; Yuan, Y.; et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 2019, 51, 1106–1112. [Google Scholar] [CrossRef]
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 2019, 51, 1099–1105. [Google Scholar] [CrossRef]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liu, Y.; Zhao, X.; Zhang, S.; Ma, H. Exploring and applying genes to enhance the resistance to Fusarium head blight in wheat. Front. Plant Sci. 2022, 13, 1026611. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, X.; Xia, X.; Wang, Y.; Dong, Y.; Wu, L.; Jiang, P.; Zhang, X.; Jiang, C.; Ma, H.; et al. A phase-separated protein hub modulates resistance to Fusarium head blight in wheat. Cell Host Microbe 2024, 32, 710–726.e10. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Manuka, R.; Penna, S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol. Plant. 2021, 171, 739–755. [Google Scholar] [CrossRef]
- Nägele, T.; Gibon, Y.; Le, H.R. Plant sugar metabolism, transport and signalling in challenging environments. Physiol. Plant. 2022, 174, e13768. [Google Scholar] [CrossRef]
- Mishra, B.S.; Sharma, M.; Laxmi, A. Role of sugar and auxin crosstalk in plant growth and development. Physiol. Plant. 2022, 174, e13546. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Doidy, J.; Grace, E.; Kühn, C.; Simon-Plas, F.; Casieri, L.; Wipf, D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012, 17, 413–422. [Google Scholar] [CrossRef]
- Doidy, J.; Vidal, U.; Lemoine, R. Sugar transporters in Fabaceae, featuring SUT MST and SWEET families of the model plant Medicago truncatula and the agricultural crop Pisum sativum. PLoS ONE 2019, 14, e0223173. [Google Scholar] [CrossRef]
- Kanwar, P.; Jha, G. Alterations in plant sugar metabolism: Signatory of pathogen attack. Planta 2019, 249, 305–318. [Google Scholar] [CrossRef]
- Liu, Y.H.; Song, Y.H.; Ruan, Y.L. Sugar conundrum in plant-pathogen interactions: Roles of invertase and sugar transporters depend on pathosystems. J. Exp. Bot. 2022, 73, 1910–1925. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant. 2021, 171, 620–637. [Google Scholar] [CrossRef]
- Hu, Z.; Tang, Z.; Zhang, Y.; Niu, L.; Yang, F.; Zhang, D.; Hu, Y. Rice SUT and SWEET Transporters. Int. J. Mol. Sci. 2021, 22, 11198. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, Y.; Yang, X.; Tang, L.; Wan, C.; Liu, J.; Chen, C.; Zhang, H.; He, C.; Liu, C.; et al. MATE transporter GFD1 cooperates with sugar transporters, mediates carbohydrate partitioning and controls grain-filling duration, grain size and number in rice. Plant Biotechnol. J. 2023, 21, 621–634. [Google Scholar] [CrossRef]
- Truernit, E.; Schmid, J.; Epple, P.; Illig, J.; Sauer, N. The sink-specific and stress-regulated Arabidopsis STP4 gene: Enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 1996, 8, 2169–2182. [Google Scholar] [CrossRef]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef]
- Büttner, M. The Arabidopsis sugar transporter (AtSTP) family: An update. Plant Biol. 2010, 12, 35–41. [Google Scholar] [CrossRef]
- Lemonnier, P.; Gaillard, C.; Veillet, F.; Verbeke, J.; Lemoine, R.; Coutos-Thévenot, P.; La, C.S. Expression of Arabidopsis sugar transport protein STP13 differentially affects glucose transport activity and basal resistance to Botrytis cinerea. Plant Mol. Biol. 2014, 85, 473–484. [Google Scholar] [CrossRef]
- Norholm, M.H.; Nour-Eldin, H.H.; Brodersen, P.; Mundy, J.; Halkier, B.A. Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS Lett. 2006, 580, 2381–2387. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Saijo, Y.; Nakagami, H.; Takano, Y. Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science 2016, 354, 1427–1430. [Google Scholar] [CrossRef] [PubMed]
- Fotopoulos, V.; Gilbert, M.J.; Pittman, J.K.; Marvier, A.C.; Buchanan, A.J.; Sauer, N.; Hall, J.L.; Williams, L.E. The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, Atbetafruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol. 2003, 132, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef]
- Huai, B.; Yang, Q.; Wei, X.; Pan, Q.; Kang, Z.; Liu, J. TaSTP13 contributes to wheat susceptibility to stripe rust possibly by increasing cytoplasmic hexose concentration. BMC Plant Biol. 2020, 20, 49. [Google Scholar] [CrossRef]
- Huai, B.; Yuan, P.; Ma, X.; Zhang, X.; Jiang, L.; Zheng, P.; Yao, M.; Chen, Z.; Chen, L.; Shen, Q.; et al. Sugar transporter TaSTP3 activation by TaWRKY19/61/82 enhances stripe rust susceptibility in wheat. New Phytol. 2022, 236, 266–282. [Google Scholar] [CrossRef]
- Huai, B.; Yang, Q.; Qian, Y.; Qian, W.; Kang, Z.; Liu, J. ABA-Induced Sugar Transporter TaSTP6 Promotes Wheat Susceptibility to Stripe Rust. Plant Physiol. 2019, 181, 1328–1343. [Google Scholar] [CrossRef]
- Liu, H.; Li, C.; Qiao, L.; Hu, L.; Wang, X.; Wang, J.; Ruan, X.; Yang, G.; Yin, G.; Wang, C.; et al. The Sugar Transporter family in wheat (Triticum aestivum L.): Genome-wide identification, classification, and expression profiling during stress in seedlings. PeerJ 2021, 9, e11371. [Google Scholar] [CrossRef]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Kong, W.; Li, Y. A Novel Insight into Functional Divergence of the MST Gene Family in Rice Based on Comprehensive Expression Patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; International Wheat Genome Sequencing Consortium; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Hao, Y.; Pan, Y.; Chen, W.; Rashid, M.A.R.; Li, M.; Che, N.; Duan, X.; Zhao, Y. Contribution of Duplicated Nucleotide-Binding Leucine-Rich Repeat (NLR) Genes to Wheat Disease Resistance. Plants 2023, 12, 2794. [Google Scholar] [CrossRef]
- Zhou, C.; Dong, Z.; Zhang, T.; Wu, J.; Yu, S.; Zeng, Q.; Han, D.; Tong, W. Genome-Scale Analysis of Homologous Genes among Subgenomes of Bread Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2020, 21, 3015. [Google Scholar] [CrossRef]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H., Jr. The major facilitator superfamily (MFS) revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef]
- Sun, L.; Zeng, X.; Yan, C.; Sun, X.; Gong, X.; Rao, Y.; Yan, N. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 2012, 490, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Poschet, G.; Hannich, B.; Büttner, M. Identification and characterization of AtSTP14, a novel galactose transporter from Arabidopsis. Plant Cell Physiol. 2010, 51, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Rottmann, T.; Klebl, F.; Schneider, S.; Kischka, D.; Rüscher, D.; Sauer, N.; Stadler, R. Sugar transporter STP7 specificity for L-arabinose and D-xylose contrasts with the typical hexose transporters STP8 and STP12. Plant Physiol. 2018, 176, 2330–2350. [Google Scholar] [CrossRef]
- Schneidereit, A.; Scholz-Starke, J.; Büttner, M. Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol. 2003, 133, 182–190. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.Z.; Song, L.F.; Zou, J.J.; Su, Z.; Wu, W.H. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008, 148, 1201–1211. [Google Scholar] [CrossRef]
- Bai, G.; Su, Z.; Cai, J. Wheat resistance to Fusarium head blight. Can. J. Plant Pathol. 2018, 40, 336–346. [Google Scholar] [CrossRef]
- Ma, H.; Liu, Y.; Zhang, S.; Sha, J.; Sun, Y.; Hu, Z.; Gong, L.; Dai, Y.; Gao, Y.; Wang, Y.; et al. Wheat resistance to Fusarium head blight and breeding strategies. Crop Health 2025, 3, 9. [Google Scholar] [CrossRef]
- Agriopoulou, S.; Stamatelopoulou, E.; Varzakas, T. Advances in Occurrence, Importance, and Mycotoxin Control Strategies: Prevention and Detoxification in Foods. Foods 2020, 9, 137. [Google Scholar] [CrossRef]
- He, Y.; Wu, L.; Liu, X.; Jiang, P.; Yu, L.; Qiu, J.; Wang, G.; Zhang, X.; Ma, H. TaUGT6, a Novel UDP-Glycosyltransferase Gene Enhances the Resistance to FHB and DON Accumulation in Wheat. Front. Plant Sci. 2020, 16, 574775. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kenneth, J.L.; Thomas, D.S. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, C.; Shen, S.; Zhou, S.; Li, Y.; Mao, Y.; Zhou, J.; Shi, Y.; An, L.; Zhou, Q.; Peng, W.; et al. Rice metabolic regulatory network spanning the entire life cycle. Mol. Plant 2022, 15, 258–275. [Google Scholar] [CrossRef]






| Gene Name | Fielder | Yangmai158 | Selected Genes | ||||
|---|---|---|---|---|---|---|---|
| 0 vs. 5 M | 0 vs. 15 M | 0 vs. 30 M | 0 vs. 5 M | 0 vs. 15 M | 0 vs. 30 M | ||
| TaSTP1-2A | 0.80 | 1.72 * | 1.77 * | 1.26 | 1.28 | 1.45 * | √ |
| TaSTP1-2D | 1.42 ** | 0.97 | 0.67 ** | 1.86 * | 2.63 ** | 1.96 ** | √ |
| TaSTP3-2A | 0.85 | 1.79 ** | 3.76 * | 1.55 ** | 1.80 * | 3.18 ** | √ |
| TaSTP3-2B | 1.11 | 1.96 * | 2.78 ** | 1.29 * | 2.27 ** | 3.33 ** | √ |
| TaSTP3-2D | 2.39 | 2.37 * | 6.11 ** | 2.10 * | 2.54 ** | 4.20 ** | √ |
| TaSTP3-3A.1 | 0.90 | 1.90 * | 2.21 * | 0.71 * | 1.22 * | 2.27 * | √ |
| TaSTP3-3B.1 | 0.95 | 0.94 | 0.72 ** | 1.32 ** | 1.55 ** | 1.32 * | √ |
| TaSTP6-2A | 1.54 | 2.54 ** | 1.23 | 1.03 | 1.88 ** | 3.39 ** | √ |
| TaSTP6-2B | 0.56 * | 0.37 ** | 0.32 ** | 3.08 ** | 5.38 ** | 5.71 ** | √ |
| TaSTP6-2D | 1.66 * | 7.71 * | 5.14 ** | 1.15 | 1.78 ** | 0.70 | √ |
| TaSTP7-5A.1 | 1.13 | 0.97 | 0.96 | 1.22 | 1.21 | 1.42 ** | |
| TaSTP7-5A.2 | 1.58 ** | 1.16 * | 0.69 * | 1.27 | 1.32 | 1.23 | |
| TaSTP7-5B.1 | 1.38 * | 1.39 ** | 1.35 * | 1.31 ** | 1.74 ** | 2.38 ** | √ |
| TaSTP7-5B.2 | 1.46 ** | 1.14 * | 1.05 | 1.50 ** | 1.44 ** | 2.10 ** | √ |
| TaSTP7-5D.1 | 1.43 * | 1.07 | 0.58 * | 1.53 ** | 1.25 * | 2.37 ** | √ |
| TaSTP10-1A.1 | 0.64 | 1.73 | 1.36 | 1.24 | 1.16 | 0.90 | |
| TaSTP11-6A | 0.96 | 1.34 | 1.21 | 1.05 | 0.58 * | 0.41 ** | √ |
| TaSTP13-4A | 2.32 ** | 6.07 ** | 4.31 ** | 2.02 * | 3.47 ** | 4.37 * | √ |
| TaSTP13-4B | 2.29 ** | 4.34 ** | 5.00 ** | 3.34 ** | 7.35 ** | 12.76 ** | √ |
| TaSTP13-4B.1 | 0.56 * | 0.39 ** | 0.43 * | 1.35 | 0.84 | 0.67 | |
| TaSTP13-4D | 1.46 ** | 2.61 ** | 3.22 ** | 2.19 ** | 3.96 ** | 5.61 ** | √ |
| TaSTP13-4D.1 | 0.75 | 0.68 | 0.30 ** | 0.95 | 0.60 * | 0.65 * | |
| TaSTP13-6A | 0.75 * | 0.96 * | 0.69 * | 1.55 ** | 2.28 ** | 1.01 | √ |
| TaSTP13-6B | 0.89 | 1.12 | 1.15 | 1.32 ** | 1.51 ** | 1.08 | √ |
| TaSTP14-5A | 0.74 * | 0.77 ** | 0.81 * | 1.05 | 0.94 | 0.73 ** | |
| TaSTP14-4B | 0.85 | 0.93 | 0.58 * | 0.62 | 0.48 | 0.58 | |
| TaSTP14-4D | 1.17 | 1.02 | 0.34 ** | 1.07 | 1.12 | 0.90 | |
| TaSTP15-2A | 2.07 * | 1.83 * | 2.00 * | 3.59 ** | 6.49 ** | 3.71 ** | √ |
| TaSTP15-2B | 0.58 ** | 0.86 * | 0.94 | 0.63 ** | 0.76 * | 0.98 | |
| TaSTP16-2B | 1.91 ** | 4.95 ** | 2.95 * | 1.62 | 2.75 ** | 5.10 ** | √ |
| TaSTP17-2A | 1.59 | 0.93 | 0.66 | 1.68 * | 1.12 | 1.34 * | |
| TaSTP17-2B.1 | 0.69 * | 0.69 * | 0.34 ** | 1.09 | 0.86 | 0.62 * | |
| TaSTP17-2D | 1.19 | 0.91 | 0.36 ** | 1.23 | 1.00 | 1.50 * | |
| TaSTP18-2A | 1.11 | 0.86 | 0.75 | 0.86 | 1.35 | 0.92 | √ |
| TaSTP18-2B | 1.02 | 0.84 * | 0.87 | 0.88 | 1.00 | 1.25 | |
| TaSTP18-2D | 1.04 | 1.06 | 1.12 | 0.77 * | 0.95 | 0.85 | |
| TaSTP19-4A | 3.82 ** | 3.47 ** | 2.66 ** | 1.51 * | 0.52 * | 0.09 * | √ |
| TaSTP19-7A | 0.58* | 0.67 * | 0.85 | 1.19 | 0.60 * | 0.39 ** | √ |
| TaSTP19-7D | 0.59 ** | 1.29 | 1.71 * | 0.60 | 0.53 * | 0.47 * | √ |
| TaSTP25-5A | 0.58 * | 0.71 | 1.07 | 1.32 | 1.39 | 1.13 | √ |
| TaSTP25-5B | 0.89 | 1.01 | 0.89 | 1.16 | 1.21 * | 1.03 | √ |
| TaSTP25-5D | 0.89 * | 1.03 | 0.91 * | 1.30 ** | 1.86 ** | 1.57 ** | √ |
| TaSTP26-5A | 2.01 ** | 5.83** | 6.03 ** | 3.01 ** | 13.69 ** | 28.11 ** | √ |
| TaSTP26-5B | 0.75 | 0.55** | 0.39 ** | 0.86 | 0.95 | 0.72 ** | √ |
| TaSTP26-5D | 1.48 | 3.40** | 2.38 ** | 2.93 ** | 1.54 ** | 0.71 ** | √ |
| TaSTP27-1A | 0.88 | 0.75 | 0.72 | 3.09 * | 5.39 * | 3.93 ** | √ |
| TaSTP28-1B.1 | 0.87 * | 0.91 | 0.67 | 2.05 ** | 1.84 * | 1.43 ** | |
| TaSTP28-3A | 2.53 * | 8.45* | 21.33 ** | 0.77 | 1.78 * | 3.71 * | √ |
| TaSTP28-3B | 1.30 * | 1.27* | 1.06 | 1.27 ** | 1.60 ** | 3.44 ** | √ |
| TaSTP28-3D | 0.97 | 2.07** | 2.93 ** | 1.88 ** | 1.99 * | 2.00 | √ |
| Gene Name | Fielder | Yangmai158 | Selected Genes | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 vs. 2 h | 0 vs. 8 h | 0 vs. 24 h | 0 vs. 48 h | 0 vs. 2 h | 0 vs. 8 h | 0 vs. 24 h | 0 vs. 48 h | ||
| TaSTP1-2A | 1.82 ** | 4.28 ** | 2.93 ** | 2.84 ** | 2.19 * | 2.84 ** | 2.04 * | 2.15 ** | |
| TaSTP1-2D | 0.91 | 0.91 | 1.03 | 1.61 * | 1.41 * | 1.22 * | 1.09 | 1.33 * | √ |
| TaSTP3-2A | 2.45 * | 4.69 * | 1.93 * | 7.59 * | 23.10 | 24.85 * | 7.37 ** | 8.02 * | √ |
| TaSTP3-2B | 8.91 ** | 6.09 ** | 2.59 ** | 6.69 ** | 10.01 ** | 8.85 ** | 4.91 ** | 5.72 ** | √ |
| TaSTP3-2D | 5.25 ** | 4.22 ** | 1.70 ** | 4.57 ** | 10.43 ** | 7.58 ** | 4.03 ** | 6.34 ** | √ |
| TaSTP3-3A.1 | 6.10 ** | 4.82 * | 1.63 | 7.20 ** | 14.20 ** | 9.49 * | 4.13 * | 5.90 ** | |
| TaSTP3-3B.1 | 7.20 ** | 7.40 ** | 2.45 ** | 7.96 ** | 14.40 ** | 11.06 ** | 4.75 ** | 4.90 ** | |
| TaSTP6-2A | 0.94 | 0.93 | 1.30 ** | 2.70 ** | 6.05 ** | 1.70 ** | 1.14 | 2.82 ** | √ |
| TaSTP6-2B | 1.04 | 0.62 ** | 1.34 ** | 1.31 | 21.37 ** | 2.88 ** | 1.97 ** | 5.56 ** | √ |
| TaSTP6-2D | 3.85 ** | 3.13 ** | 1.39 | 5.75 ** | 20.91 ** | 4.45 ** | 3.75 ** | 10.61 * | √ |
| TaSTP7-5B.1 | 1.35 * | 2.02 ** | 1.30 * | 0.50 ** | 1.27 * | 1.43 | 0.65 | 0.85 | |
| TaSTP7-5D.1 | 0.25 * | 0.21 * | 0.32 * | 0.03 * | 2.46 ** | 0.43 ** | 0.63 * | 0.91 ** | |
| TaSTP7-5B.2 | 0.39 * | 0.41 * | 0.41 * | 0.34 ** | 1.41 ** | 0.66 ** | 1.02 | 0.87 * | |
| TaSTP11-6A | 1.83 ** | 5.96 * | 4.67 ** | 2.56 | 1.27 | 4.42 ** | 3.14 * | 1.66 ** | |
| TaSTP13-4A | 1.04 | 1.07 | 0.78 | 0.66 | 1.10 | 0.60 | 0.56 | 0.72 | √ |
| TaSTP13-4B | 4.96 ** | 3.19 ** | 1.20 * | 5.92 ** | 10.47 ** | 4.47** | 4.14 ** | 7.21 ** | √ |
| TaSTP13-4D | 3.48 * | 2.42 ** | 1.21 * | 4.84 ** | 5.93 ** | 4.48 ** | 3.87 ** | 7.12 ** | √ |
| TaSTP13-6A | 1.06 | 1.08 | 1.14 | 1.97 * | 5.95 ** | 3.19 ** | 2.33 * | 5.73 ** | √ |
| TaSTP13-6B | 1.04 | 1.85 * | 1.82 * | 2.22 ** | 4.66 * | 2.86 ** | 3.55 ** | 2.67 ** | √ |
| TaSTP15-2A | 1.78 ** | 2.06 ** | 1.72 ** | 1.36 ** | 1.23 * | 0.64 ** | 0.60 ** | 1.32 * | √ |
| TaSTP16-2B | 0.68 ** | 0.58 ** | 0.83 ** | 0.65 * | 0.76 * | 0.81 * | 0.92 | 0.81 * | |
| TaSTP18-2A | 1.14 | 0.16 ** | 0.22 ** | 0.22 ** | 0.21 ** | 0.21 ** | 0.12 ** | 0.42 ** | |
| TaSTP19-4A | 1.62 * | 0.89 | 1.86 ** | 2.12 ** | 3.16 * | 7.42 ** | 8.19 ** | 1.13 ** | √ |
| TaSTP19-7D | 1.80 ** | 2.18 * | 1.58 * | 2.03 * | 8.19 ** | 4.33 ** | 5.80 ** | 2.44 ** | √ |
| TaSTP19-7A | 1.37 * | 0.99 | 1.70 * | 2.18 * | 2.32 ** | 2.27 ** | 3.59 ** | 0.32 ** | √ |
| TaSTP25-5A | 2.55 ** | 4.37 ** | 3.44 ** | 4.83 * | 3.16 ** | 3.86 ** | 2.05 ** | 2.72 ** | |
| TaSTP25-5B | 1.29 * | 2.11 ** | 1.88 ** | 2.47 ** | 2.68 * | 2.21 ** | 1.09 | 2.42 * | |
| TaSTP25-5D | 1.37 ** | 2.22 ** | 1.96 ** | 2.30** | 3.35 ** | 2.66 ** | 1.47 ** | 2.34 ** | |
| TaSTP26-5A | 1.30 | 1.35 * | 0.85 | 0.88 | 2.30 ** | 1.45 ** | 0.96 | 1.16 | √ |
| TaSTP26-5B | 0.66 ** | 0.71 * | 0.52 ** | 0.51 ** | 1.61 * | 1.33 | 1.07 | 0.84 | √ |
| TaSTP26-5D | 0.49 ** | 1.23 ** | 0.94 * | 1.03 | 2.39 ** | 2.02 ** | 1.57 ** | 1.36 ** | √ |
| TaSTP27-1A | 1.54 * | 2.31 ** | 3.54 ** | 3.54 ** | 7.34 ** | 4.03 ** | 3.32 * | 4.44 ** | √ |
| TaSTP28-3A | 4.52 ** | 8.81 ** | 3.28 ** | 4.18 ** | 7.45 * | 10.12 ** | 2.62 * | 6.46 ** | √ |
| TaSTP28-3B | 1.47* | 3.61 ** | 2.23 * | 5.06 ** | 7.43 ** | 4.04 * | 3.03 * | 3.70 ** | |
| TaSTP28-3D | 2.15 * | 5.58 ** | 1.92 ** | 4.61 ** | 3.05 * | 8.13 ** | 4.41 ** | 3.97 ** | √ |
| Gene Name | Fielder | Yangmai 15 | ||||
|---|---|---|---|---|---|---|
| 0 vs. 0.5 h | 0 vs. 2 h | 0 vs. 6 h | 0 vs. 0.5 h | 0 vs. 2 h | 0 vs. 6 h | |
| TaSTP1-2D | 0.34 ** | 0.91 | 2.71 ** | 1.31 * | 0.79 * | 1.85 * |
| TaSTP3-2A | 0.38 ** | 0.78 ** | 2.10 * | 4.66 ** | 9.71 ** | 53.65 ** |
| TaSTP3-2B | 0.41 ** | 2.76 * | 8.17 ** | 4.76 ** | 12.85 ** | 94.78 ** |
| TaSTP3-2D | 1.09 | 2.65 ** | 7.21 ** | 2.99 ** | 6.59 * | 24.64 ** |
| TaSTP6-2A | 0.30 ** | 2.68 * | 1.21 * | 2.02 * | 2.28 ** | 3.93 ** |
| TaSTP6-2B | 0.13 ** | 0.66 * | 0.40 * | 3.40 ** | 6.36 ** | 21.23 ** |
| TaSTP6-2D | 0.85 | 1.25 | 1.38 | 2.93 * | 4.20 ** | 14.85 ** |
| TaSTP13-4A | 1.00 | 2.41 ** | 3.37 ** | 3.09 ** | 4.62 | 20.61 ** |
| TaSTP13-4B | 1.23 | 4.07 ** | 4.80 ** | 5.15 ** | 12.43 ** | 26.07 ** |
| TaSTP13-4D | 0.88 | 3.98 ** | 5.82 ** | 2.53 ** | 7.36 ** | 29.16 ** |
| TaSTP13-6A | 0.27 * | 0.20 ** | 0.22 ** | 1.07 | 1.11 * | 1.19 |
| TaSTP13-6B | 0.41 ** | 0.36 ** | 0.15 ** | 1.74 * | 1.16 | 1.10 * |
| TaSTP15-2A | 0.34 * | 0.56 ** | 0.49 * | 1.47 * | 1.26 | 2.76 ** |
| TaSTP19-4A | 1.39 | 17.39 ** | 2.13 * | 1.37 | 1.67 | 36.28 ** |
| TaSTP19-7A | 1.59 | 18.13 * | 1.57 ** | 1.28 * | 1.27 | 23.24 ** |
| TaSTP19-7D | 0.28 * | 3.99 * | 7.75 ** | 0.6 | 2.01 * | 12.23 ** |
| TaSTP26-5A | 1.60 ** | 7.82 ** | 2.82 ** | 7.95 ** | 12.01 ** | 4.59 ** |
| TaSTP26-5B | 1.12 | 0.40 * | 1.88 | 1.03 | 1.01 | 0.77 |
| TaSTP26-5D | 1.06 | 1.23 * | 0.41 ** | 4.00 ** | 1.33 * | 0.31 ** |
| TaSTP27-1A | 0.59 ** | 0.49 ** | 0.32 ** | 1.85 | 3.69 ** | 15.27 ** |
| TaSTP28-3A | 5.68 | 141.87 * | 206.36 ** | 0.85 | 14.34 ** | 59.54 ** |
| TaSTP28-3D | 0.38 * | 11.95 ** | 11.67 ** | 1.1 | 18.55 ** | 34.79 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Sha, J.; Zhang, S.; Sun, Y.; Hu, Z.; Ma, H.; Ma, H. Genome-Wide Identification and Expression Profiling of Sugar Transport Protein Response to Fusarium Head Blight in Wheat (Triticum aestivum L.). Plants 2025, 14, 2976. https://doi.org/10.3390/plants14192976
Liu Y, Sha J, Zhang S, Sun Y, Hu Z, Ma H, Ma H. Genome-Wide Identification and Expression Profiling of Sugar Transport Protein Response to Fusarium Head Blight in Wheat (Triticum aestivum L.). Plants. 2025; 14(19):2976. https://doi.org/10.3390/plants14192976
Chicago/Turabian StyleLiu, Yongjiang, Jianfeng Sha, Suhong Zhang, Yawen Sun, Zhiruo Hu, Haigang Ma, and Hongxiang Ma. 2025. "Genome-Wide Identification and Expression Profiling of Sugar Transport Protein Response to Fusarium Head Blight in Wheat (Triticum aestivum L.)" Plants 14, no. 19: 2976. https://doi.org/10.3390/plants14192976
APA StyleLiu, Y., Sha, J., Zhang, S., Sun, Y., Hu, Z., Ma, H., & Ma, H. (2025). Genome-Wide Identification and Expression Profiling of Sugar Transport Protein Response to Fusarium Head Blight in Wheat (Triticum aestivum L.). Plants, 14(19), 2976. https://doi.org/10.3390/plants14192976






