Genome-Wide Characterization of the Von Willebrand Factor a Gene Family in Wheat: Highlights Their Functional Roles in Growth and Biotic Stress Response
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Property Analysis of TavWA Genes
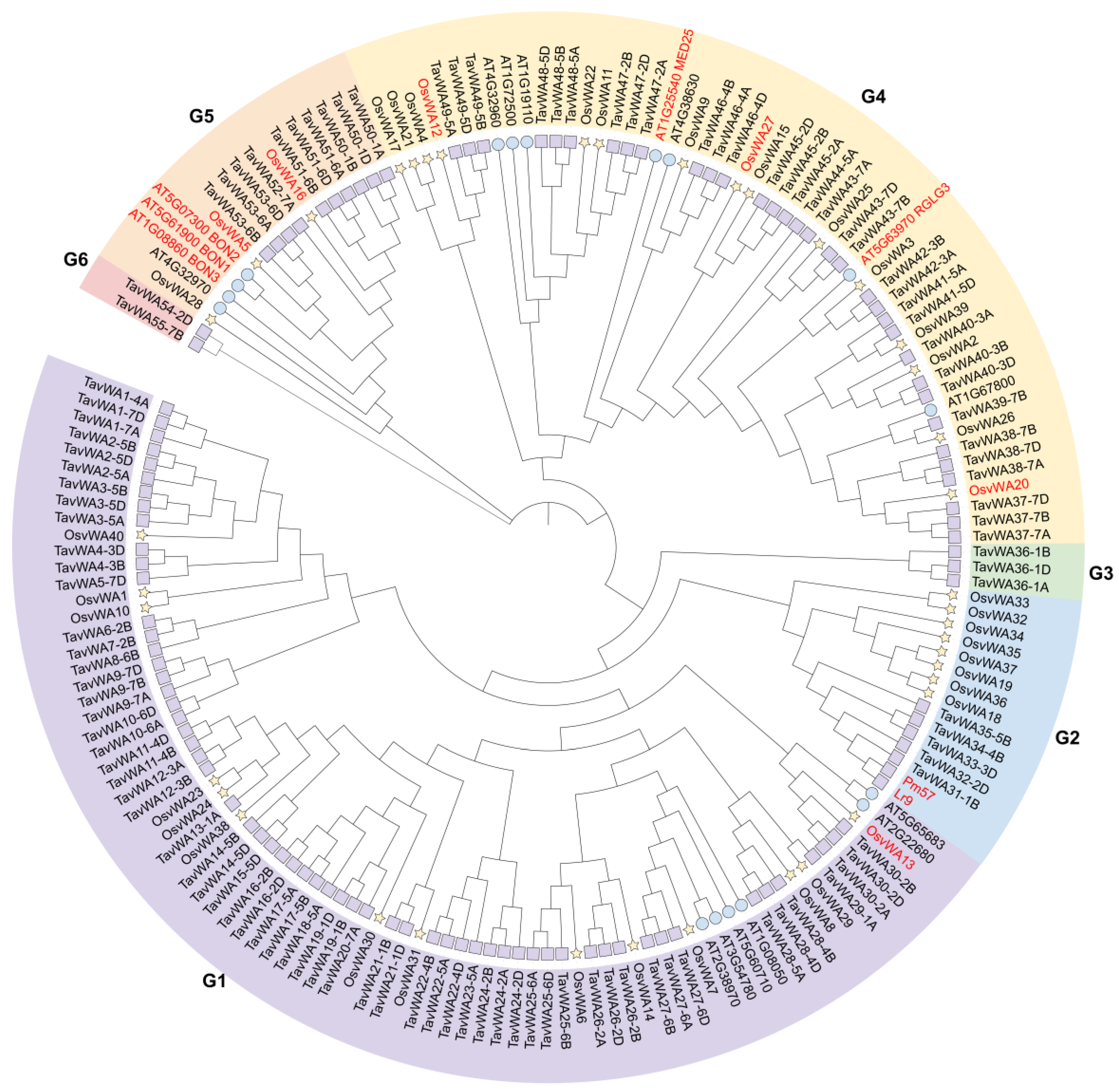
2.2. Phylogenetic Relation and Classification of TavWAs
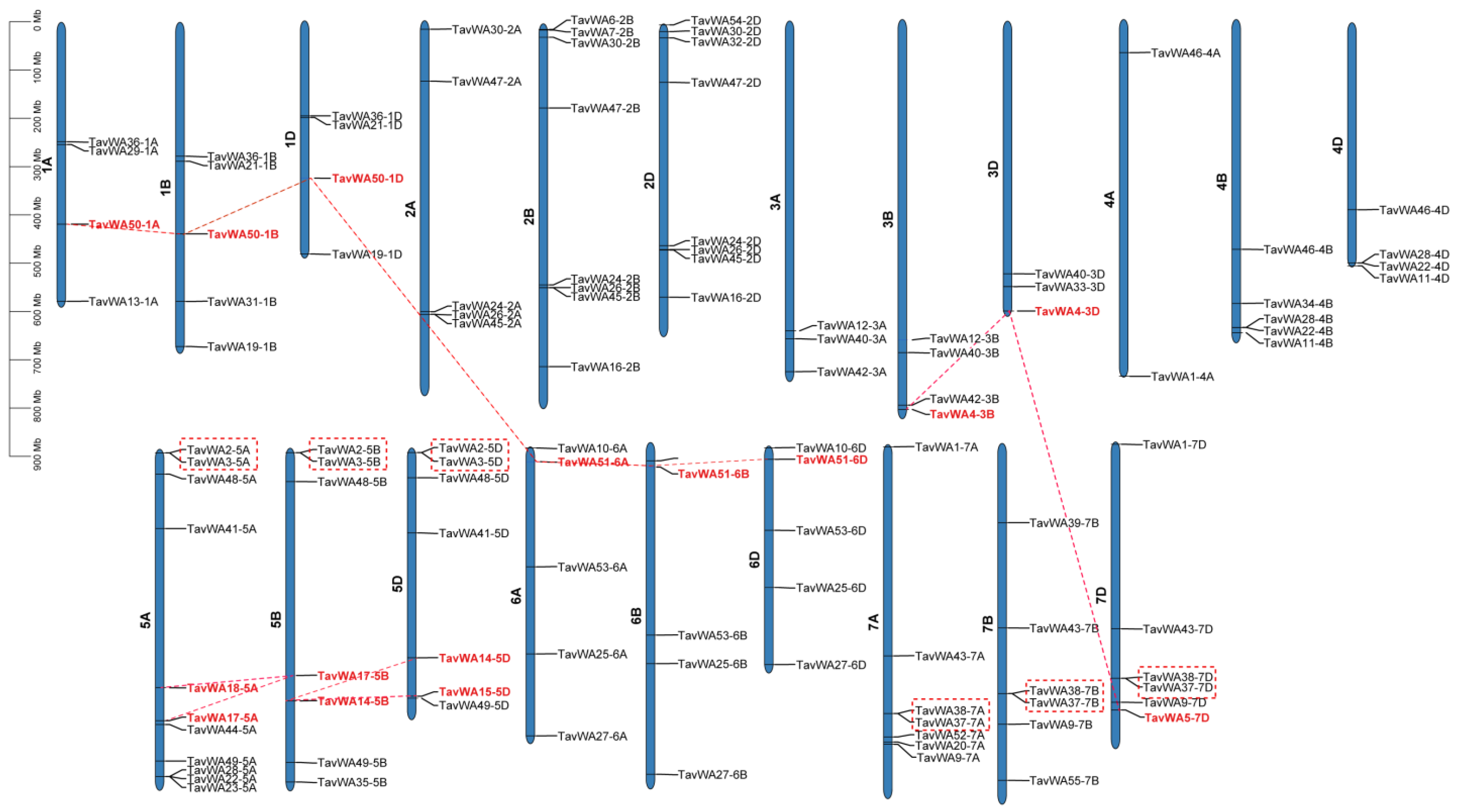
2.3. Chromosomal Distribution and Gene Duplication of TavWA Genes
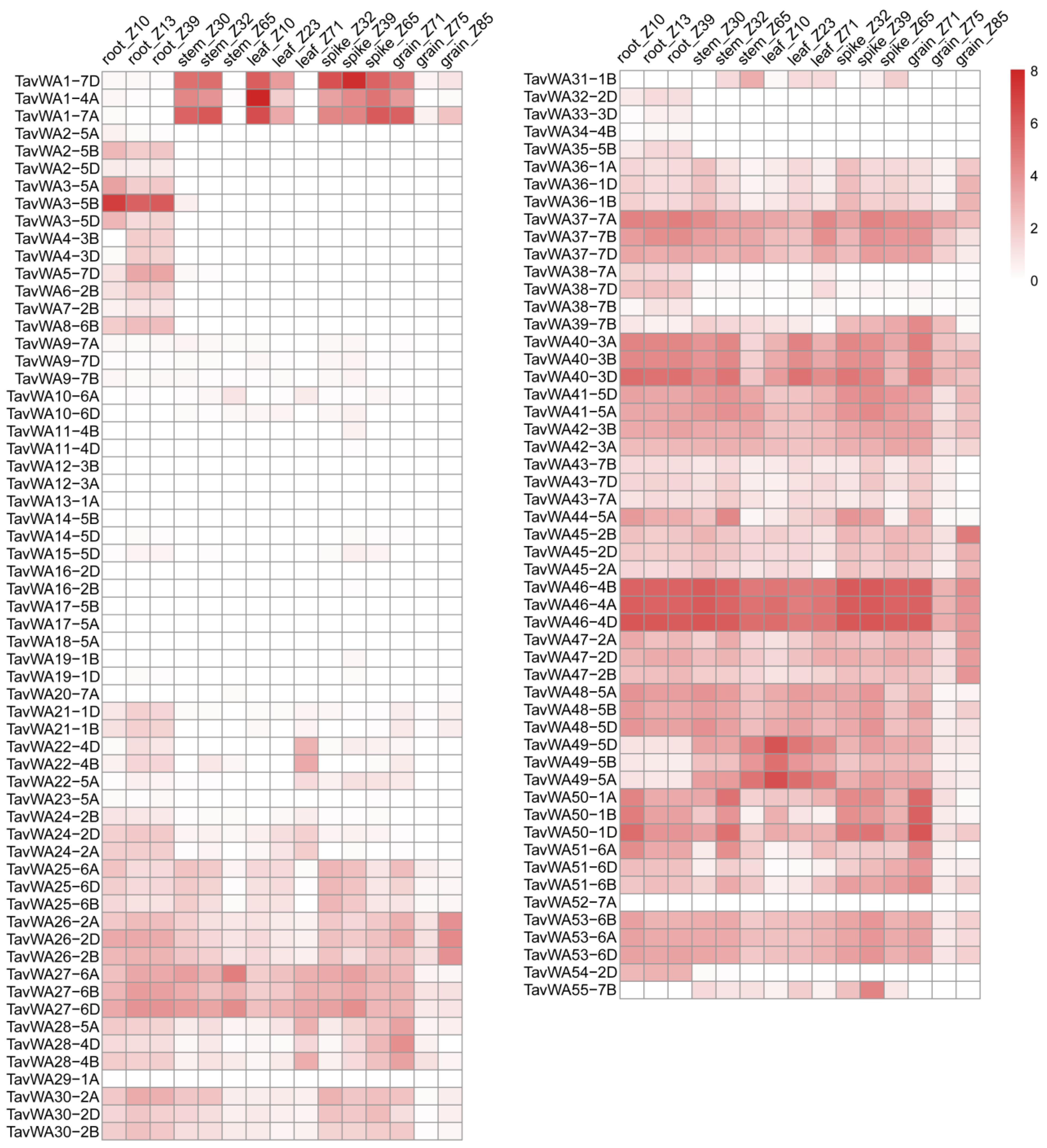
2.4. Expression Analysis of TavWA Genes in Various Wheat Tissues
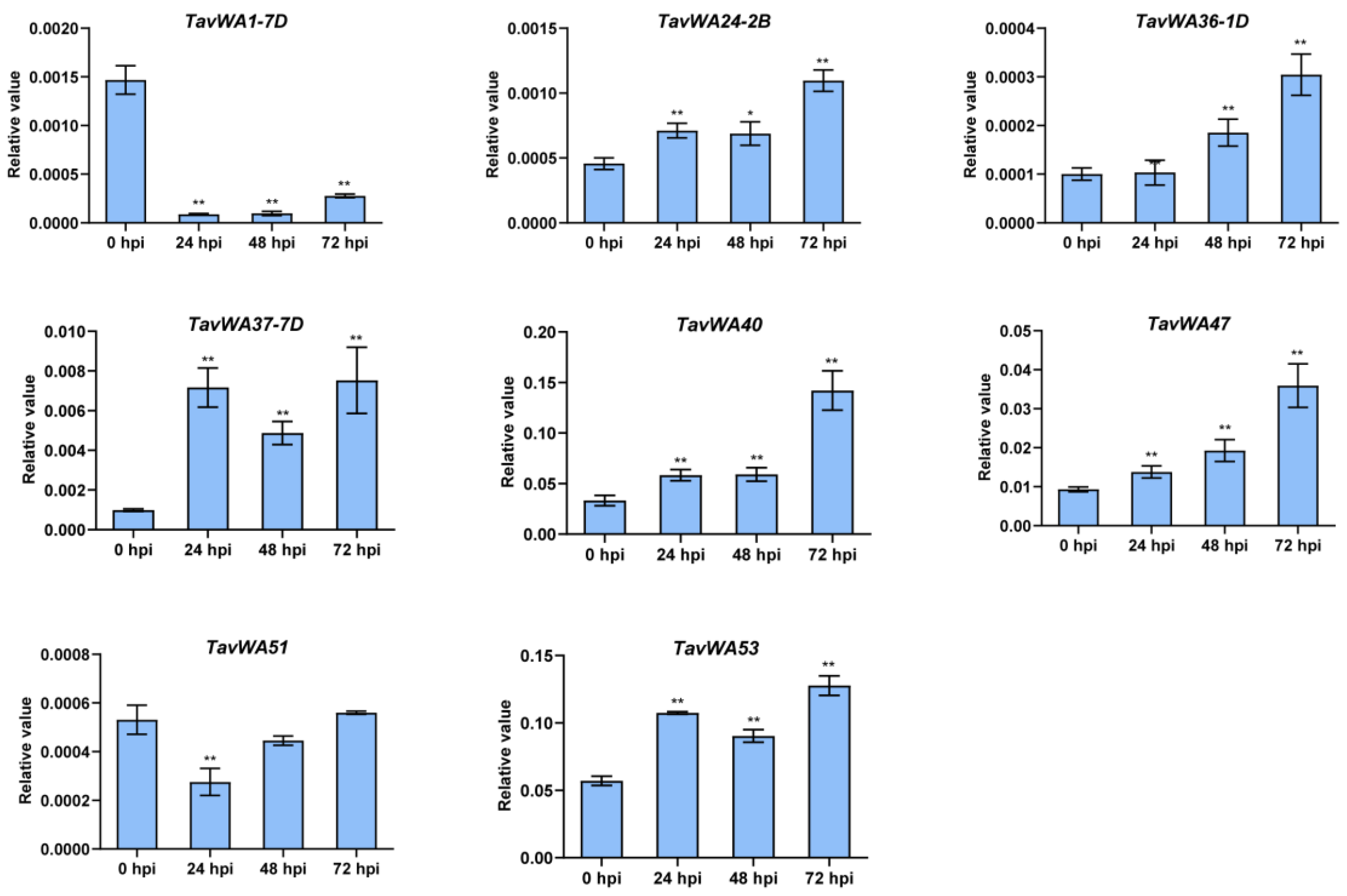
2.5. Expression Patterns of TavWA Genes Under Biotic Stresses
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. Database Search and Structural Analysis of vWA Domain-Containing Proteins in Wheat
4.3. Physicochemical Property Analysis and Subcellular Localization of TavWA Proteins
4.4. Physical Locations, Multiple Sequence Alignment, and Construction of Phylogenetic Tree
4.5. Gene Duplication and Ka/Ks Analysis
4.6. Expression Analysis of TavWA Genes During Plant Development and Biotic Stress
4.7. Plant Materials and Stress Treatments
4.8. RNA Extraction and RT-qPCR Analysis
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Colombatti, A.; Bonaldo, P.; Doliana, R. Type a modules: Interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix 1993, 13, 297–306. [Google Scholar] [CrossRef]
- Whittaker, C.A.; Hynes, R.O. Distribution and evolution of von willebrand/integrin a domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 2002, 13, 3369–3387. [Google Scholar] [CrossRef] [PubMed]
- Tuckwell, D. Evolution of von Willebrand factor A (vWA) domains. Biochem. Soc. Trans. 1999, 27, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Karkute, S.G.; Kumar, V.; Tasleem, M.; Mishra, D.C.; Chaturvedi, K.K.; Rai, A.; Sevanthi, A.M.; Gaikwad, K.; Sharma, T.R.; Solanke, A.U. Genome-wide analysis of von willebrand factor a gene family in rice for its role in imparting biotic stress resistance with emphasis on rice blast disease. Rice Sci. 2022, 29, 375–384. [Google Scholar] [CrossRef]
- Bharati, K.P.; Prashanth, U.R. Von willebrand disease: An overview. Indian J. Pharm. Sci. 2011, 73, 7–16. [Google Scholar] [CrossRef]
- Yang, S.; Yang, H.; Grisafi, P.; Sanchatjate, S.; Fink, G.R.; Sun, Q.; Hua, J. The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant. J. 2006, 45, 166–179. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, P.; Zhang, X.; Ren, D.; Yang, S. BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 2011, 67, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Hussein, N.K.; Sabr, L.J.; Lobo, E.; Booth, J.; Ariens, E.; Detchanamurthy, S.; Schenk, P.M. Suppression of Arabidopsis mediator subunit-encoding MED18 confers broad resistance against DNA and RNA viruses while MED25 is required for virus defense. Front. Plant Sci. 2020, 11, 162. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Q.; Cui, S.; Ren, J.; Qian, W.; Yang, Y.; He, S.; Chu, J.; Sun, X.; Yan, C.; et al. Hijacking of the jasmonate pathway by the mycotoxin fumonisin B1 (FB1) to initiate programmed cell death in Arabidopsis is modulated by RGLG3 and RGLG4. J. Exp. Bot. 2015, 66, 2709–2721. [Google Scholar] [CrossRef]
- Yin, X.; Zou, B.; Hong, X.; Gao, M.; Yang, W.; Zhong, X.; He, Y.; Kuai, P.; Lou, Y.; Huang, J.; et al. Rice copine genes OsBON1 and OsBON3 function as suppressors of broad-spectrum disease resistance. Plant Biotechnol. J. 2018, 16, 1476–1487. [Google Scholar] [CrossRef]
- Ren, Y.; Tian, X.; Li, S.; Mei, E.; He, M.; Tang, J.; Xu, M.; Li, X.; Wang, Z.; Li, C.; et al. Oryza sativa mediator subunit OsMED25 interacts with OsBZR1 to regulate brassinosteroid signaling and plant architecture in rice. J. Integr. Plant Biol. 2020, 62, 793–811. [Google Scholar] [CrossRef]
- Thangasamy, S.; Chen, P.; Lai, M.; Chen, J.; Jauh, G. Rice LGD1 containing RNA binding activity affects growth and development through alternative promoters. Plant J. 2012, 71, 288–302. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Yoo, J.H.; Yoo, S.C.; Cho, S.H.; Koh, H.J.; Seo, H.S.; Paek, N.C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef]
- Li, Y.; Ren, M.; Wu, Y.; Wang, L.; Zhao, K.; Gao, H.; Li, M.; Liu, Y.; Zhu, J.; Xu, J.; et al. A root system architecture regulator modulates OsPIN2 polar localization in rice. Nat. Commun. 2025, 16, 15. [Google Scholar] [CrossRef]
- Liu, Z.; Qiu, J.; Shen, Z.; Wang, C.; Jiang, N.; Shi, H.; Kou, Y. The E3 ubiquitin ligase OsRGLG5 targeted by the Magnaporthe oryzae effector AvrPi9 confers basal resistance against rice blast. Plant Commun. 2023, 4, 100626. [Google Scholar] [CrossRef]
- Jing, T.; Wu, Y.; Yu, Y.; Li, J.; Mu, X.; Xu, L.; Wang, X.; Qi, G.; Tang, J.; Wang, D.; et al. Copine proteins are required for brassinosteroid signaling in maize and Arabidopsis. Nat. Commun. 2024, 15, 2028. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Abrouk, M.; Gourdoupis, S.; Koo, D.H.; Karafiatova, M.; Molnar, I.; Holusova, K.; Dolezel, J.; Athiyannan, N.; Cavalet-Giorsa, E.; et al. An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet. 2023, 55, 914–920. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, Z.; Miao, J.; Liu, Q.; Ma, C.; Tian, X.; He, J.; Bi, H.; Yao, W.; Li, T.; et al. Pm57 from Aegilops searsii encodes a tandem kinase protein and confers wheat powdery mildew resistance. Nat. Commun. 2024, 15, 4796. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Li, Y.; Peng, L.; Shen, C.; Lu, Y.; Teng, W.; Liu, Y.; Wang, Y.; Zhu, W.; Liu, C.; et al. TavWA1 is critical for wheat growth by modulating cell morphology and arrangement. J. Integr. Plant Biol. 2025, 67, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xiao, H.; Wang, Y.; Li, W.; Li, L.; Dong, L.; Zhao, X.; Li, M.; Lu, P.; Zhang, H.; et al. WPA1 encodes a vWA domain protein that regulates wheat plant architecture. Crop J. 2024, 12, 992–1000. [Google Scholar] [CrossRef]
- Zhou, C.; Xiong, H.; Jia, Y.; Guo, H.; Fu, M.; Xie, Y.; Zhao, L.; Gu, J.; Li, H.; Li, Y.; et al. Identification of a von Willebrand factor type A protein affecting both grain and flag leaf morphologies in wheat. Sci. China Life Sci. 2024, 67, 2283–2286. [Google Scholar] [CrossRef]
- Bai, S.; Wang, G.; Song, R.; Liu, Y.; Hua, L.; Yang, J.; Zhang, L.; Ur Rehman, S.; Hao, X.; Hou, L.; et al. Mutations in wheat TaAPA2 gene result in pleiotropic effects on plant architecture. Sci. China Life Sci. 2024, 67, 2039–2042. [Google Scholar] [CrossRef]
- Mao, L. One bird, multiple stones: The race to find a gene of dominant negative effect in wheat. Crop J. 2024, 12, 951–952. [Google Scholar] [CrossRef]
- Ma, C.; Tian, X.; Dong, Z.; Li, H.; Chen, X.; Liu, W.; Yin, G.; Ma, S.; Zhang, L.; Cao, A.; et al. An Aegilops longissima NLR protein with integrated CC-BED module mediates resistance to wheat powdery mildew. Nat. Commun. 2024, 15, 8281. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Feng, J.; Zhang, Q.; Wang, Y.; Guan, Y.; Wang, R.; Shi, F.; Zeng, F.; Wang, Y.; Chen, M.; et al. Integrative gene duplication and genome-wide analysis as an approach to facilitate wheat reverse genetics: An example in the TaCIPK family. J. Adv. Res. 2024, 61, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yao, Y.; Xin, M.; Peng, H.; Ni, Z.; Sun, Q. Shaping polyploid wheat for success: Origins, domestication, and the genetic improvement of agronomic traits. J. Integr. Plant Biol. 2022, 64, 536–563. [Google Scholar] [CrossRef]
- Bi, H.; Liu, Z.; Liu, S.; Qiao, W.; Zhang, K.; Zhao, M.; Wang, D. Genome-wide analysis of wheat xyloglucan endotransglucosylase/hydrolase (XTH) gene family revealed TaXTH17 involved in abiotic stress responses. BMC Plant Biol. 2024, 24, 640. [Google Scholar] [CrossRef]
- Berkman, P.J.; Skarshewski, A.; Manoli, S.; Lorenc, M.T.; Stiller, J.; Smits, L.; Lai, K.; Campbell, E.; Kubaláková, M.; Simková, H.; et al. Sequencing wheat chromosome arm 7BS delimits the 7BS/4AL translocation and reveals homoeologous gene conservation. Theor. Appl. Genet. 2012, 124, 423–432. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, H.; Fu, X.; Zhou, N.; Liu, M.; Bai, S.; Zhao, X.; Cheng, R.; Li, S.; Zhang, D. Identification and map-based cloning of an EMS-induced mutation in wheat gene TaSP1 related to spike architecture. Theor. Appl. Genet. 2024, 137, 119. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jambunathan, N.; McNellis, T.W. Transgenic expression of the von Willebrand A domain of the BONZAI1/ COPINE1 protein triggers a lesion-mimic phenotype in Arabidopsis. Planta 2005, 221, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Fang, N.; Wei, X.; Shen, L.; Yu, Y.; Li, M.; Yin, C.; He, W.; Guan, C.; Chen, H.; Zhang, H.; et al. Fine mapping of a panicle blast resistance gene pb-bd1 in japonica landrace bodao and its application in rice breeding. Rice 2019, 12, 18. [Google Scholar] [CrossRef]
- Reveguk, T.; Fatiukha, A.; Potapenko, E.; Reveguk, I.; Sela, H.; Klymiuk, V.; Li, Y.; Pozniak, C.; Wicker, T.; Coaker, G.; et al. Tandem kinase proteins across the plant kingdom. Nat. Genet. 2025, 57, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids. Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, R.; Xu, D.; Bi, H.; Xia, Z.; Peng, H. Genome-wide identification and analysis of the AP2 transcription factor gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 1286. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Steuernagel, B.; Witek, K.; Krattinger, S.G.; Ramirez-Gonzalez, R.H.; Wulff, B. Physical and transcriptional organisation of the bread wheat intracellular immune receptor repertoire. bioRxiv 2018. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Wang, C.; Liu, M.; Li, H.; Fu, Y.; Wang, Y.; Nie, Y.; Liu, X.; Ji, W. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genom. 2014, 15, 898. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Li, H.; Men, W.; Ma, C.; Liu, Q.; Dong, Z.; Tian, X.; Wang, C.; Liu, C.; Gill, H.S.; Ma, P.; et al. Wheat powdery mildew resistance gene Pm13 encodes a mixed lineage kinase domain-like protein. Nat. Commun. 2024, 15, 2449. [Google Scholar] [CrossRef]
- Zhao, Y.; He, J.; Liu, M.; Miao, J.; Ma, C.; Feng, Y.; Qian, J.; Li, H.; Bi, H.; Liu, W. The SPL transcription factor TaSPL6 negatively regulates drought stress response in wheat. Plant Physiol. Biochem. 2024, 206, 108264. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(t) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, L.; Yang, Z.; Han, K.; Ma, C.; Ren, Y.; Jia, R.; Li, H.; Liu, Q.; Zhao, Y.; Liu, W. Genome-Wide Characterization of the Von Willebrand Factor a Gene Family in Wheat: Highlights Their Functional Roles in Growth and Biotic Stress Response. Plants 2025, 14, 2965. https://doi.org/10.3390/plants14192965
Tao L, Yang Z, Han K, Ma C, Ren Y, Jia R, Li H, Liu Q, Zhao Y, Liu W. Genome-Wide Characterization of the Von Willebrand Factor a Gene Family in Wheat: Highlights Their Functional Roles in Growth and Biotic Stress Response. Plants. 2025; 14(19):2965. https://doi.org/10.3390/plants14192965
Chicago/Turabian StyleTao, Luna, Zheng Yang, Kai Han, Chao Ma, Yueming Ren, Ranran Jia, Huanhuan Li, Qianwen Liu, Yue Zhao, and Wenxuan Liu. 2025. "Genome-Wide Characterization of the Von Willebrand Factor a Gene Family in Wheat: Highlights Their Functional Roles in Growth and Biotic Stress Response" Plants 14, no. 19: 2965. https://doi.org/10.3390/plants14192965
APA StyleTao, L., Yang, Z., Han, K., Ma, C., Ren, Y., Jia, R., Li, H., Liu, Q., Zhao, Y., & Liu, W. (2025). Genome-Wide Characterization of the Von Willebrand Factor a Gene Family in Wheat: Highlights Their Functional Roles in Growth and Biotic Stress Response. Plants, 14(19), 2965. https://doi.org/10.3390/plants14192965





