Integrating Genetic Mapping and BSR-Seq Analysis to Identify Candidate Genes Controlling Fruitfulness in Camellia sinensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Method
2.2.1. Phenotypic Data Collection
2.2.2. Data Statistics and Analysis
2.2.3. QTL Mapping Analysis
2.2.4. Hybrid Pollination Treatment and Sampling
2.2.5. Pollen Tube Fluorescence Microscopy Observation
2.2.6. BSR-Seq Analysis
Sequence Data Analysis
Differential Expression Analysis and Enrichment Analysis
BSR-Seq Analysis for Fruit Number Traits
2.2.7. Combined Analysis of QTL and BSR-Seq
2.2.8. Validation of Differential Genes by qRT-PCR
2.2.9. Functional Analysis of the CsETR2
Overexpression of CsETR2 in Arabidopsis thaliana
Yeast One-Hybrid Assay
Dual-Luciferase Assay
3. Results
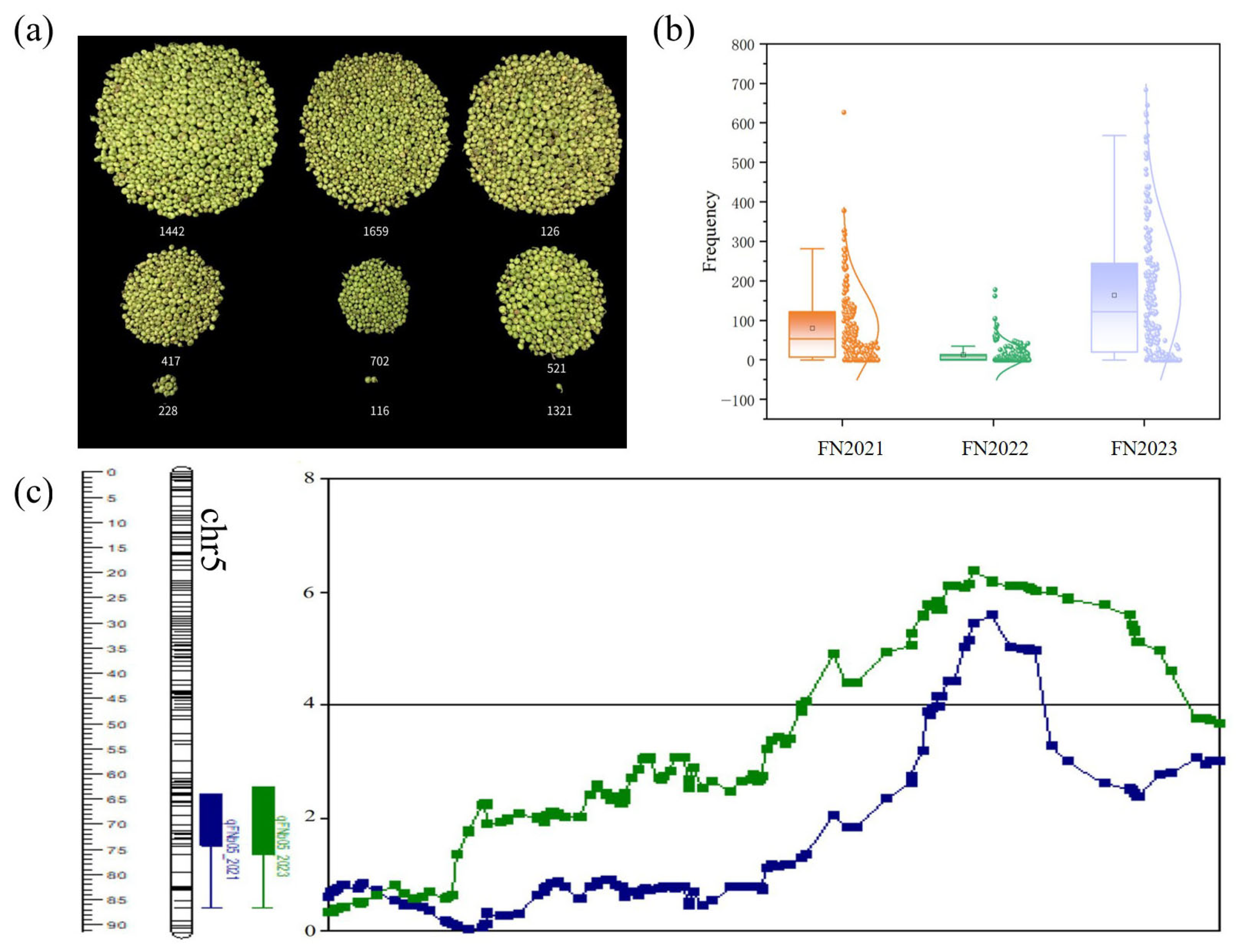
3.1. Tea Plant Fruit Number Related Parameter Phenotypic Data Collection
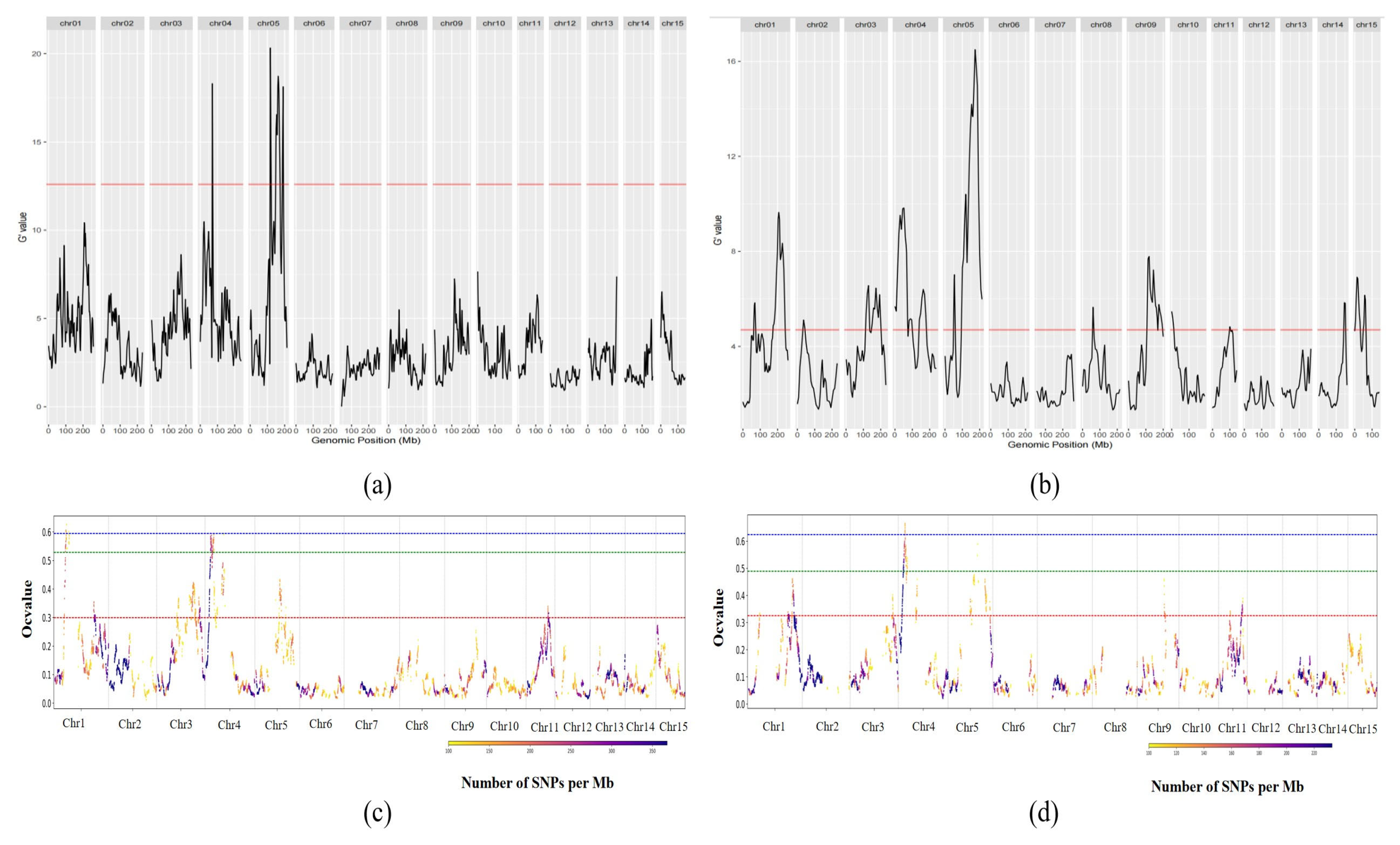
3.2. Analysis of QTL Mapping Results
3.3. BSR-Seq Result Analysis
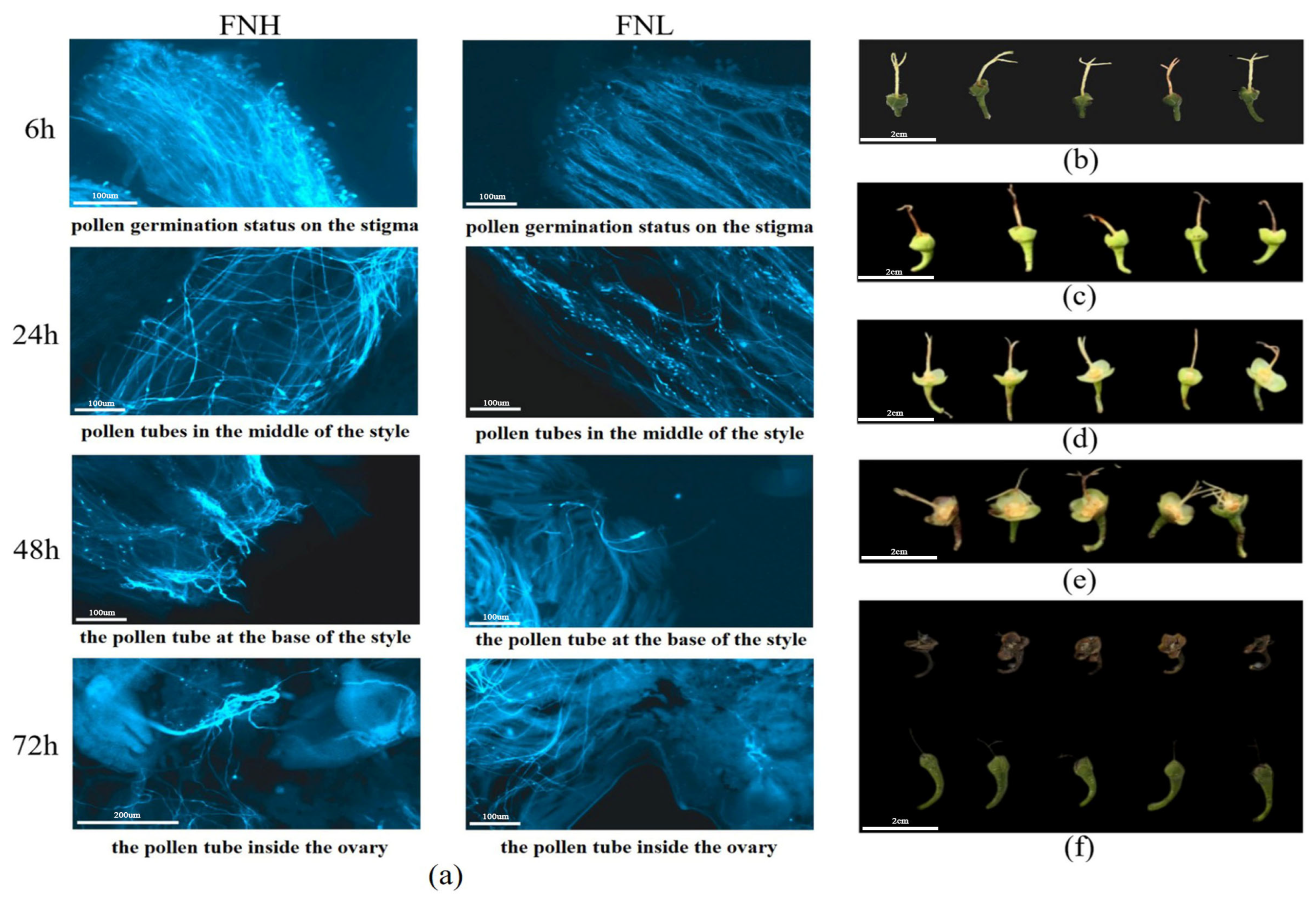
3.3.1. Fluorescence Microscopic Observation of Pollen Tube Germination and BSR-Seq Sequencing Data Analysis
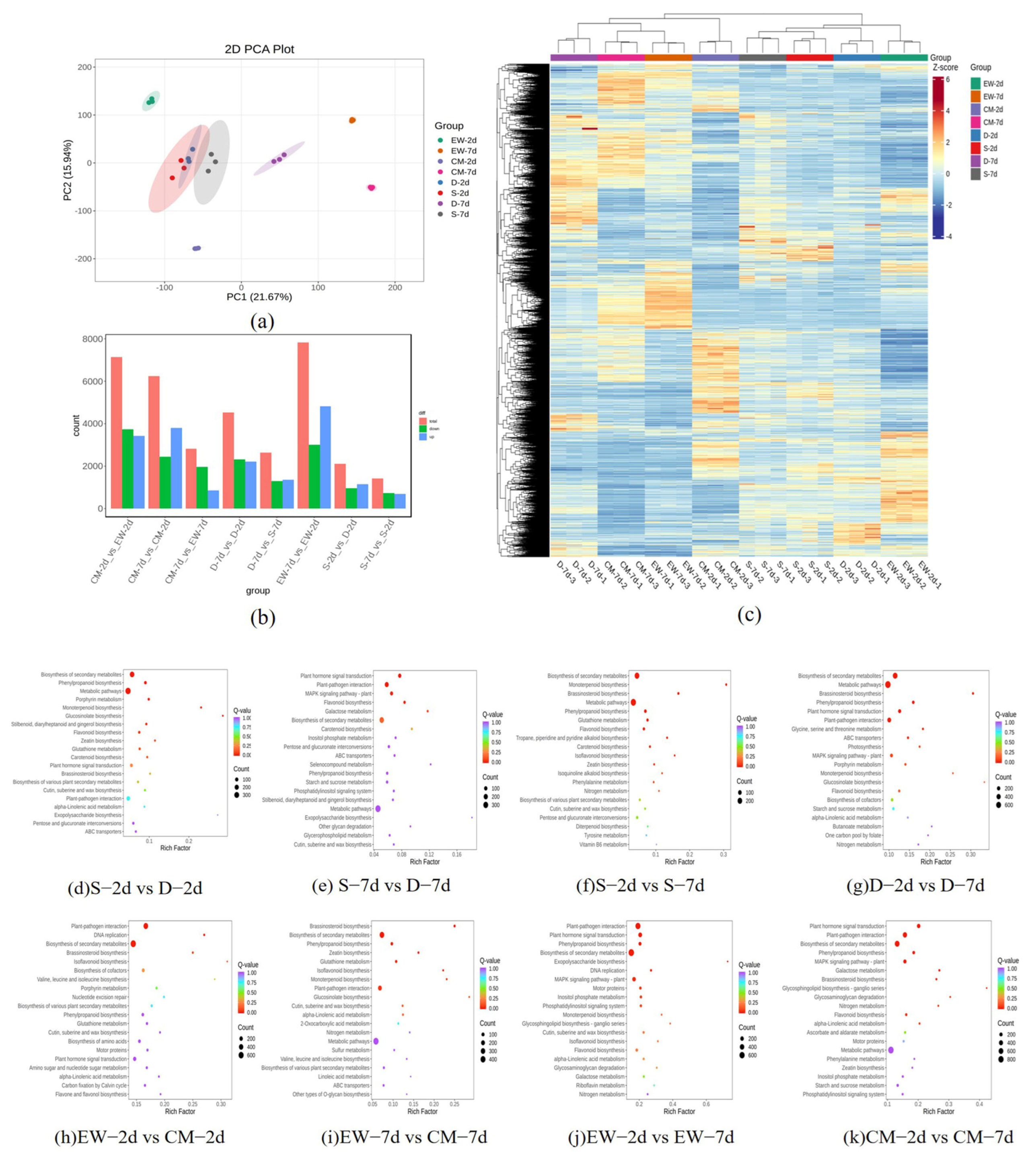
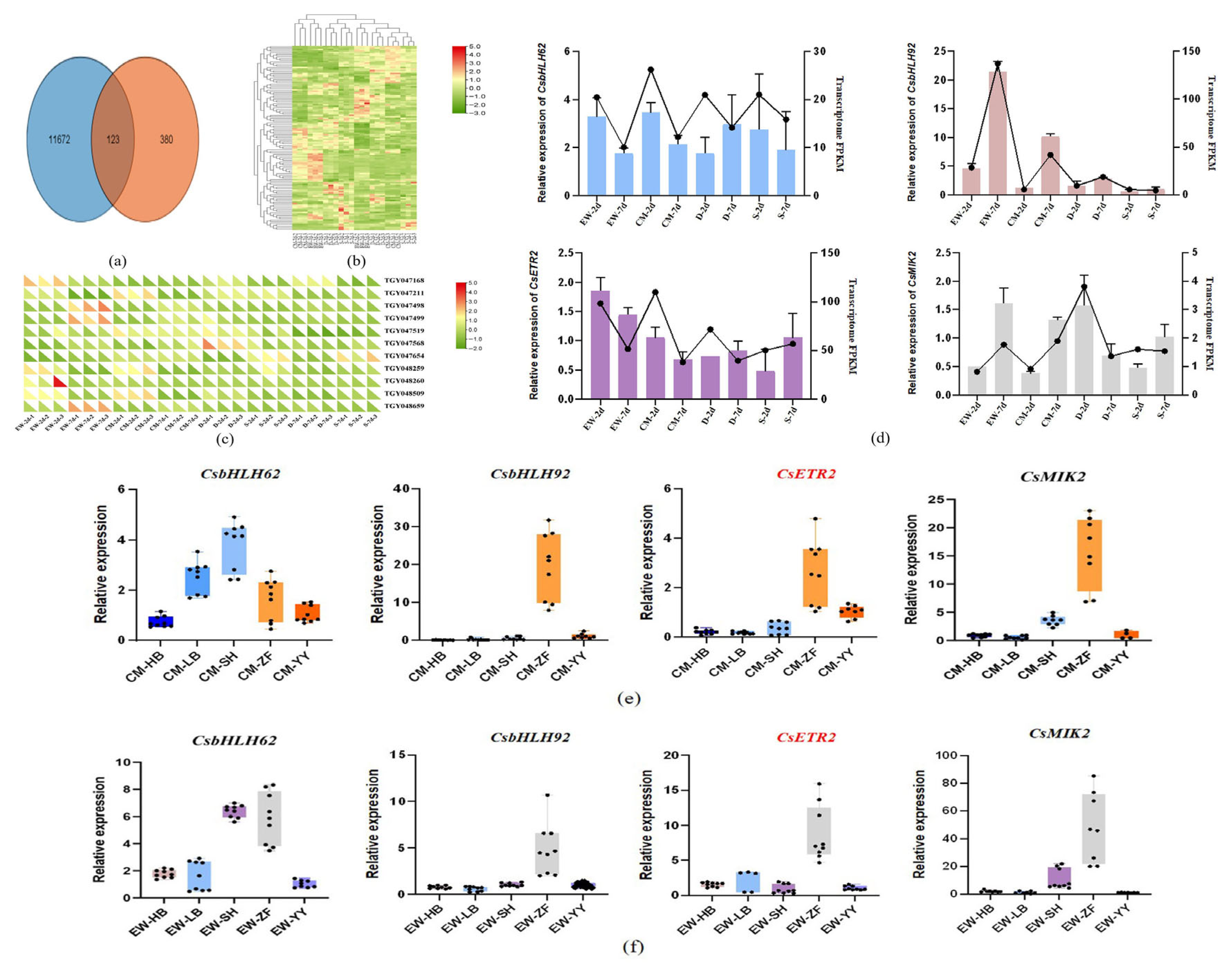
3.3.2. Differential Expression Analysis and Functional Annotation
3.3.3. BSR-Seq and Candidate Gene Screening
3.3.4. Candidate Gene qRT-PCR Validation
3.3.5. Candidate Gene Quantitative Analysis by qRT-PCR in Various Tissues of Tea Plants
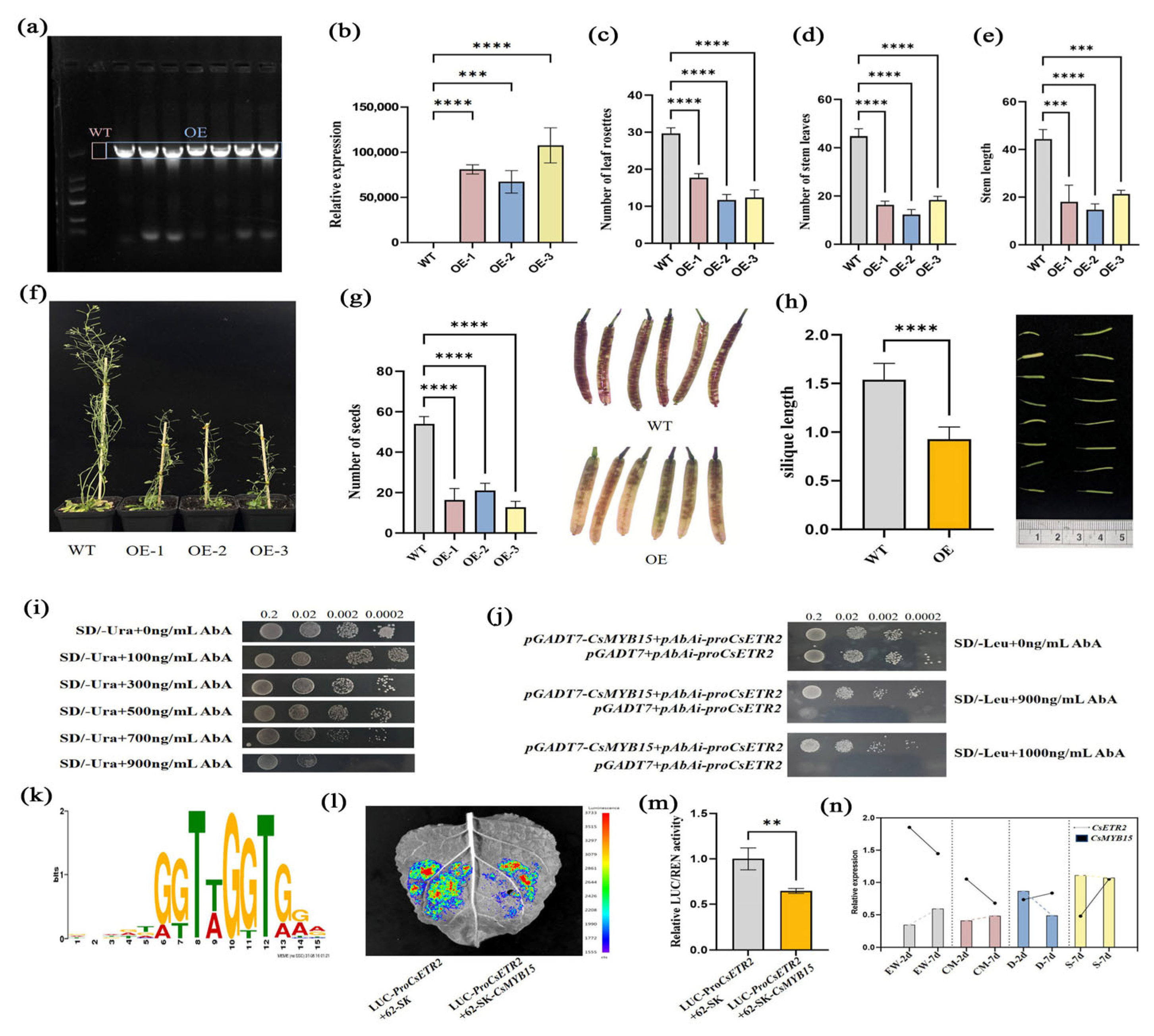
3.3.6. Overexpression of CsETR2 Affects the Growth, Development, and Seed Setting of Arabidopsis thaliana
3.3.7. CsMYB15 Was Able to Bind the Promoter of CsETR2
4. Discussion
4.1. Variation and Genetic Characteristics of Tea Plant Fruit Number
4.2. The QTL Mapping of Fruit Number Traits
4.3. The Application of BSR-Seq in Genetic Mapping of Tea Plants
4.4. CsBHLH92 May Be Involved in Regulating Tea Plant Fruit Set
4.5. Functional Characterization of CsETR2 in Tea Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, L.; Cui, D.; Wang, L.; Liu, Q.; Zhang, D.; Hu, X.; Fu, Y.; Chen, S.; Zou, Y.; Chen, W.; et al. Genetic analysis of the early bud flush trait of tea plants (Camellia sinensis) in the cultivar ‘Emei Wenchun’ and its open-pollinated offspring. Hortic. Res. 2022, 9, uhac086. [Google Scholar] [CrossRef]
- Wei, K.; Wang, X.; Hao, X.; Qian, Y.; Li, X.; Xu, L.; Ruan, L.; Wang, Y.; Zhang, Y.; Bai, P.; et al. Development of a genome-wide 200K SNP array and its application for high-density genetic mapping and origin analysis of Camellia sinensis. Plant Biotechnol. J. 2021, 20, 414–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wei, X.; Zhang, J.; Cui, D.; Zhang, P.; Chen, S.; Zou, Y.; Chen, W.; Tang, D.; Liu, C.; et al. Variation analysis and quantitative trait loci mapping of 16 free amino acid traits in the tea plant (Camellia sinensis). BMC Plant Biol. 2025, 25, 194. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qian, Y.; Wu, L.; Wei, K.; Wang, L. The MADS-box transcription factor CsAGL9 plays essential roles in seed setting in Camellia sinensis. Plant Physiol. Biochem. 2024, 207, 108301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R Package for Bulk Segregant Analysis with Next-Generation Sequencing. Plant Genome 2018, 11, 180006. [Google Scholar] [CrossRef]
- Van Ooijen, J.; Van Ooijen, J.W.; Ooijen, J.; Hoorn, J.; Duin, J.; Jw, V.T.V. MapQTL®6. Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma BV: Wageningen, The Netherlands, 2009. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Alinezhad Jahromi, H.; Zarei, A.; Mohammadkhani, A. Analysis the effects of pollen grain sources on the fruits set and their characteristics of ‘Clementine’ mandarin using microscopic and molecular approaches. Sci. Hortic. 2019, 249, 347–354. [Google Scholar] [CrossRef]
- Deng, L.; Wang, T.; Hu, J.; Yang, X.; Yao, Y.; Jin, Z.; Huang, Z.; Sun, G.; Xiong, B.; Liao, L.; et al. Effects of Pollen Sources on Fruit Set and Fruit Characteristics of ‘Fengtangli’ Plum (Prunus salicina Lindl.) Based on Microscopic and Transcriptomic Analysis. Int. J. Mol. Sci. 2022, 23, 12959. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, L.; Duan, Y.; Zhang, Z.; Zhang, L.; Chen, S.; Cai, C.; Duan, S.; Zhang, K.; Li, G.; Cheng, F. OcBSA: An NGS-based bulk segregant analysis tool for outcross populations. Mol. Plant 2024, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.; Yuan, D.Y.; Zou, F.; Gao, C.; Yang, Y.; Zhang, L.; Tan, X.F. Self-Sterility in Camellia oleifera May Be Due to the Prezygotic Late-Acting Self-Incompatibility. PLoS ONE 2014, 9, e99639. [Google Scholar] [CrossRef]
- Wilkins, K.A.; Bancroft, J.; Bosch, M.; Ings, J.; Smirnoff, N.; Franklin-Tong, V.E. Reactive oxygen species and nitric oxide mediate actin reorganization and programmed cell death in the self-incompatibility response of papaver. Plant Physiol. 2011, 156, 404–416. [Google Scholar] [CrossRef]
- Ahmad, M.H.; Rao, M.J.; Hu, J.; Xu, Q.; Liu, C.; Cao, Z.; Larkin, R.M.; Deng, X.; Bosch, M.; Chai, L. Systems and breakdown of self-incompatibility. Crit. Rev. Plant Sci. 2022, 41, 32. [Google Scholar] [CrossRef]
- Gnanesh, B.N.; Mondal, R.; Arunakumar, G.S.; Manojkumar, H.B.; Singh, P.; Bhavya, M.R.; Sowbhagya, P.; Burji, S.M.; Mogili, T.; Sivaprasad, V. Genome size, genetic diversity, and phenotypic variability imply the effect of genetic variation instead of ploidy on trait plasticity in the cross-pollinated tree species of mulberry. PLoS ONE 2023, 18, e0289766. [Google Scholar] [CrossRef]
- Janko, K.; Bartoš, O.; Kočí, J.; Roslein, J.; Drdová, E.J.; Kotusz, J.; Eisner, J.; Mokrejš, M.; Štefková-Kašparová, E. Genome Fractionation and Loss of Heterozygosity in Hybrids and Polyploids: Mechanisms, Consequences for Selection, and Link to Gene Function. Mol. Biol. Evol. 2021, 38, 5255–5274. [Google Scholar] [CrossRef]
- Kong, W.; Zhu, Q.; Zhang, Q.A.-O.; Zhu, Y.; Yang, J.; Chai, K.; Lei, W.; Jiang, M.; Zhang, S.; Lin, J.; et al. 5mC DNA methylation modification-mediated regulation in tissue functional differentiation and important flavor substance synthesis of tea plant (Camellia sinensis L.). Hortic. Res. 2023, 10, uhad126. [Google Scholar] [CrossRef]
- Wei, J.; Fang, Y.; Jiang, H.; Wu X-t Zuo J-h Xia X-c et, a.l. Combining QTL mapping and gene co-expression network analysis for prediction of candidate genes and molecular network related to yield in wheat. BMC Plant Biol. 2022, 22, 288. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, R.; Zhao, Y.; Yao, J.; Li, W.; Yang, Z.; Sun, F.; Yang, X. Identifying QTL and candidate genes for prolificacy in maize. Crop J. 2023, 11, 9. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Sederoff, R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: Mapping strategy and RAPD markers. Genetics 1994, 137, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yeh, C.T.; Tang, H.M.; Nettleton, D.; Schnable, P.S. Gene Mapping via Bulked Segregant RNA-Seq (BSR-Seq). PLoS ONE 2012, 7, e36406. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, L.; Qiu, D.; Cao, Z.; Wang, K.; Li, Z.; Wang, X.; Wang, J.; Ma, Q.; Cao, D.; et al. PSW1, an LRR receptor kinase, regulates pod size in peanut. Plant Biotechnol. J. 2023, 21, 2113–2124. [Google Scholar] [CrossRef]
- Ma, C.; Rehman, A.; Li, H.G.; Zhao, Z.B.; Sun, G.; Du, X.M. Mapping of dwarfing QTL of Ari1327, a semi-dwarf mutant of upland cotton. BMC Plant Biol. 2022, 22, 5. [Google Scholar] [CrossRef]
- Aurélie, G.; Gendrot, G.; Chamot, S.; Widiez, T.; Nathalie, D. ZmZHOUPI, an endosperm-specific bHLH transcription factor involved in maize seed development. Plant J. 2015, 84, 574–586. [Google Scholar]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell: Pecific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006, 46, 768–779. [Google Scholar] [CrossRef]
- Dany, H.; Hidenori, S.; Keqiang, W. Antagonistic Actions of HLH/bHLH Proteins Are Involved in Grain Length and Weight in Rice. PLoS ONE 2012, 7, e31325. [Google Scholar]
- Lu, M.; Mingqian, L.; Qin, L.; Yubing, H.; Yanan, T.; Huadong, Z. The brassinosteroid signaling-related ILI–OsAIF–OsbHLH92 transcription factor module antagonistically controls leaf angle and grain size in rice. Plant Physiol. 2024, 197, kiae668. [Google Scholar] [CrossRef]
- Teng, S.; Liu, Q.; Chen, G.; Chang, Y.; Cui, X.; Wu, J.; Ai, P.; Sun, X.; Zhang, Z.; Lu, T. OsbHLH92, in the noncanonical brassinosteroid signaling pathway, positively regulates leaf angle and grain weight in rice. New Phytol. 2023, 240, 1066–1081. [Google Scholar] [CrossRef]
- Müller, R.; Sisler, E.C.; Serek, M. Stress induced ethylene production, ethylene binding, and the response to the ethylene action inhibitor 1-MCP in miniature roses. Sci. Hortic. 2000, 83, 51–59. [Google Scholar] [CrossRef]
- Vriezen, W.H.; Feron, R.; Maretto, F.; Keijman, J.; Mariani, C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 2008, 177, 60–76. [Google Scholar] [CrossRef]
- Wang, H.; Schauer, N.; Usadel, B.; Frasse, P.; Zouine, M.; Hernould, M.; Latche, A.; Pech, J.-C.; Fernie, A.R.; Bouzayen, M. Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 2009, 21, 1428–1452. [Google Scholar] [CrossRef] [PubMed]
- Pascual, L.; Blanca, J.M.; Cañizares, J.; Nuez, F. Transcriptomic analysis of tomato carpel development reveals alterations in ethylene and gibberellin synthesis during pat3/pat4 parthenocarpic fruit set. BMC Plant Biol. 2009, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Arciga-Reyes, L.; Zhong, S.; Alexander, L.; Hackett, R.; Wilson, I.; Grierson, D. SlTPR1, a tomato tetratricopeptide repeat protein, interacts with the ethylene receptors NR and LeETR1, modulating ethylene and auxin responses and development. J. Exp. Bot. 2008, 59, 4271–4287. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, T.; Higashi, K.; Kamada, H.; Ezura, H. Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J. Plant Physiol. 2003, 160, 1335–1340. [Google Scholar] [CrossRef]
- Wuriyanghan, H.; Zhang, B.; Cao, W.H.; Ma, B.; Lei, G.; Liu, Y.F.; Wei, W.; Wu, H.J.; Chen, L.J.; Chen, H.W.; et al. The Ethylene Receptor ETR2 Delays Floral Transition and Affects Starch Accumulation in Rice. Plant Cell 2009, 21, 1473–1494. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kan, S.; Tang, D.; Chen, W.; Gu, Y.; Zhao, S.; Long, L.; Zhang, J.; Tan, X.; Tan, L.; Tang, Q. Integrating Genetic Mapping and BSR-Seq Analysis to Identify Candidate Genes Controlling Fruitfulness in Camellia sinensis. Plants 2025, 14, 2963. https://doi.org/10.3390/plants14192963
Kan S, Tang D, Chen W, Gu Y, Zhao S, Long L, Zhang J, Tan X, Tan L, Tang Q. Integrating Genetic Mapping and BSR-Seq Analysis to Identify Candidate Genes Controlling Fruitfulness in Camellia sinensis. Plants. 2025; 14(19):2963. https://doi.org/10.3390/plants14192963
Chicago/Turabian StyleKan, Shizhuo, Dandan Tang, Wei Chen, Yuxin Gu, Shenxin Zhao, Lu Long, Jing Zhang, Xiaoqin Tan, Liqiang Tan, and Qian Tang. 2025. "Integrating Genetic Mapping and BSR-Seq Analysis to Identify Candidate Genes Controlling Fruitfulness in Camellia sinensis" Plants 14, no. 19: 2963. https://doi.org/10.3390/plants14192963
APA StyleKan, S., Tang, D., Chen, W., Gu, Y., Zhao, S., Long, L., Zhang, J., Tan, X., Tan, L., & Tang, Q. (2025). Integrating Genetic Mapping and BSR-Seq Analysis to Identify Candidate Genes Controlling Fruitfulness in Camellia sinensis. Plants, 14(19), 2963. https://doi.org/10.3390/plants14192963






