The Emerging Role of the Salt Tolerance-Related Protein in the Abiotic Stress Response of Arabidopsis thaliana
Abstract
1. Introduction
2. Role in Abiotic Stress Tolerance
3. Molecular Features
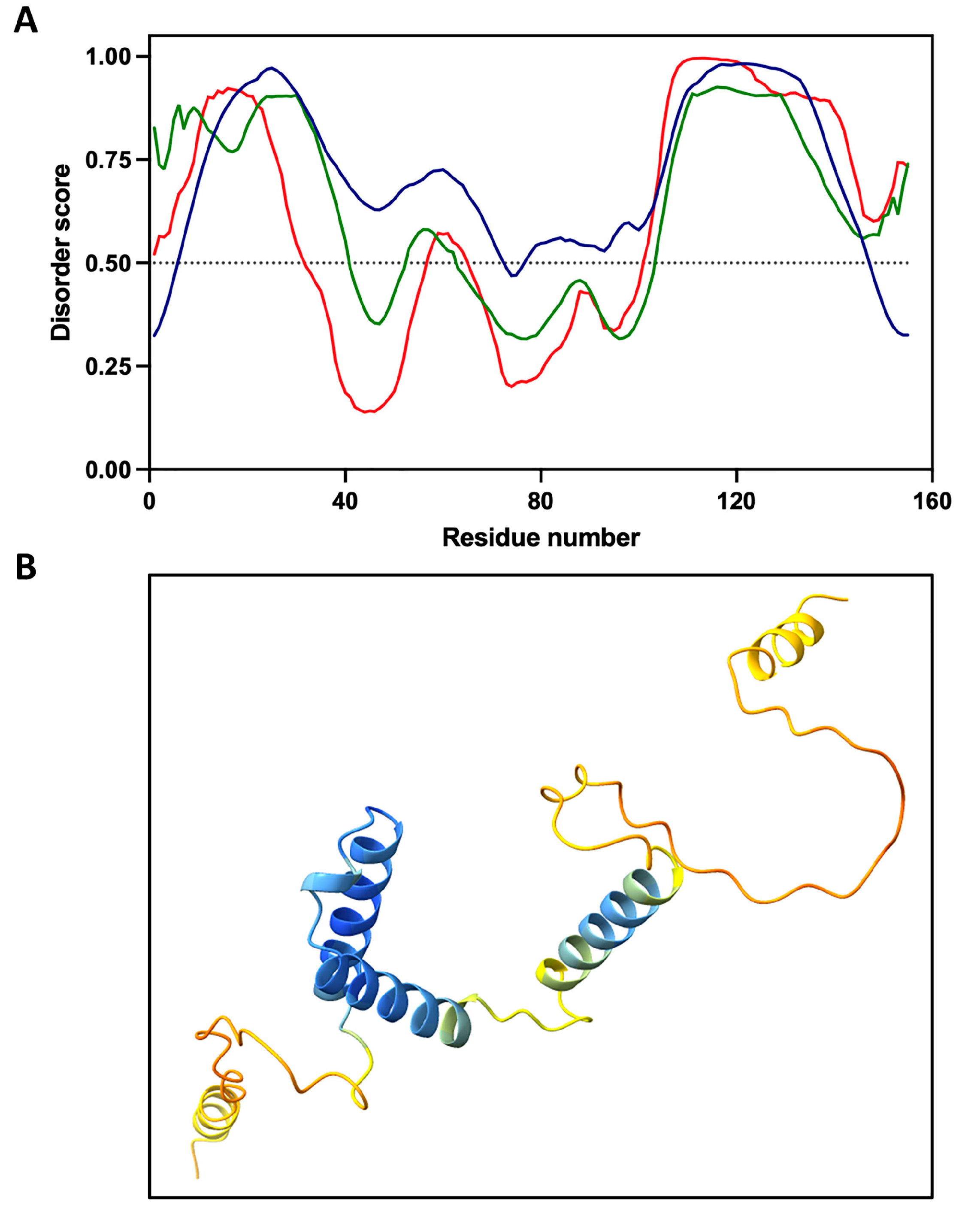
4. Regulation of STRP Expression
5. Post-Translational Modifications
6. Subcellular Localization
7. Mechanism of Action of STRP: Hypothesis and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| STRP | Salt Tolerance-Related Protein |
| ABA | Abscisic Acid |
| GRAVY | Grand Average of Hydropathy |
| NRP1 | Nodulin-Related Protein 1 |
| LEA | Late Embryogenesis Abundant |
| IDP | Intrinsically Disordered Protein |
| pLDDT | Predicted Local Distance Difference Test |
| UTR | Untranslated Region |
| cis-nat-siRNA | cis-natural antisense small interfering RNA |
| UPS | Ubiquitin Proteasome System |
| SCF | SKP1, Cullin, F-box |
| NatB | Nα-terminal acetyltransferase B |
| Nt | N-terminal |
| HSP | Heat Shock Protein |
| DEK3 | DEK domain-containing protein 3 |
| LLPS | Liquid–Liquid Phase Separation |
| SG | Stress Granules |
References
- Kopecká, R.; Kameniarová, M.; Černý, M.; Brzobohatý, B.; Novák, J. Abiotic Stress in Crop Production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef]
- Licaj, I.; Fiorillo, A.; Di Meo, M.C.; Varricchio, E.; Rocco, M. Effect of Polyethylene Glycol-Simulated Drought Stress on Stomatal Opening in “Modern” and “Ancient” Wheat Varieties. Plants 2024, 13, 1575. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Tackling abiotic stress in plants: Recent insights and trends. Stress Biol. 2025, 5, 8. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. The Stress Concept in Plants: An Introduction. Ann. N. Y. Acad. Sci. 1998, 851, 187–198. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant Responses to Drought, Salinity and Extreme Temperatures: Towards Genetic Engineering for Stress Tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, Salt, and Temperature Stress-Induced Metabolic Rearrangements and Regulatory Networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; van Zanten, M.; Sasidharan, R. Mechanisms of plant acclimation to multiple abiotic stresses. Commun. Biol. 2025, 8, 655. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Visconti, S.; D’Ambrosio, C.; Fiorillo, A.; Arena, S.; Muzi, C.; Zottini, M.; Aducci, P.; Marra, M.; Scaloni, A.; Camoni, L. Overexpression of 14-3-3 proteins enhances cold tolerance and increases levels of stress-responsive proteins of Arabidopsis plants. Plant Sci. 2019, 289, 110215. [Google Scholar] [CrossRef]
- Ruszczyńska, M.; Sytykiewicz, H. New Insights into Involvement of Low Molecular Weight Proteins in Complex Defense Mechanisms in Higher Plants. Int. J. Mol. Sci. 2024, 25, 8531. [Google Scholar] [CrossRef]
- Du, J.; Huang, Y.P.; Xi, J.; Cao, M.J.; Ni, W.S.; Chen, X.; Zhu, J.K.; Oliver, D.J.; Xiang, C.B. Functional gene mining for salt tolerance genes with the power of Arabidopsis. Plant J. 2008, 56, 653–664. [Google Scholar] [CrossRef]
- Fiorillo, A.; Mattei, M.; Aducci, P.; Visconti, S.; Camoni, L. The Salt Tolerance Related Protein (STRP) Mediates Cold Stress Responses and Abscisic Acid Signalling in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 1251. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, A.; Manai, M.; Visconti, S.; Camoni, L. The Salt Tolerance-Related Protein (STRP) Is a Positive Regulator of the Response to Salt Stress in Arabidopsis thaliana. Plants 2023, 12, 1704. [Google Scholar] [CrossRef] [PubMed]
- Theologis, A.; Ecker, J.R.; Palm, C.J.; Federspiel, N.A.; Kaul, S.; White, O.; Alonso, J.; Altafi, H.; Araujo, R.; Bowman, C.L.; et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 2000, 408, 816–820. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Rutschow, H.; Ytterberg, A.J.; Friso, G.; Nilsson, R.; van Wijk, K.J. Quantitative proteomics of a chloroplast SRP54 sorting mutant and its genetic interactions with CLPC1 in Arabidopsis. Plant Physiol. 2008, 148, 156–175. [Google Scholar] [CrossRef]
- Waidmann, S.; Kusenda, B.; Mayerhofer, J.; Mechtler, K.; Jonak, C. A DEK Domain-Containing Protein Modulates Chromatin Structure and Function in Arabidopsis. Plant Cell 2014, 26, 4328–4344. [Google Scholar] [CrossRef]
- Rocco, M.; Arena, S.; Renzone, G.; Scippa, S.; Lomaglio, T.; Verrillo, F.; Scaloni, A.; Marra, M. Proteomic analysis of temperature stress-responsive proteins in Arabidopsis thaliana rosette leaves. Mol. Biosyst. 2013, 9, 1257–1267. [Google Scholar] [CrossRef] [PubMed]
- McBride, Z.; Chen, D.; Reick, C.; Xie, J.; Szymanski, D.B. Global Analysis of Membrane-associated Protein Oligomerization Using Protein Correlation Profiling. Mol. Cell Proteom. 2017, 16, 1972–1989. [Google Scholar] [CrossRef]
- Aslam, M.M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Ahmad Saqib, H.S.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of Abscisic Acid-Mediated Drought Stress Responses in Plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar] [CrossRef]
- Rai, G.K.; Khanday, D.M.; Choudhary, S.M.; Kumar, P.; Kumari, S.; Martínez-Andújar, C.; Martínez-Melgarejo, P.A.; Rai, P.K.; Pérez-Alfocea, F. Unlocking nature’s stress buster: Abscisic acid’s crucial role in defending plants against abiotic stress. Plant Stress 2024, 11, 100359. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 190245. [Google Scholar] [CrossRef]
- Leonhardt, N.; Kwak, J.M.; Robert, N.; Waner, D.; Leonhardt, G.; Schroeder, J.I. Microarray Expression Analyses of Arabidopsis Guard Cells and Isolation of a Recessive Abscisic Acid Hypersensitive Protein Phosphatase 2C Mutant. Plant Cell 2004, 16, 596–615. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, S.; Yu, D. Identification of an Arabidopsis Nodulin-related protein in heat stress. Mol. Cells 2010, 29, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Berardini, T.Z.; Ebert, D.; Li, Q.; Mi, H.; Muruganujan, A.; Prithvi, T.; Reiser, L.; Sawant, S.; Thomas, P.D.; et al. PhyloGenes: An Online Phylogenetics and Functional Genomics Resource for Plant Gene Function Prediction. Plant Direct 2020, 4, e00293. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Christov, N.K.; Tsuda, S.; Imai, R. Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol. 2014, 55, 136–147. [Google Scholar] [CrossRef]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef]
- Tompa, P.; Schad, E.; Tantos, A.; Kalmar, L. Intrinsically disordered proteins: Emerging interaction specialists. Curr. Opin. Struct. Biol. 2015, 35, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Han, K. Transient Secondary Structures as General Target-Binding Motifs in Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2018, 19, 3614. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Garay-Arroyo, A.; Colmenero-Flores, J.M.; Garciarrubio, A.; Covarrubias, A.A. Highly Hydrophilic Proteins in Prokaryotes and Eukaryotes Are Common during Conditions of Water Deficit. J. Biol. Chem. 2000, 275, 5668–5674. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Erdős, G.; Pajkos, M.; Dosztányi, Z. IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Petryszak, R.; Keays, M.; Tang, Y.A.; Fonseca, N.A.; Barrera, E.; Burdett, T.; Füllgrabe, A.; Fuentes, A.M.; Jupp, S.; Koskinen, S.; et al. Expression Atlas update—An integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016, 44, D746–D752. [Google Scholar] [CrossRef]
- George, N.; Fexova, S.; Fuentes, A.M.; Madrigal, P.; Bi, Y.; Iqbal, H.; Kumbham, U.; Nolte, N.F.; Zhao, L.; Thanki, A.S.; et al. Expression Atlas update: Insights from sequencing data at both bulk and single cell level. Nucleic Acids Res. 2024, 52, D107–D114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, F.; Yu, Y.; Feng, L.; Jia, J.; Liu, B.; Li, B.; Guo, H.; Zhai, J. A Comprehensive Online Database for Exploring ∼20,000 Public Arabidopsis RNA-Seq Libraries. Mol. Plant 2020, 13, 1231–1233. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Du, Y.; Cao, L.; Wang, S.; Guo, L.; Tan, L.; Liu, H.; Feng, Y.; Wu, W. Differences in alternative splicing and their potential underlying factors between animals and plants. J. Adv. Res. 2024, 64, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Hardy, E.C.; Balcerowicz, M. Untranslated yet indispensable—UTRs act as key regulators in the environmental control of gene expression. J. Exp. Bot. 2024, 75, 4314–4331. [Google Scholar] [CrossRef]
- Tiwari, B.; Habermann, K.; Arif, M.A.; Weil, H.L.; Garcia-Molina, A.; Kleine, T.; Mühlhaus, T.; Frank, W. Identification of small RNAs during cold acclimation in Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 298. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Montes, C.; Olatunji, D.; Malik, S.; Ji, C.; Clark, N.M.; Pu, Y.; Kelley, D.R.; Walley, J.W. Quantitative proteomics reveals extensive lysine ubiquitination and transcription factor stability states in Arabidopsis. Plant Cell 2024, 37, koae310. [Google Scholar] [CrossRef]
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Su, Y.; Ngolong Ngea, G.L.; Wang, K.; Lu, Y.; Godana, E.A.; Ackah, M.; Yang, Q.; Zhang, H. Deciphering the mechanism of E3 ubiquitin ligases in plant responses to abiotic and biotic stresses and perspectives on PROTACs for crop resistance. Plant Biotechnol. J. 2024, 22, 2811–2843. [Google Scholar] [CrossRef]
- Bienvenut, W.V.; Sumpton, D.; Martinez, A.; Lilla, S.; Espagne, C.; Meinnel, T.; Giglione, C. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol. Cell Proteom. 2012, 11, M111.015131. [Google Scholar] [CrossRef]
- Huber, M.; Bienvenut, W.V.; Linster, E.; Stephan, I.; Armbruster, L.; Sticht, C.; Layer, D.; Lapouge, K.; Meinnel, T.; Sinning, I.; et al. NatB-Mediated N-Terminal Acetylation Affects Growth and Biotic Stress Responses. Plant Physiol. 2020, 182, 792–806. [Google Scholar] [CrossRef]
- Ferrández-Ayela, A.; Micol-Ponce, R.; Sánchez-García, A.B.; Alonso-Peral, M.M.; Micol, J.L.; Ponce, M.R. Mutation of an Arabidopsis NatB N-Alpha-Terminal Acetylation Complex Component Causes Pleiotropic Developmental Defects. PLoS ONE 2013, 8, e80697. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J. Emerging Functions for N-Terminal Protein Acetylation in Plants. Trends Plant Sci. 2015, 20, 599. [Google Scholar] [CrossRef] [PubMed]
- Linster, E.; Wirtz, M. N-terminal acetylation: An essential protein modification emerges as an important regulator of stress responses. J. Exp. Bot. 2018, 69, 4555–4568. [Google Scholar] [CrossRef]
- McTiernan, N.; Kjosås, I.; Arnesen, T. Illuminating the impact of N-terminal acetylation: From protein to physiology. Nat. Commun. 2025, 16, 703. [Google Scholar] [CrossRef]
- Huber, M.; Armbruster, L.; Etherington, R.D.; De La Torre, C.; Hawkesford, M.J.; Sticht, C.; Gibbs, D.J.; Hell, R.; Wirtz, M. Disruption of the Nα-Acetyltransferase NatB Causes Sensitivity to Reductive Stress in Arabidopsis thaliana. Front. Plant Sci. 2022, 12, 799954. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Banik, S.; Dutta, D. Membrane Proteins in Plant Salinity Stress Perception, Sensing, and Response. J. Membr. Biol. 2023, 256, 109–124. [Google Scholar] [CrossRef]
- Niu, Y.; Xiang, Y. An Overview of Biomembrane Functions in Plant Responses to High-Temperature Stress. Front. Plant Sci. 2018, 9, 915. [Google Scholar] [CrossRef]
- Ruiz-Lopez, N.; Pérez-Sancho, J.; Del Valle, A.E.; Haslam, R.P.; Vanneste, S.; Catalá, R.; Perea-Resa, C.; Damme, D.V.; García-Hernández, S.; Albert, A.; et al. Synaptotagmins at the endoplasmic reticulum-plasma membrane contact sites maintain diacylglycerol homeostasis during abiotic stress. Plant Cell 2021, 33, 2431–2453. [Google Scholar] [CrossRef]
- Wei, X.; Liu, S.; Sun, C.; Xie, G.; Wang, L. Convergence and Divergence: Signal Perception and Transduction Mechanisms of Cold Stress in Arabidopsis and Rice. Plants 2021, 10, 1864. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, A.S. Protein disorder in plant stress adaptation: From late embryogenesis abundant to other intrinsically disordered proteins. Int. J. Mol. Sci. 2024, 25, 1178. [Google Scholar] [CrossRef]
- Kovacs, D.; Agoston, B.; Tompa, P. Disordered plant LEA proteins as molecular chaperones. Plant Signal. Behav. 2008, 3, 710. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.V.; Rausch, S.; Hundertmark, M.; Gibon, Y.; Hincha, D.K. The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim. Biophys. Acta Proteins Proteom. 2015, 1854, 1517–1525. [Google Scholar] [CrossRef]
- Bhadouriya, S.L.; Mehrotra, S.; Basantani, M.K.; Loake, G.J.; Mehrotra, R. Role of Chromatin Architecture in Plant Stress Responses: An Update. Front. Plant Sci. 2021, 11, 603380. [Google Scholar] [CrossRef]
- Song, Z.T.; Liu, J.X.; Han, J.J. Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 2021, 63, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Tissier, A.F.; Signer, E.R. Cloning and Characterization of an Arabidopsis thaliana Topoisomerase I Gene. Plant Physiol. 1992, 99, 1493–1501. [Google Scholar] [CrossRef]
- Schubert, V.; Weissleder, A.; Ali, H.; Fuchs, J.; Lermontova, I.; Meister, A.; Schubert, I. Cohesin Gene Defects May Impair Sister Chromatid Alignment and Genome Stability in Arabidopsis thaliana. Chromosoma 2009, 118, 591–605. [Google Scholar] [CrossRef]
- Pradillo, M.; Knoll, A.; Oliver, C.; Varas, J.; Corredor, E.; Puchta, H.; Santos, J.L. Involvement of the Cohesin Cofactor PDS5 (SPO76) during Meiosis and DNA Repair in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1034. [Google Scholar] [CrossRef]
- Doskočilová, A.; Kohoutová, L.; Volc, J.; Kourová, H.; Benada, O.; Chumová, J.; Plíhal, O.; Petrovská, B.; Halada, P.; Bögre, L.; et al. NITRILASE1 Regulates the Exit from Proliferation, Genome Stability and Plant Development. New Phytol. 2013, 198, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.X.; Liu, Z.W.; Li, Y.Q.; Li, L.; Wang, B.; Chen, S.; He, X.J. Arabidopsis PWWP Domain Proteins Mediate H3K27 Trimethylation on FLC and Regulate Flowering Time. J. Integr. Plant Biol. 2018, 60, 362–368. [Google Scholar] [CrossRef]
- Qian, S.; Lv, X.; Scheid, R.N.; Lu, L.; Yang, Z.; Chen, W.; Liu, R.; Boersma, M.D.; Denu, J.M.; Zhong, X.; et al. Dual Recognition of H3K4me3 and H3K27me3 by a Plant Histone Reader SHL. Nat. Commun. 2018, 9, 2425. [Google Scholar] [CrossRef] [PubMed]
- Colville, A.; Alhattab, R.; Hu, M.; Labbé, H.; Xing, T.; Miki, B. Role of HD2 Genes in Seed Germination and Early Seedling Growth in Arabidopsis. Plant Cell Rep. 2011, 30, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome Changes for Arabidopsis in Response to Salt, Osmotic, and Cold Stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef]
- Palmer, S.R.; Villa, R.D.; Graether, S.P. Sequence composition versus sequence order in the cryoprotective function of an intrinsically disordered stress-response protein. Protein Sci. 2019, 28, 1448–1459. [Google Scholar] [CrossRef]
- Lv, A.; Su, L.; Liu, X.; Xing, Q.; Huang, B.; An, Y.; Zhou, P. Characterization of dehydrin protein, CdDHN4-L and CdDHN4-S, and their differential protective roles against abiotic stress in vitro. BMC Plant Biol. 2018, 18, 299. [Google Scholar] [CrossRef]
- Artur, M.A.; Rienstra, J.; Dennis, T.J.; Farrant, J.M.; Ligterink, W.; Hilhorst, H. Structural plasticity of intrinsically disordered LEA proteins from Xerophyta schlechteri provides protection in vitro and in vivo. Front. Plant Sci. 2019, 10, 469003. [Google Scholar] [CrossRef]
- Lv, A.; Su, L.; Wen, W.; Fan, N.; Zhou, P.; An, Y. Analysis of the function of the alfalfa Mslea-D34 gene in abiotic stress responses and flowering time. Plant Cell Physiol. 2021, 62, 28–42. [Google Scholar] [CrossRef]
- Pougy, K.C.; Brito, B.A.; Melo, G.S.; Pinheiro, A.S. Phase Separation as a Key Mechanism in Plant Development, Environmental Adaptation, and Abiotic Stress Response. J. Biol. Chem. 2025, 301, 108548. [Google Scholar] [CrossRef]

| Year | Discovery | References |
|---|---|---|
| 2008 | First studies on the protective role of STRP in salt stress | [13] |
| 2013 | STRP levels increase under temperature stress | [20] |
| 2014 | The wheat STRP homolog WCI16 is a LEA-like protein | [28] |
| 2015 | STRP is part of the DEK3 interactome | [19] |
| 2020 | Protective role in cold stress and involvement in ABA signaling | [14] |
| 2020 | STRP is N-terminally acetylated by the NatB complex | [52] |
| 2023 | Overexpression of STRP and antioxidant role under salt stress | [15] |
| 2024 | STRP is ubiquitinated at multiple Lys residues | [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiorillo, A.; Manai, M.; Falliti, E.; Visconti, S.; Camoni, L. The Emerging Role of the Salt Tolerance-Related Protein in the Abiotic Stress Response of Arabidopsis thaliana. Plants 2025, 14, 2954. https://doi.org/10.3390/plants14192954
Fiorillo A, Manai M, Falliti E, Visconti S, Camoni L. The Emerging Role of the Salt Tolerance-Related Protein in the Abiotic Stress Response of Arabidopsis thaliana. Plants. 2025; 14(19):2954. https://doi.org/10.3390/plants14192954
Chicago/Turabian StyleFiorillo, Anna, Michela Manai, Elisa Falliti, Sabina Visconti, and Lorenzo Camoni. 2025. "The Emerging Role of the Salt Tolerance-Related Protein in the Abiotic Stress Response of Arabidopsis thaliana" Plants 14, no. 19: 2954. https://doi.org/10.3390/plants14192954
APA StyleFiorillo, A., Manai, M., Falliti, E., Visconti, S., & Camoni, L. (2025). The Emerging Role of the Salt Tolerance-Related Protein in the Abiotic Stress Response of Arabidopsis thaliana. Plants, 14(19), 2954. https://doi.org/10.3390/plants14192954








