Population Genetic Analysis Reveals Recent Demographic Expansion and Local Differentiation of Areca Palm Velarivirus 1 in Hainan Island
Abstract
1. Introduction
2. Results
2.1. APV1 Exhibits High Haplotype Diversity but Low Nucleotide Diversity
2.2. Neutrality Tests and Mismatch Distribution Support APV1 Population Expansion
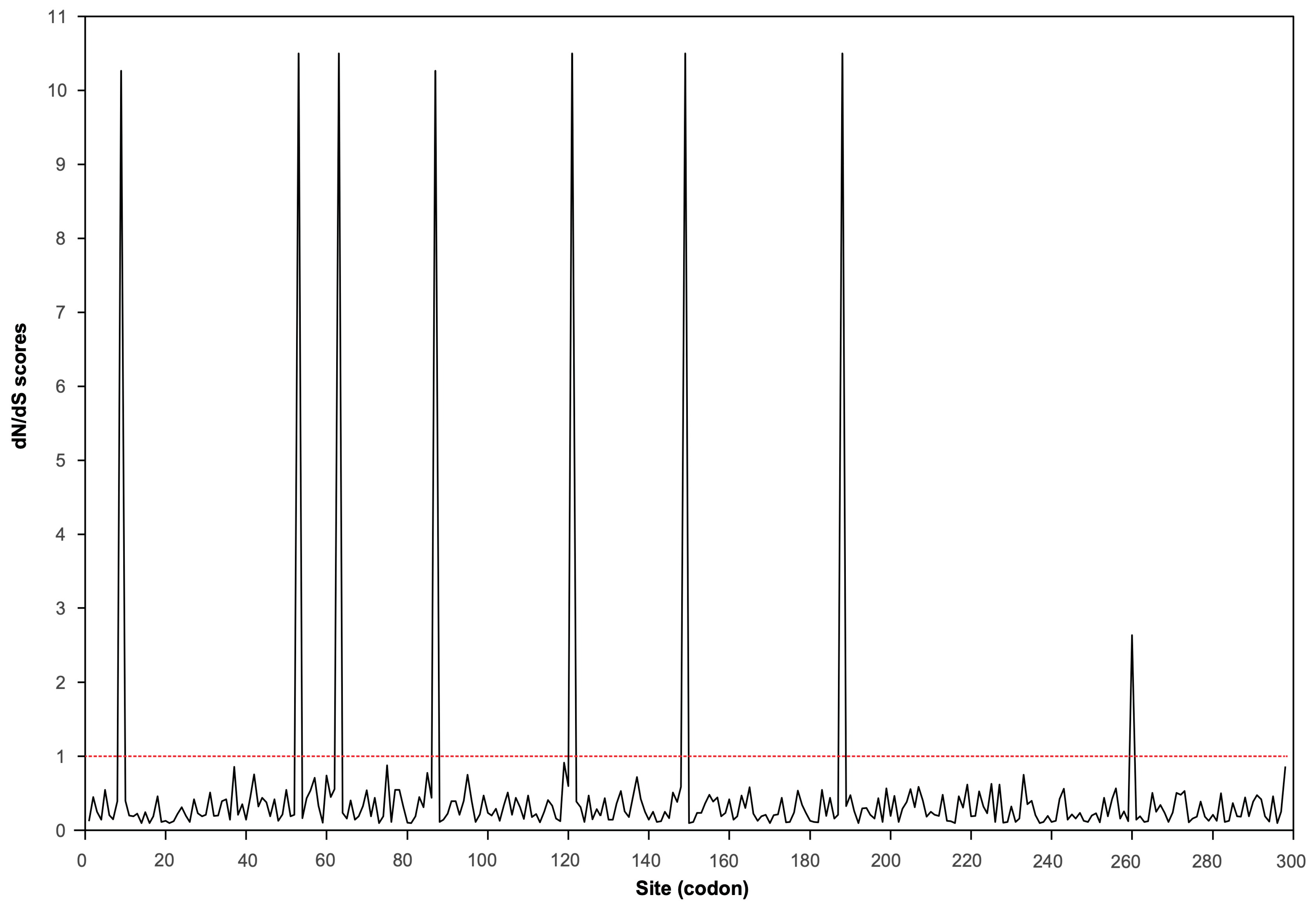
2.3. The APV1 CP Gene Is Under Strong Purifying Selection with Episodic Adaptation
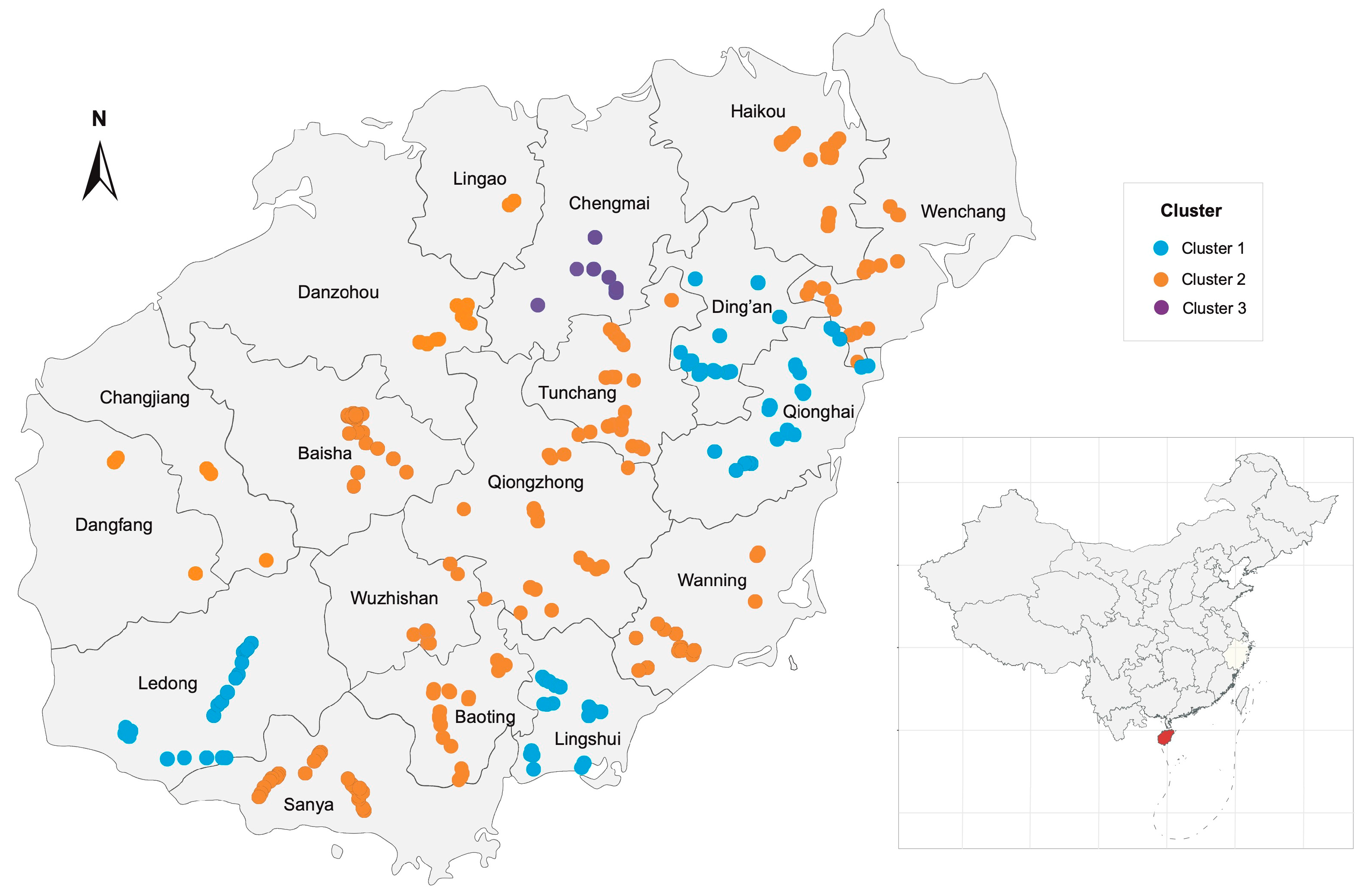
2.4. APV1 Populations Exhibit Significant Geographic Structuring
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Processing
4.2. Virus Isolates and Genetic Diversity Analysis
4.3. Demographic History of the APV1 Population
4.4. Detecting Natural Selection
4.5. Testing Population Differentiation
4.6. Phylogeny-Geography Correlation of APV1
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, L.U.; Zhao, R.; Wang, H.; Huang, X. The recent advances of the causal agent of yellow leaf disease (YLD) on areca palm (Areca catechu L.). Trop. Plants 2023, 2, 7. [Google Scholar] [CrossRef]
- Wang, C.; Yin, Z.; Luo, R.; Qian, J.; Fu, C.; Wang, Y.; Xie, Y.; Liu, Z.; Qiu, Z.; Pei, H. Spatiotemporal evolution and impact mechanisms of areca palm plantations in China (1987–2022). Forests 2024, 15, 1679. [Google Scholar] [CrossRef]
- Yu, H.; Qi, S.; Chang, Z.; Rong, Q.; Akinyemi, I.A.; Wu, Q. Complete genome sequence of a novel velarivirus infecting areca palm in China. Arch. Virol. 2015, 160, 2367–2370. [Google Scholar] [CrossRef]
- Nayar, R.; Seliskar, C.E. Mycoplasma like organisms associated with yellow leaf disease of Areca catechu L. Eur. J. For. Pathol. 1978, 8, 125–128. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, R.; Zhang, H.; Cao, X.; Li, Z.; Zhang, Z.; Zhai, J.; Huang, X. Prevalence of yellow leaf disease (YLD) and its associated areca palm velarivirus 1 (APV1) in betel palm (Areca catechu) plantations in Hainan, China. Plant Dis. 2020, 104, 2556–2562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, X.; Cao, X.; Khan, L.U.; Zhao, R.; Wang, H.; Huang, X. Transmission of areca palm velarivirus 1 by mealybugs causes yellow leaf disease in betel palm (Areca catechu). Phytopathology 2022, 112, 700–707. [Google Scholar] [CrossRef]
- Fuchs, M.; Bar-Joseph, M.; Candresse, T.; Maree, H.J.; Martelli, G.P.; Melzer, M.J.; Menzel, W.; Minafra, A.; Sabanadzovic, S.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Closteroviridae. J. Gen. Virol. 2020, 101, 364–365. [Google Scholar] [CrossRef]
- Cao, X.; Zhao, R.; Wang, H.; Zhang, H.; Zhao, X.; Khan, L.U.; Huang, X. Genomic diversity of areca palm velarivirus 1 (APV1) in areca palm (Areca catechu) plantations in Hainan, China. BMC Genom. 2021, 22, 725. [Google Scholar] [CrossRef]
- Niu, X.; Xu, Z.; Tian, Y.; Xiao, S.; Xie, Y.; Du, Z.; Qin, W.; Gao, F. A putative ormycovirus that possibly contributes to the yellow leaf disease of areca palm. Forests 2024, 15, 1025. [Google Scholar] [CrossRef]
- Khan, L.U.; Cao, X.; Zhao, R.; Tan, H.; Xing, Z.; Huang, X. Effect of temperature on yellow leaf disease symptoms and its associated areca palm velarivirus 1 titer in areca palm (Areca catechu L.). Front. Plant Sci. 2022, 13, 1023386. [Google Scholar] [CrossRef] [PubMed]
- García-Andrés, S.; Accotto, G.P.; Navas-Castillo, J.; Moriones, E. Founder effect, plant host, and recombination shape the emergent population of begomoviruses that cause the tomato yellow leaf curl disease in the Mediterranean basin. Virology 2007, 359, 302–312. [Google Scholar] [CrossRef]
- Delatte, H.; Holota, H.; Moury, B.; Reynaud, B.; Lett, J.M. Evidence for a founder effect after introduction of tomato yellow leaf curl virus–mild in an insular environment. J. Mol. Evol. 2007, 65, 112–118. [Google Scholar] [CrossRef]
- Lin, H.X.; Rubio, L.; Smythe, A.B.; Falk, B.W. Molecular population genetics of cucumber mosaic virus in California: Evidence for founder effects and reassortment. J. Virol. 2004, 78, 6666–6675. [Google Scholar] [CrossRef]
- Zwart, M.P.; Elena, S.F. Matters of size: Genetic bottlenecks in virus infection and their potential impact on evolution. Annu. Rev. Virol. 2015, 2, 161–179. [Google Scholar] [CrossRef]
- Jin, K.X.; Sun, F.S.; Chen, M.R.; Luo, D.Q.; Cai, X.Z. Yellows disease of betel nut palm in Hainan, China. Sci. Silvae Sin. 1995, 31, 556–558+560. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Carlos Sánchez-DelBarrio, J.; Guirao-Rico, S.; Sanz, P.L.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Harpending, H.C. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 1994, 66, 591–600. [Google Scholar] [PubMed]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Balloux, F.; Lugon-Moulin, N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002, 11, 155–165. [Google Scholar] [CrossRef]
- Gao, F.; Jin, J.; Zou, W.; Liao, F.; Shen, J. Geographically driven adaptation of chilli veinal mottle virus revealed by genetic diversity analysis of the coat protein gene. Arch. Virol. 2016, 161, 1329–1333. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Parker, J.; Rambaut, A.; Pybus, O.G. Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2008, 8, 239–246. [Google Scholar] [CrossRef]


| Virus | n | h | Haplotype Diversity | Nucleotide Diversity | Tajima’s D | Fu’s FS |
|---|---|---|---|---|---|---|
| APV1 | 364 | 287 | 0.997 ± 0.001 | 0.017 ± 0.0003 | −2.266 *** | −23.697 ** |
| GLV7 | 17 | 13 | 0.949 ± 0.008 | 0.067 ± 0.008 | −0.818 ns | 3.442 ns |
| LCV1 | 57 | 50 | 0.999 ± 0.004 | 0.206 ± 0.007 | 2.179 ns | −2.915 ns |
| Model | np | ln L | Model Compared | LRT p-Value | Positively Selected Sites |
|---|---|---|---|---|---|
| M1a | 64 | −5473.213 | Not allowed | ||
| M2a | 66 | −5473.108 | M1a vs. M2a | < 0.001 | [] |
| M7 | 64 | −5476.803 | Not allowed | ||
| M8 | 66 | −5472.930 | M7 vs. M8 | < 0.001 | 9 *, 53 **, 63 **, 87 *, 121 **, 149 **, 188 ** |
| M8a | 65 | −5473.183 | M8a vs. M8 | < 0.001 | Not allowed |
| Population | KST | Z | Snn | FST |
|---|---|---|---|---|
| Cluster 1 vs. Cluster 2 | 0.021 *** | 28821.739 *** | 0.877 *** | 0.055 *** |
| Cluster 1 vs. Cluster 3 | 0.057 *** | 2455.444 *** | 0.946 *** | 0.153 *** |
| Cluster 2 vs. Cluster 3 | 0.018 *** | 19545.859 *** | 0.957 *** | 0.117 *** |
| Statistic | Observed Mean (95% CI) | Null Mean (95% CI) | p-Value |
|---|---|---|---|
| AI | 4.964 (3.889–6.081) | 17.694 (16.640–18.678) | <0.001 |
| PS | 40.706 (37.000–43.000) | 93.567 (91.058–96.190) | <0.001 |
| MC (Cluster 1) | 17.946 (18.000–19.000) | 2.504 (2.220–3.019) | <0.010 ** |
| MC (Cluster 2) | 39.070 (39.00–40.000) | 9.009 (7.365–11.975) | <0.010 ** |
| MC (Cluster 3) | 5.680 (3.000–10.000) | 1.326 (1.026–1.995) | <0.010 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, X.; Xu, Z.; Lin, Z.; Tang, Q.; Du, Z.; Gao, F. Population Genetic Analysis Reveals Recent Demographic Expansion and Local Differentiation of Areca Palm Velarivirus 1 in Hainan Island. Plants 2025, 14, 2952. https://doi.org/10.3390/plants14192952
Niu X, Xu Z, Lin Z, Tang Q, Du Z, Gao F. Population Genetic Analysis Reveals Recent Demographic Expansion and Local Differentiation of Areca Palm Velarivirus 1 in Hainan Island. Plants. 2025; 14(19):2952. https://doi.org/10.3390/plants14192952
Chicago/Turabian StyleNiu, Xiaoqing, Zhongtian Xu, Zhaowei Lin, Qinghua Tang, Zhenguo Du, and Fangluan Gao. 2025. "Population Genetic Analysis Reveals Recent Demographic Expansion and Local Differentiation of Areca Palm Velarivirus 1 in Hainan Island" Plants 14, no. 19: 2952. https://doi.org/10.3390/plants14192952
APA StyleNiu, X., Xu, Z., Lin, Z., Tang, Q., Du, Z., & Gao, F. (2025). Population Genetic Analysis Reveals Recent Demographic Expansion and Local Differentiation of Areca Palm Velarivirus 1 in Hainan Island. Plants, 14(19), 2952. https://doi.org/10.3390/plants14192952





