Cloning and Functional Analysis of ClVND1, a Member of the OsNAC7 Subfamily of the NAC Family in Chrysanthemum lavandulifolium
Abstract
1. Introduction
2. Results
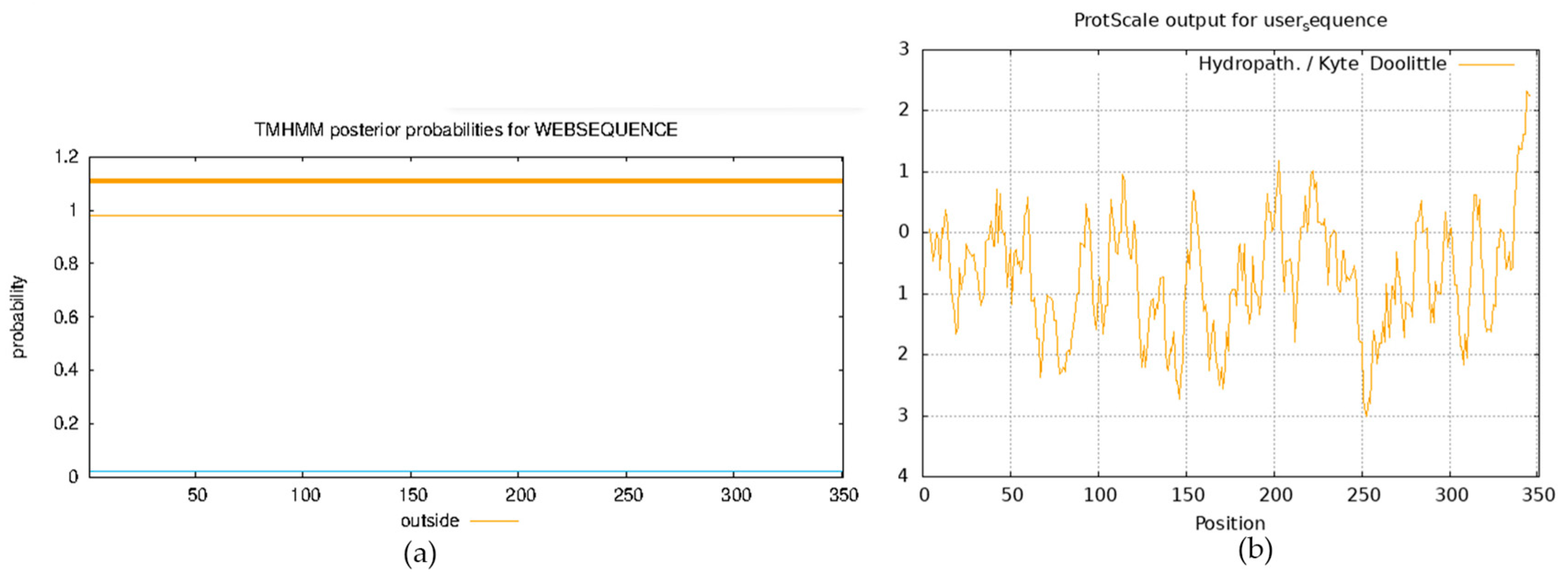
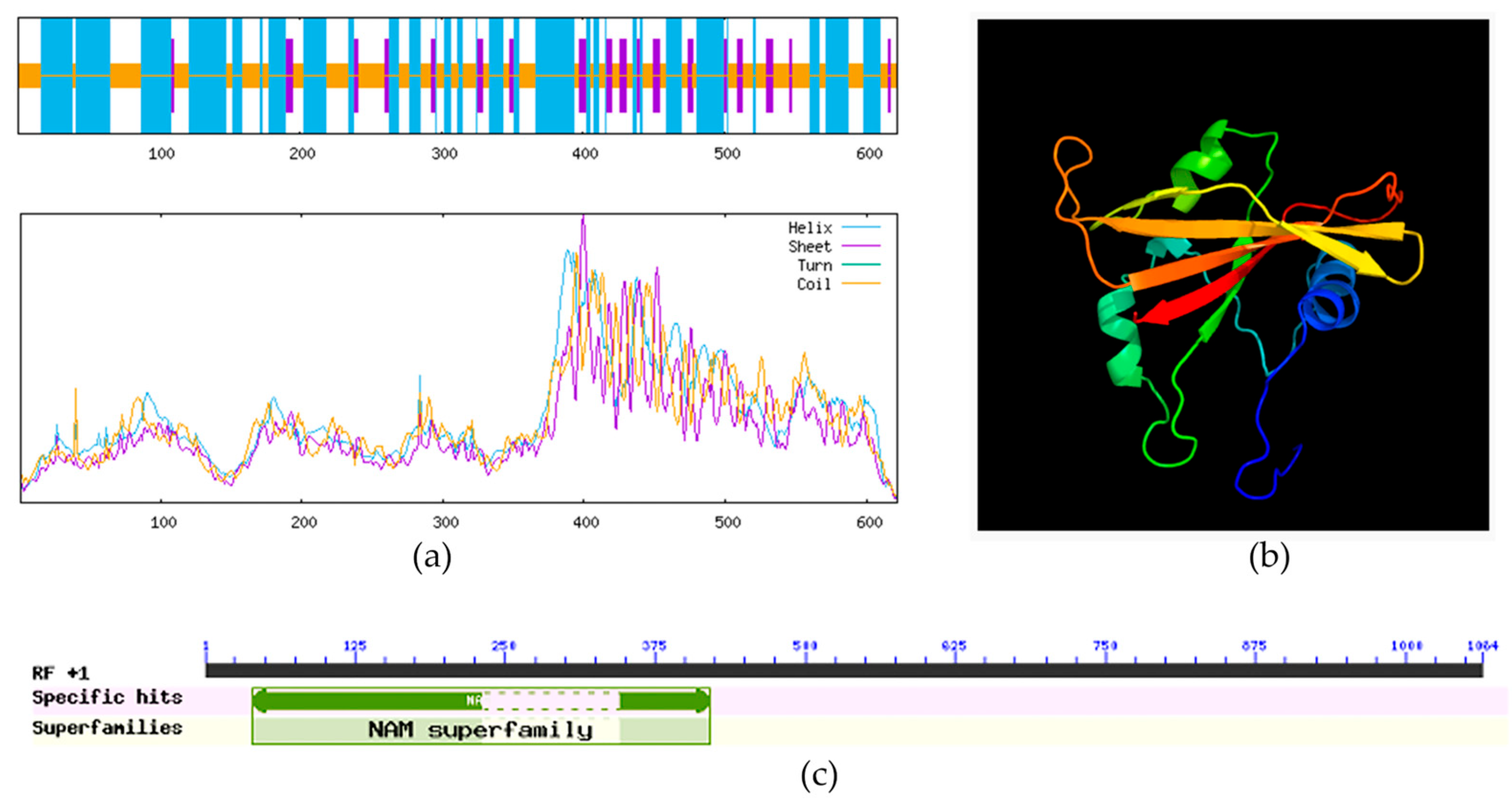
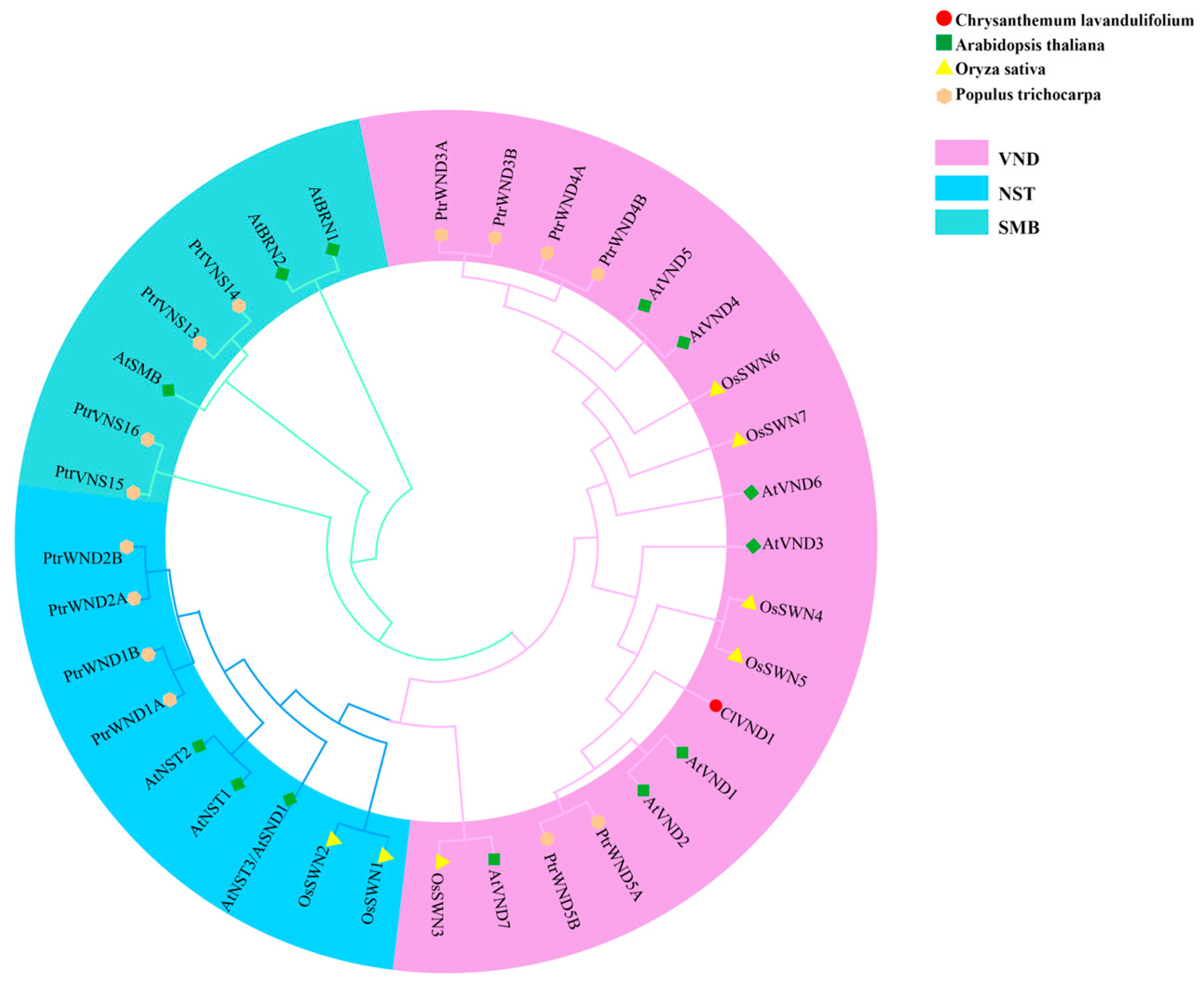
2.1. Bioinformatic Characterization of the ClVND1 Gene
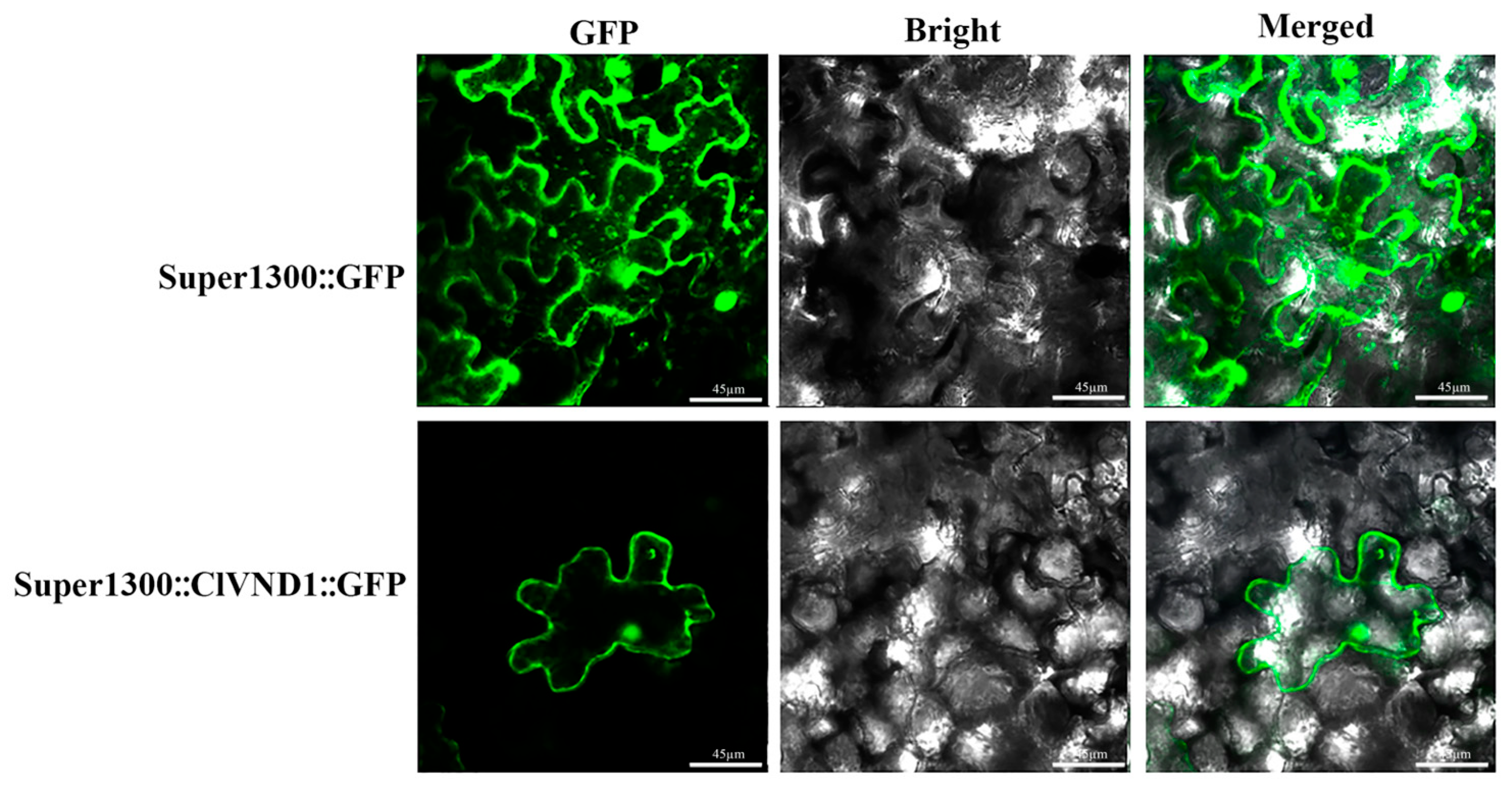
2.2. Analysis of Subcellular Localization in Nicotiana benthamiana
2.3. Identification of Positive Transgenic Lines and Validation by qRT-PCR
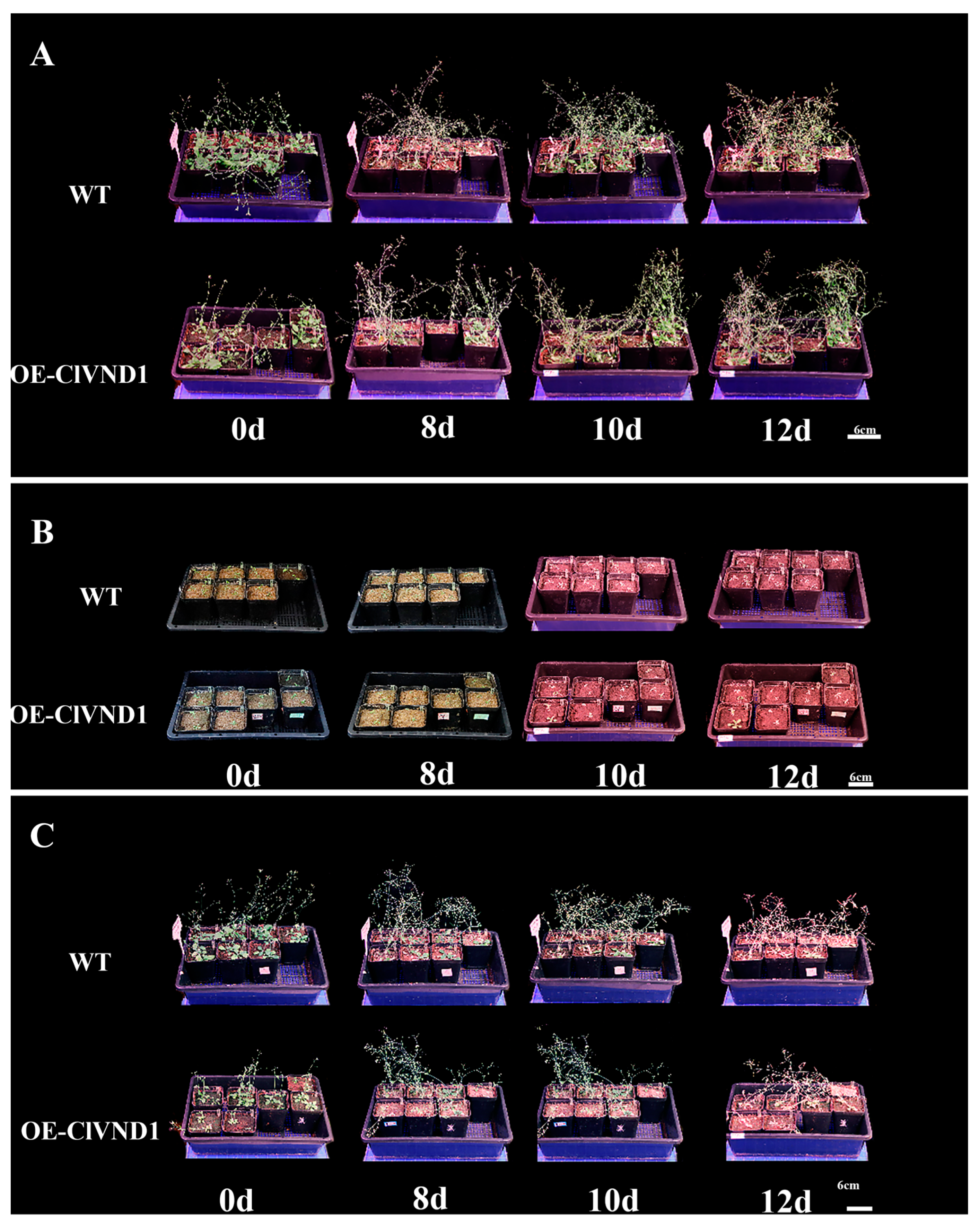
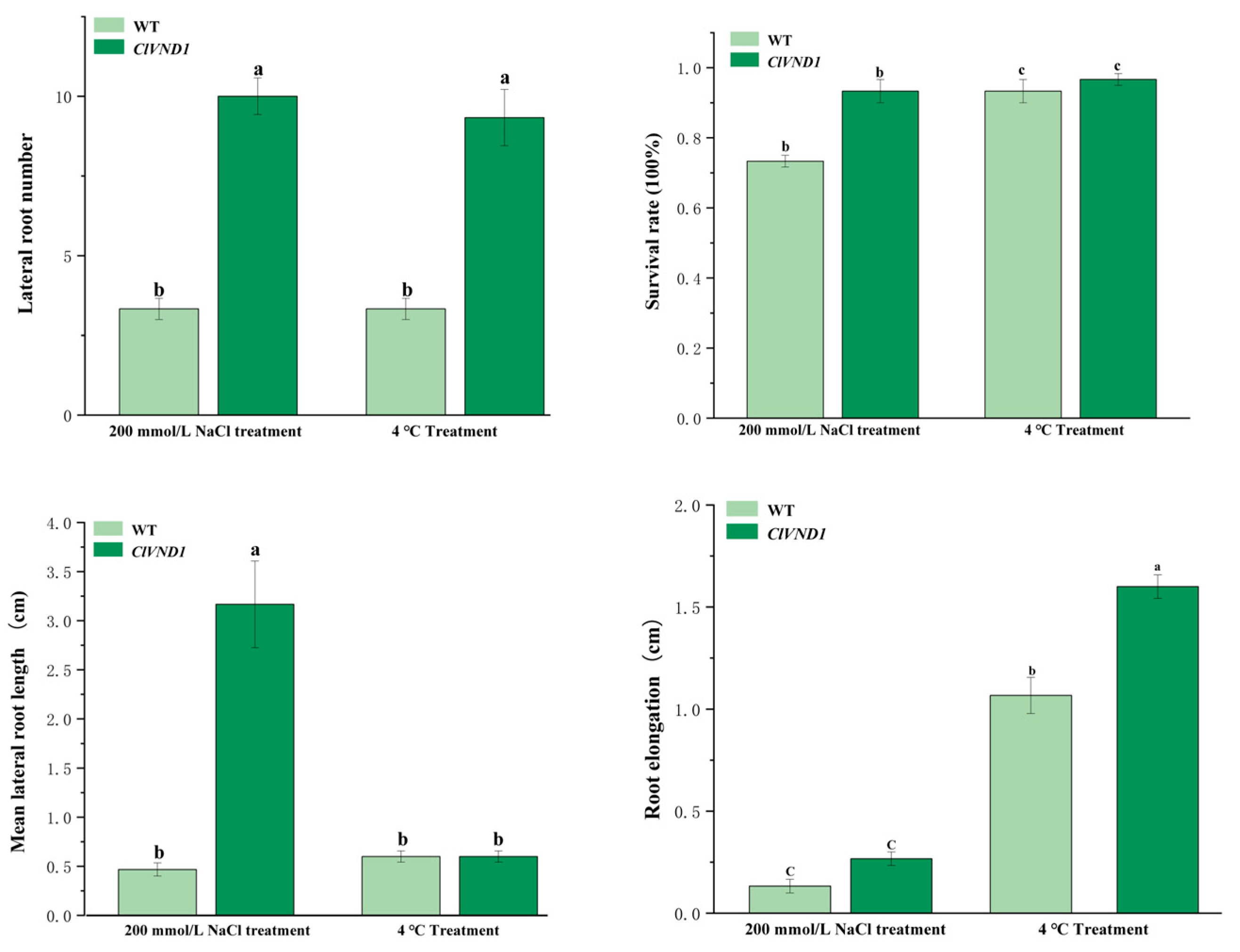
2.4. Response of Transgenic A. thaliana to NaCl and Low-Temperature (4 °C) Stress Treatments
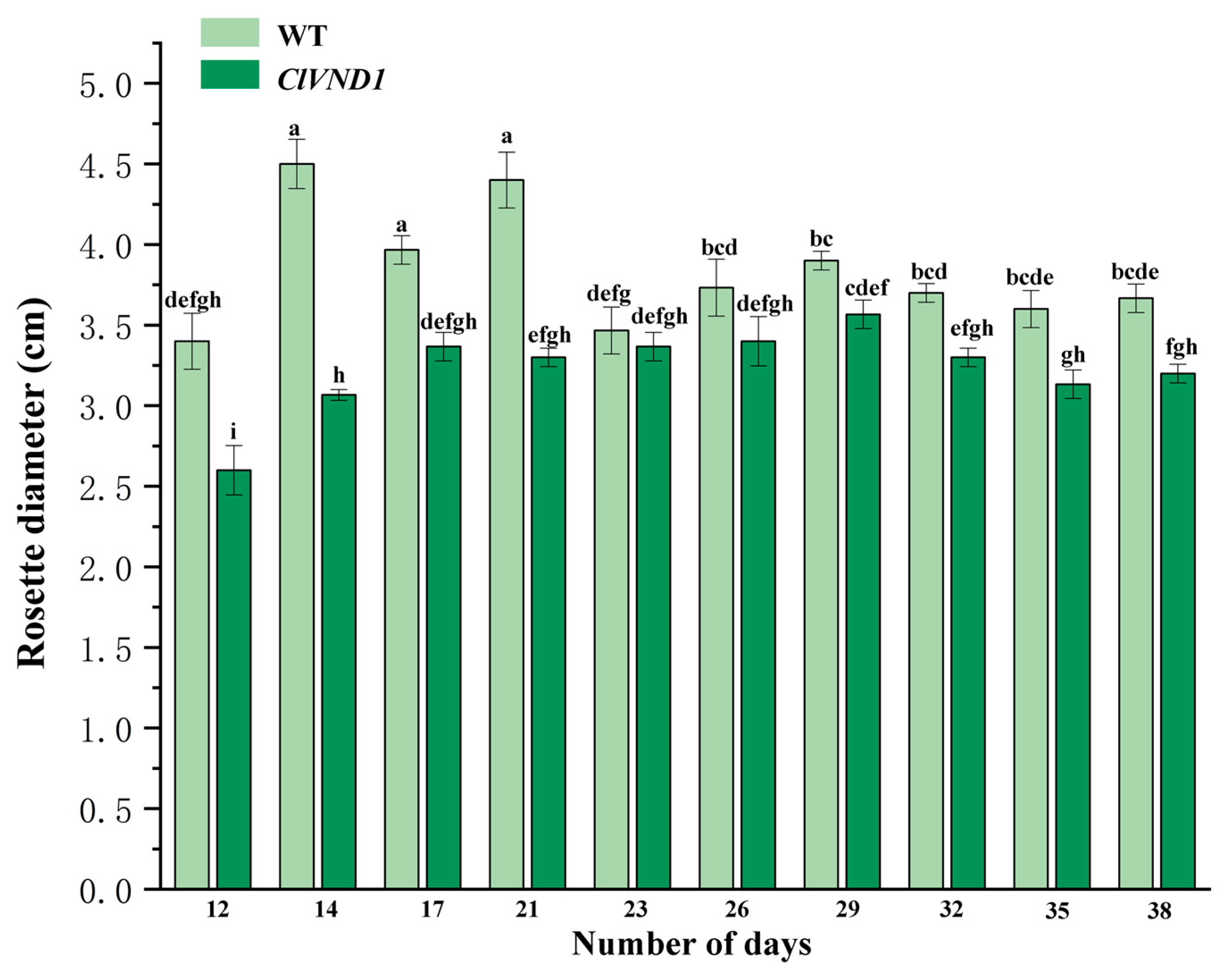
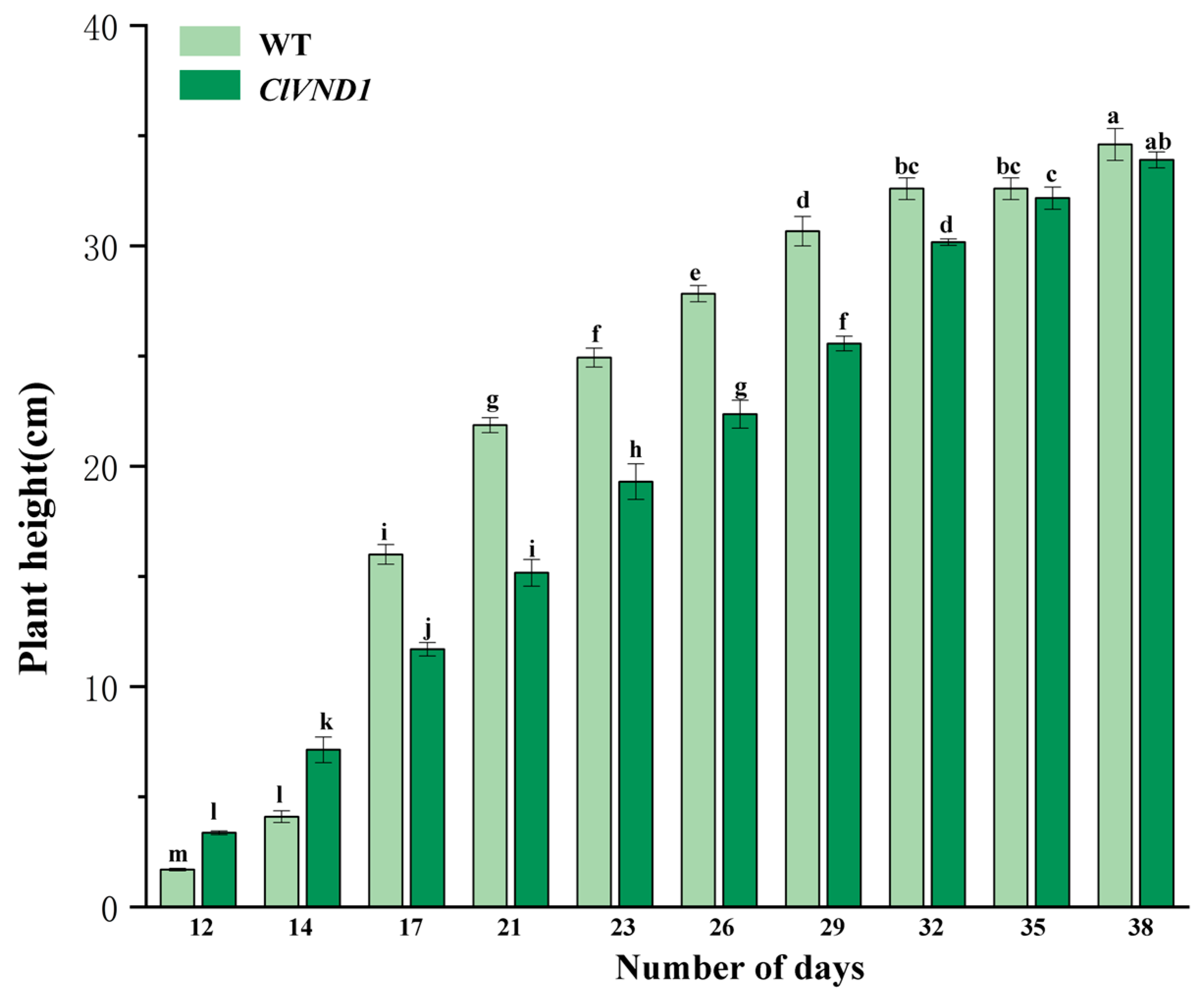
2.5. Observation of the Growth and Development of Transgenic A. thaliana
2.6. Paraffin Section Analysis of Stem Segments in Wild-Type and Transgenic Arabidopsis thaliana
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.2. Test Method
4.2.1. Target Gene Fragment
4.2.2. Bioinformatics Analysis
4.2.3. Observation of the Subcellular Localization Signal
4.2.4. Construction of the Super1300 Overexpression Vector
4.2.5. Identification of Transgenic Plants
4.2.6. Quantitative Real-Time PCR Assay
4.2.7. Transformation and Screening of A. thaliana
4.2.8. Stress Treatment and Phenotypic Observation
4.2.9. Observation of the Growth and Development of Transgenic A. thaliana
4.2.10. Paraffin Sectioning
4.2.11. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

Appendix B
References
- Sun, L.J.; Li, D.Y.; Zhang, H.J.; Song, F.M. The role of NAC transcription factors in Plant disease resistance and abiotic stress resistance. Genetics 2012, 34, 993–1002. [Google Scholar]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Demura, T.; Ye, Z.-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]
- Ko, J.H.; Yang, S.H.; Park, A.H.; Lerouxel, O.; Han, K.H. AtNAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 2007, 50, 1035–1048. [Google Scholar] [CrossRef]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef]
- Soyano, T.; Thitamadee, S.; Machida, Y.; Chua, N.H. ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 2018, 20, 3359–3373. [Google Scholar] [CrossRef]
- Bennett, T.; Van Den Toorn, A.; Sanchez-Perez, G.F.; Campilho, A.; Willemsen, V.; Snel, B.; Scheres, B. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 2010, 22, 640–654. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar] [CrossRef]
- Fendrych, M.; Van Hautegem, T.; Van Durme, M.; Olvera-Carrillo, Y.; Huysmans, M.; Karimi, M.; Lippens, S.; Guérin, C.J.; Krebs, M.; Schumacher, K.; et al. Programmed cell death controlled by AtNAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 2014, 24, 931–940. [Google Scholar] [CrossRef]
- Zhou, J.L.; Zhong, R.Q.; Ye, Z.H. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS ONE 2014, 9, e105726. [Google Scholar] [CrossRef]
- Endo, H.; Yamaguchi, M.; Tamura, T.; Nakano, Y.; Nishikubo, N.; Yoneda, A.; Kato, K.; Kubo, M.; Kajita, S.; Katayama, Y.; et al. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 2015, 56, 242–254. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Ye, Z.H. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal. Behav. 2015, 10, e989746. [Google Scholar] [CrossRef]
- Fang, S.; Shang, X.G.; Yao, Y.; Li, W.X.; Guo, W.Z. NST- and SND-subgroup NAC proteins coordinately act to regulate secondary cell wall formation in cotton. Plant Sci. 2012, 301, 110657. [Google Scholar] [CrossRef]
- Sun, L.; Liu, L.P.; Wang, Y.Z.; Yang, L.; Wang, M.J.; Liu, J.X. NAC103, a NAC family transcription factor, regulates ABA response during seed germination and seedling growth in Arabidopsis. Planta 2020, 252, 95. [Google Scholar] [CrossRef]
- Yang, J.H.; Lee, K.H.; Du, Q.; Yang, S.; Yuan, B.J.; Qi, L.Y.; Wang, H.Z. A membrane-associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytol. 2020, 226, 59–74. [Google Scholar] [CrossRef]
- Wen, J.; Wang, C.T.; Yang, Y.P. Research progress on the Regulation of Secondary Cell Wall Thickening in the xylem of plants. J. Southwest For. Univ. (Nat. Sci.) 2021, 41, 182–188. [Google Scholar]
- Zhong, R.Q.; Ye, Z.H. Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci. 2014, 229, 193–207. [Google Scholar] [CrossRef]
- Chen, X.; Lu, S.C.; Wang, Y.F.; Zhang, X.; Lv, B.; Luo, L.Q.; Xi, D.D.; Shen, J.B.; Ma, H.; Ming, F. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015, 82, 302–314. [Google Scholar] [CrossRef]
- Dong, F.; Huang, H.; Liu, J.; Zhang, M.; Zhou, Y.; Dai, S. Cloning and function analysis of ClNAC9 from Chrysanthemum lavandulifolium. Can. J. Plant Sci. 2018, 98, 1265–1279. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, G.Y.; Chen, S.Y. Structure and regulation of plant transcription factors. Chin. Sci. Bull. 2020, 45, 1465–1474. [Google Scholar]
- Huang, T.L.; Zhang, R.; He, Y.X.; Yang, R.Z.; Song, W.W.; Lai, Z.X.; Li, N.; Liu, S.C. Identification of NAC family members and expression analysis of their coding genes under NaCl stress in pepper. J. Plant Resour. Environ. 2023, 32, 12–24. [Google Scholar]
- Zhang, Y.L.; Zhang, C.; Chen, X.X.; Liu, A.X.; Sun, L.J. Identification and functional prediction of tomato NAC family genes. Anhui Agric. Sci. 2014, 20, 6549–6552+6561. [Google Scholar]
- Zhou, J.J.; Fang, Z.J.; Han, Q.J.; Pan, J.L.; Lian, Y.Y.; Gu, H.Y.; Sun, L.J. Identification and functional prediction of NAC family genes in maize. Genom. Appl. Biol. 2017, 36, 16. [Google Scholar]
- Sun, X.; Shangguan, L.F.; Fang, J.G.; Song, C.N.; Wang, C.; Mu, X. Bioinformatics analysis of NAC transcription factor family in grape. Genom. Appl. Biol. 2011, 30, 14. [Google Scholar]
- Wang, Y.; Bai, X. Bioinformatics analysis of soybean NAC gene family. Soybean Sci. 2014, 33, 325–333. [Google Scholar]
- He, L.; Bian, J.; Xu, J.Y.; Yang, K.J. Novel maize NAC transcriptional repressor ZmNAC071 confers enhanced sensitivity to ABA and osmotic stress by downregulating stress-responsive genes in transgenic Arabidopsis. J. Agric. Food Chem. 2019, 67, 890–8918. [Google Scholar] [CrossRef]
- Duan, A.Q.; Yang, X.L.; Feng, K.; Liu, J.X.; Xu, Z.S.; Xiong, A.S. Genome-wide analysis of NAC transcription factors and their response to abiotic stress in celery (Apium graveolens L.). Comput. Biol. Chem. 2020, 84, 107186. [Google Scholar] [CrossRef]
- Tamura, T.; Endo, H.; Suzuki, A.; Sato, Y.; Kato, K.; Ohtani, M.; Yamaguchi, M.; Demura, T. Affinity-based high-resolution analysis of DNA binding by VASCULAR-RELATED NAC-DOMAIN7 via fluorescence correlation spectroscopy. Plant J. Cell Mol. Biol. 2019, 100, 298–313. [Google Scholar] [CrossRef]
- Tan, T.T.; Endo, H.; Sano, R.; Kurata, T.; Yamaguchi, M.; Ohtani, M.; Demura, T. Transcription factors VND1-VND3 contribute to cotyledon xylem vessel formation. Plant Physiol. 2018, 176, 773–789. [Google Scholar] [CrossRef]
- Negi, S.; Tak, H.; Ganapathi, T.R. Xylem specific activation of 5′ upstream regulatory region of two NAC transcription factors (MusaVND6 and MusaVND7) in banana is regulated by SNBE-like sites. PLoS ONE 2018, 13, e0192852. [Google Scholar] [CrossRef]
- Negi, S.; Tak, H.; Ganapathi, T.R. Cloning and functional characterization of MusaVND1 using transgenic banana plants. Transgenic Res. 2015, 24, 571–585. [Google Scholar] [CrossRef]
- Wang, H.; Li, T.; Li, W.; Wang, W.; Zhao, H. Identification and analysis of Chrysanthemum nankingense NAC transcription factors and an expression analysis of OsNAC7 subfamily members. PeerJ 2021, 9, e11505. [Google Scholar] [CrossRef]
- Deng, Z.Y.; Luo, L.; Yu, C.; Zhang, Q.X.; Sui, Y.J. The research progress of ornamental plant NAC transcription factor. J. Plant Genet. Resour. 2024, 25, 737–750. [Google Scholar]
- Wang, H.; Li, T.; Zhou, Y.Y.; Wang, W.; Zhao, H.E. Cloning and expression analysis of Chrysanthemum lavandulifolium lignin-regulated Transcription factor ClSND1-like. Northwest Acta Bot. Sin. 2021, 41, 359–367. [Google Scholar]
- Li, Y.X.; He, W.T.; Liu, Y.Y.; Mei, C.D.; Wang, H.; Song, X.B. ClBRN1 from Chrysanthemum lavandulifolium enhances the stress resistance of transgenic Arabidopsis. PeerJ 2024, 12, e18620. [Google Scholar] [CrossRef]
- Zhang, W.X.; Wang, H.; Guo, Y.N.; Hao, X.Y.; Li, Y.X.; He, W.T.; Zhao, X.; Cai, S.Y.; Song, X.B. Functional Validation of Different Alternative Splicing Variants of the Chrysanthemum lavandulifolium ClNUM1 Gene in Tobacco. Curr. Issues Mol. Biol. 2024, 46, 5242–5256. [Google Scholar] [CrossRef]
- Yang, X.F.; Kim, M.Y.; Ha, J.M.; Lee, S.H. Overexpression of the Soybean NAC Gene GmNAC109 Increases Lateral Root Formation and Abiotic Stress Tolerance in Transgenic Arabidopsis Plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Huang, S.Q.; Zhang, C.; Li, D.F.; Wu, Y.B.; Deng, J.L.; Shan, S.L.; Qi, J.M. Overexpression of CcNAC1 gene promotes early flowering and enhances drought tolerance of jute (Corchorus capsularis L.). Protoplasma 2020, 258, 337–345. [Google Scholar] [CrossRef]
- Zhou, S.Z.; Luo, T.; Liu, L. Bioinformatics analysis of NAC gene family in moss. Genom. Appl. Biol. 2022, 41, 822–833. [Google Scholar]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005, 169, 785–797. [Google Scholar] [CrossRef]
- Ohtani, M.; Nishikubo, N.; Xu, B.; Yamaguchi, M.; Mitsuda, N.; Goué, N.; Shi, F.; Ohme-Takagi, M.; Demura, T. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 2011, 67, 499–512. [Google Scholar] [CrossRef]
- Takata, N.; Awano, T.; Nakata, M.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525. [Google Scholar] [CrossRef]
- Mou, Z.L.; Fan, W.H.; Dong, X.N. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Shi, Z.; Maximova, S.; Liu, Y.; Verica, J.; Guiltinan, M. Functional analysis of the Theobroma cacao NPR1 gene in arabidopsis. BMC Plant Biol. 2010, 10, 248. [Google Scholar] [CrossRef]
- Wu, J.Y.; Yang, S.Q.; Chen, N.N.; Jiang, Q.N.; Huang, L.L.; Qi, J.X.; Xu, G.H.; Shen, L.S.; Yu, H.; Fan, X.R.; et al. Nuclear translocation of OsMADS25 facilitated by OsNAR2.1 in response to nitrate signal promotes root growth by regulating the expression of OsMADS27 and OsARF7 in rice. Plant Commun. 2023, 4, 100642. [Google Scholar] [CrossRef]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. Cell Mol. Biol. 2007, 51, 897–909. [Google Scholar] [CrossRef]
- Qi, W.W.; Gong, X.L.; Wang, Y.; Wu, Y. Research Progress on the Application of Dipping Flower Method in Plant Genetic Transformation. Mod. Agric. Sci. Technol. 2014, 9–10+12. [Google Scholar]



| Primer Name | Sequence (5′→3′) | TM/°C | Purpose |
|---|---|---|---|
| ClVND1-F | gcTCTAGAatgatcatggacacagtaga | 57.29 | Amplification of the full-length VND1 sequence with restriction sites. |
| ClVND1-R | aaCTGCAGatcaaaaatgcataatcca | 54.71 | |
| pSuper1300-F | gtgacgccatttcgccttttc | 57.69 | |
| pSuper1300-R | ggtggtgcagatgaacttcagg | 58.69 |
| Primer Name | Sequence (5′→3′) | TM/°C | Purpose |
|---|---|---|---|
| ClVND1-F | GTTTTGGAAGGCTACGGGGA | 59.96 | Real-time PCR Primers |
| ClVND1-R | GTTTGTCCCGTTGCACGTTT | 60.18 | |
| Actin8-F | TCAGCACTTTCCAGCAGATG | 55.22 | |
| Actin8-R | CTGTGGACAATGCCTGGAC | 56.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Mei, C.; Zhang, H.; Liao, Y.; Zhai, Y.; Wang, H.; Song, X. Cloning and Functional Analysis of ClVND1, a Member of the OsNAC7 Subfamily of the NAC Family in Chrysanthemum lavandulifolium. Plants 2025, 14, 2925. https://doi.org/10.3390/plants14182925
Liu Y, Mei C, Zhang H, Liao Y, Zhai Y, Wang H, Song X. Cloning and Functional Analysis of ClVND1, a Member of the OsNAC7 Subfamily of the NAC Family in Chrysanthemum lavandulifolium. Plants. 2025; 14(18):2925. https://doi.org/10.3390/plants14182925
Chicago/Turabian StyleLiu, Yueyue, Chendi Mei, Hao Zhang, Ying Liao, Yinuo Zhai, Hai Wang, and Xuebin Song. 2025. "Cloning and Functional Analysis of ClVND1, a Member of the OsNAC7 Subfamily of the NAC Family in Chrysanthemum lavandulifolium" Plants 14, no. 18: 2925. https://doi.org/10.3390/plants14182925
APA StyleLiu, Y., Mei, C., Zhang, H., Liao, Y., Zhai, Y., Wang, H., & Song, X. (2025). Cloning and Functional Analysis of ClVND1, a Member of the OsNAC7 Subfamily of the NAC Family in Chrysanthemum lavandulifolium. Plants, 14(18), 2925. https://doi.org/10.3390/plants14182925




