Effects of GS3 Editing in japonica Rice ‘Nipponbare’ on Grain Morphology, Yield Components, and Response to Heat Stress at the Reproductive Stage
Abstract
1. Introduction
2. Results
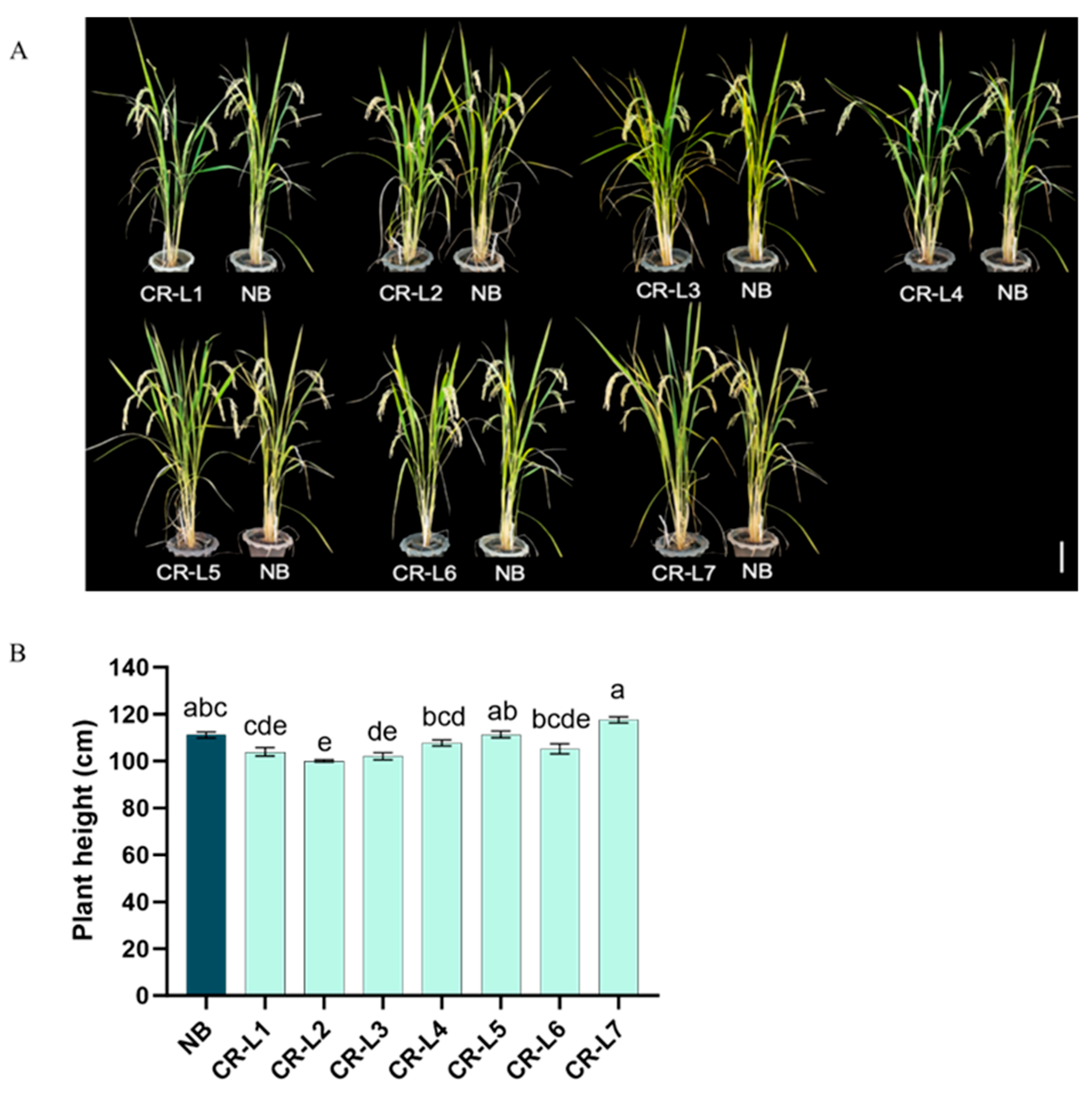
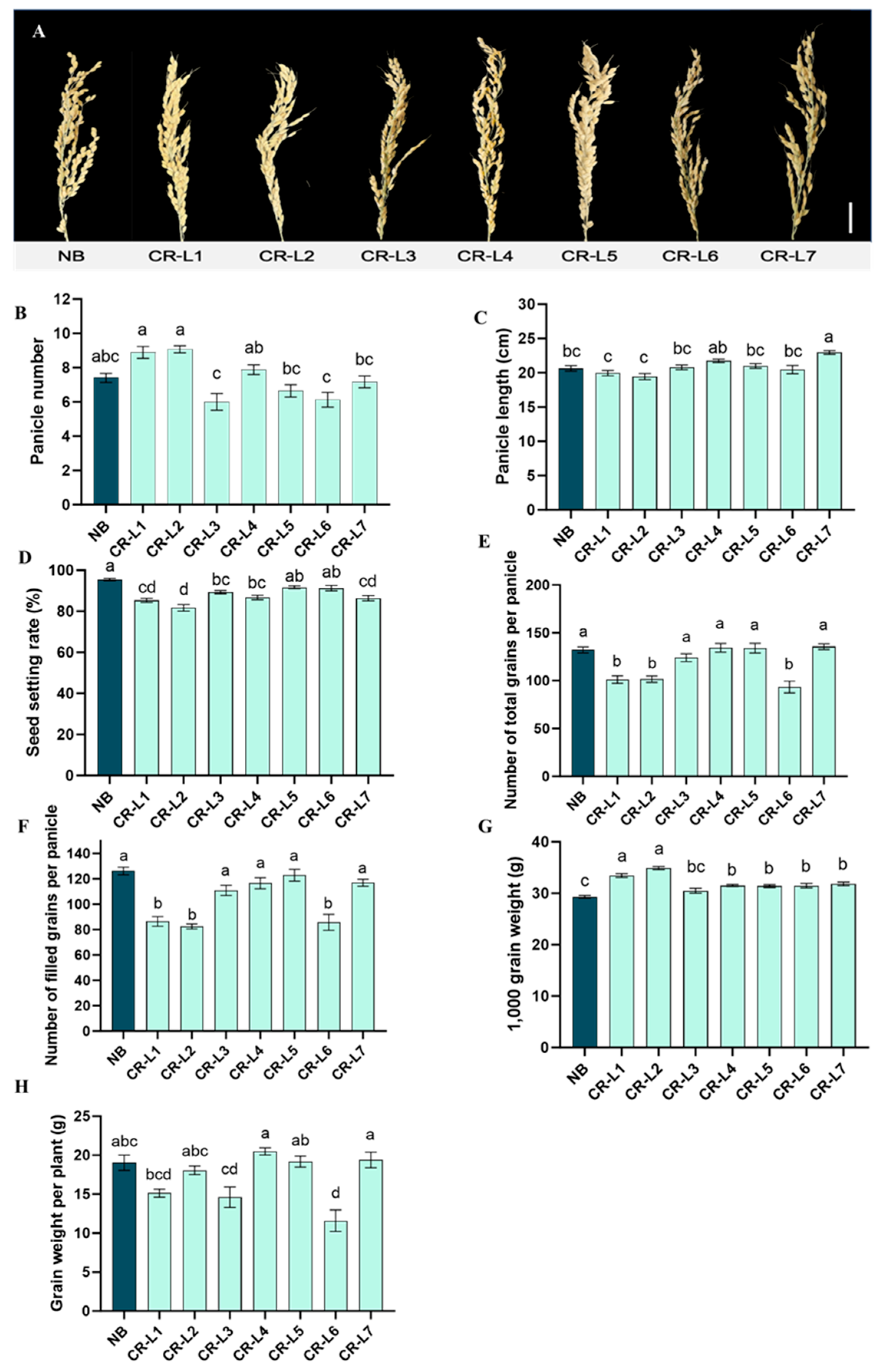
2.1. Effects of GS3 Editing on Grain Length and Yield Components
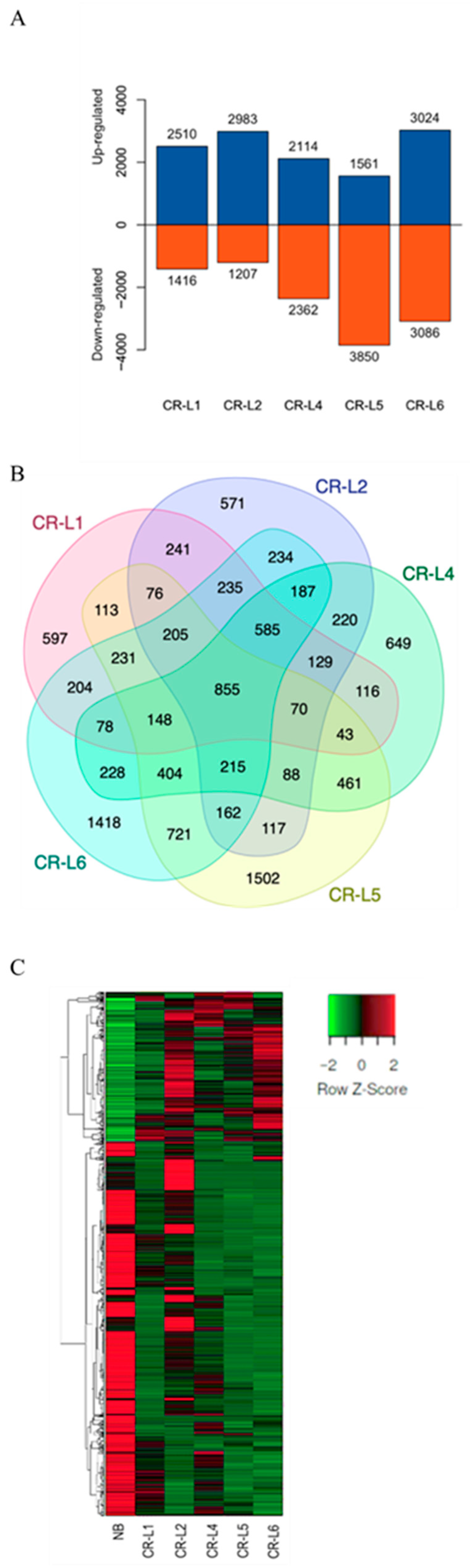
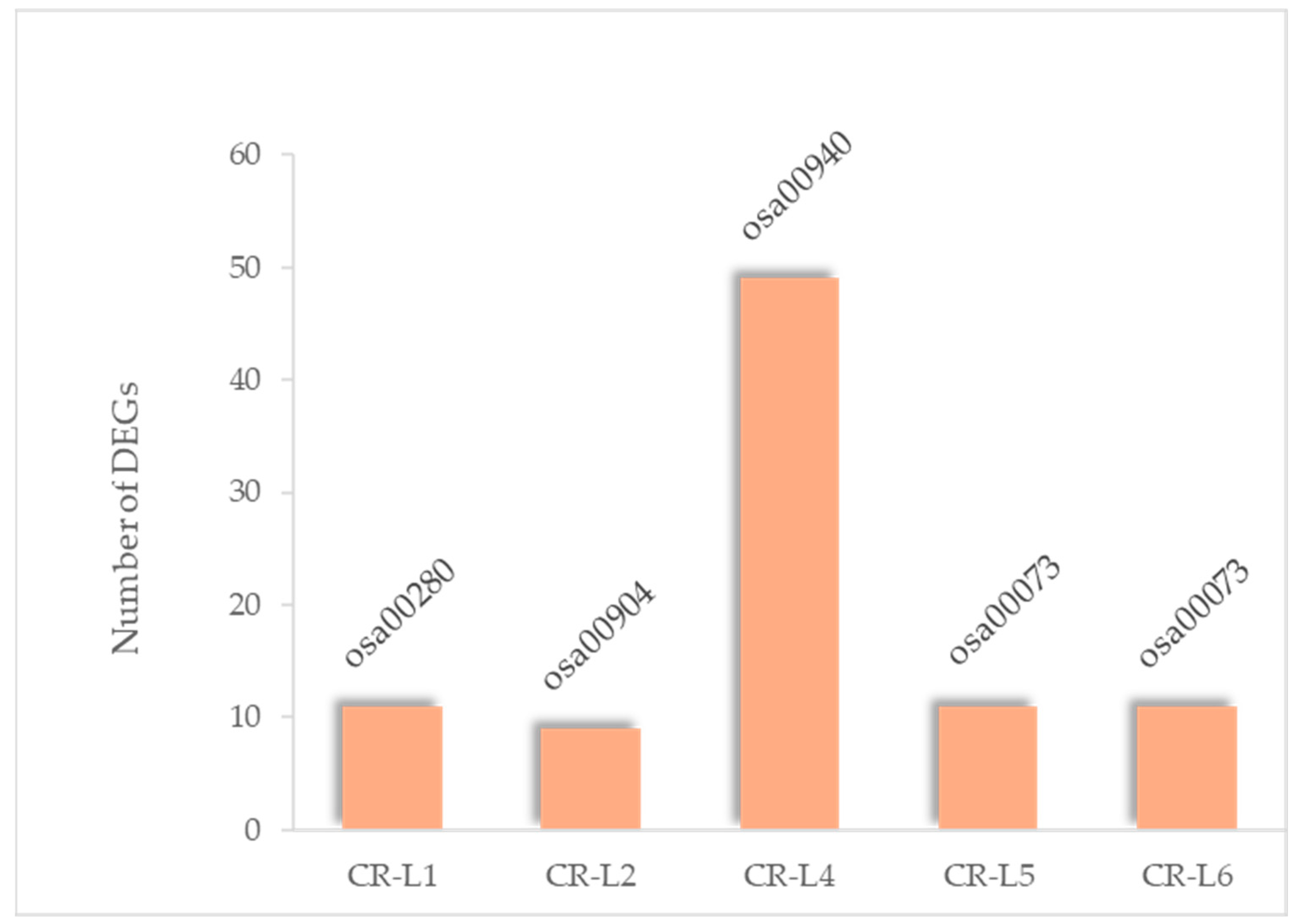
2.2. Identification of DEGs Between Wild-Type and GS3-Edited Lines and Prediction of Their Functions
2.3. Effects of GS3 on Yield and Yield Components Under High-Temperature Treatment
3. Discussion
3.1. The Pleiotropic and Allele-Specific Effects of GS3 Mutations on Agronomic Traits
3.2. Molecular Basis of Phenotypic Variation: Insights from Gene Expression
3.3. Allele-Specific Thermotolerance at the Reproductive Stage
4. Materials and Methods
4.1. Plant Materials
4.2. Construction of Vectors and Plant Transformation
4.3. Assessment of Yields and Yield Components of T3 GS3-Edited Lines
4.4. Transcriptome Analysis
4.5. High-Temperature Treatment Experiment
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar]
- Cordero-Lara, K.I. Temperate japonica rice (Oryza sativa L.) breeding: History, present and future challenges. Chil. J. Agric. Res. 2020, 80, 303–314. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, D.; Fan, C.; Zhang, C.; Zhang, C.; Liu, Z. Cell type-specific differentiation between indica and japonica rice root tip responses to different environments based on single-cell RNA sequencing. Front. Genet. 2021, 12, 659500. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Suzuki, K.; Ishikawa, H.; Nonoue, Y.; Nagata, K.; Fukuoka, S.; Tanaka, J. Genomic Regions Involved in Differences in Eating and Cooking Quality Other than Wx and Alk Genes between indica and japonica Rice Cultivars. Rice 2021, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Pacleb, M.; Jeong, O.-Y.; Lee, J.-S.; Padolina, T.; Braceros, R.; Pautin, L.; Torollo, G.; Sana, E.E.; Del-Amen, J.Y.; Baek, M.-K.; et al. Breeding Temperate Japonica Rice Varieties Adaptable to Tropical Regions: Progress and Prospects. Agronomy 2021, 11, 2253. [Google Scholar] [CrossRef]
- Nevame, A.Y.M.; Emon, R.M.; Malek, M.A.; Hasan, M.M.; Alam, M.A.; Muharam, F.M.; Aslani, F.; Rafii, M.Y.; Ismail, M.R. Relationship between high temperature and formation of chalkiness and their effects on quality of rice. Biomed. Res. Int. 2018, 2018, 1653721. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Arai-Kichise, Y.; Shiwa, Y.; Ebana, K.; Shibata-Hatta, M.; Yoshikawa, H.; Yano, M.; Wakasa, K. Genome-wide DNA polymorphisms in seven rice cultivars of temperate and tropical japonica groups. PLoS ONE 2014, 9, e86312. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Wu, J.; Itoh, T.; Numa, H.; Antonio, B.; Sasaki, T. The Nipponbare genome and the next- generation of rice genomics research in Japan. Rice 2016, 9, 33. [Google Scholar] [CrossRef]
- Ishimaru, K.; Kashiwagi, T.; Hirotsu, N.; Madoka, Y. Identification and physiological analyses of a locus for rice yield potential across the genetic background. J. Exp. Bot. 2005, 56, 2745–2753. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, H.; Ma, B.; Liu, G.; Wang, J.; Wang, J.; Gao, R.; Li, J.; Liu, J.; Xu, J.; et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 2017, 8, 14789. [Google Scholar] [CrossRef]
- Cuevas, R.P.; Pede, V.O.; McKinley, J.; Velarde, O.; Demont, M. Rice Grain Quality and Consumer Preferences: A Case Study of Two Rural Towns in the Philippines. PLoS ONE 2016, 11, e0150345. [Google Scholar] [CrossRef]
- Xu, N.; Qiu, Y.; Cui, X.; Fei, C.; Xu, Q. Enhancing grain shape, thermotolerance, and alkaline tolerance via Gγ protein manipulation in rice. Theor. Appl. Genet. 2024, 137, 154. [Google Scholar] [CrossRef]
- Ren, D.; Ding, C.; Qian, Q. Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 2023, 68, 314–350. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Wu, W.; Liu, X.; Wang, M.; Meyer, R.S.; Luo, X.; Ndjiondjop, M.N.; Tan, L.; Zhang, J.; Wu, J.; Cai, H.; et al. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 2017, 3, 17064. [Google Scholar] [CrossRef]
- Xia, D.; Zhou, H.; Liu, R.; Dan, W.; Li, P.; Wu, B.; Chen, J.; Wang, L.; Gao, G.; Zhang, Q.; et al. GL3.3, a Novel QTL Encoding a GSK3/SHAGGY-like Kinase, Epistatically Interacts with GS3 to Produce Extra-long Grains in Rice. Mol. Plant 2018, 11, 754–756. [Google Scholar] [CrossRef]
- Ying, J.Z.; Ma, M.; Bai, C.; Huang, X.H.; Liu, J.L.; Fan, Y.Y.; Song, X.J. TGW3, a Major QTL that Negatively Modulates Grain Length and Weight in Rice. Mol. Plant 2018, 11, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef]
- Zhan, P.; Ma, S.; Xiao, Z.; Li, F.; Wei, X.; Lin, S.; Wang, X.; Ji, Z.; Fu, Y.; Pan, J.; et al. Natural variations in grain length 10 (GL10) regulate rice grain size. J. Genet. Genom. 2022, 49, 405–413. [Google Scholar] [CrossRef]
- Bai, F.; Ma, H.; Cai, Y.; Shahid, M.Q.; Zheng, Y.; Lang, C.; Chen, Z.; Wu, J.; Liu, X.; Wang, L. Natural allelic variation in GRAIN SIZE AND WEIGHT 3 of wild rice regulates the grain size and weight. Plant Physiol. 2023, 193, 502–518. [Google Scholar] [CrossRef]
- Liang, X.; Liu, Y.; Tian, M.; Jiang, W.; Zheng, Y.; Chen, Z.; Liu, X.; Wang, L. The natural variation allele OsGSW3.2 in Oryza rufipogon is involved in brassinosteroid signaling and influences grain size and weight. Plant J. 2025, 121, e70110. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Fan, C.; Yu, S.; Wang, C.; Xing, Y. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 2009, 118, 465–472. [Google Scholar] [CrossRef]
- Wang, C.; Chen, S.; Yu, S. Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice. Theor. Appl. Genet. 2011, 122, 905–913. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR-Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef] [PubMed]
- Yuyu, C.; Aike, Z.; Pao, X.; Xiaoxia, W.; Yongrun, C.; Beifang, W.; Yue, Z.; Liaqat, S.; Shihua, C.; Liyong, C.; et al. Effects of GS3 and GL3. 1 for grain size editing by CRISPR/Cas9 in rice. Rice Sci. 2020, 27, 405–413. [Google Scholar] [CrossRef]
- Huang, J.; Gao, L.; Luo, S.; Liu, K.; Qing, D.; Pan, Y.; Dai, G.; Deng, G.; Zhu, C. The genetic editing of GS3 via CRISPR/Cas9 accelerates the breeding of three-line hybrid rice with superior yield and grain quality. Mol. Breed. 2022, 42, 22. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, C.; Li, Q.; Liu, Q. Genetic control of grain appearance quality in rice. Biotechnol. Adv. 2022, 60, 108014. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.R.; Zhang, H.; Gao, J.; Shan, J.X.; Ye, W.W.; Lin, H.X. TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 2022, 8, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fu, X. Can heterotrimeric G proteins improve sustainable crop production and promote a more sustainable Green Revolution? Innov. Life 2023, 1, 100024-1–100024-2. [Google Scholar]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. 2016, 43, 529–532. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y.; et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci 2017, 8, 298. [Google Scholar] [CrossRef]
- Hidangmayum, A.; Dwivedi, P.; Katiyar, D.; Hemantaranjan, A. Application of chitosan on plant responses with special reference to abiotic stress. Physiol. Mol. Biol. Plants 2019, 25, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Widiastuti, A.; Yoshino, M.; Hasegawa, M.; Nitta, Y.; Sato, T. Heat shock-induced resistance increases chitinase-1 gene expression and stimulates salicylic acid production in melon (Cucumis melo L.). Physiol. Mol. Plant Pathol. 2013, 82, 51–55. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, L.; Wang, J.; Sun, J.; Xia, X.; Geng, X.; Wang, X.; Xu, Z.; Xu, Q. Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield-and quality-associated loci. BMC Biol. 2018, 16, 102. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mc Carthy, D.J.; Smyth, G.K. Edge R: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. Shiny GO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene | Gene Annotation | RNAseq | |||||
|---|---|---|---|---|---|---|---|---|
| NB | CR-L1 | CR-L2 | CR-L4 | CR-L5 | CR-L6 | |||
| Os01g0860400 | - | Similar to Acidic endochitinase precursor (EC 3.2.1.14) | 1.96 | 0.66 | 0.67 | 0.89 | 0.93 | 0.59 |
| Os01g0860500 | C10501 | Similar to Hevamine A precursor [Includes Chitinase (EC 3.2.1.14); Lysozyme (EC 3.2.1.17)] | 8.26 | 1.28 | 0.07 | 0.27 | 0.87 | 0.18 |
| * Os01g0660200 | C10728, OsChib3a | Acidic class III chitinase OsChib3a precursor (Chitinase) (EC 3.2.1.14) | 0.58 | 0.12 | 0.03 | 0.00 | 3.42 | 0.03 |
| * Os05g0247100 | Drought-induced protein 3, Xylanase inhibitor protein 2 | Similar to Glycosyl hydrolases family 18 | 27.40 | 5.64 | 12.48 | 7.22 | 335.3 | 2.99 |
| Os07g0446800 | HEXOKINASE-1 | Similar to Hexokinase | 0.47 | 25.94 | 19.37 | 2.17 | 22.41 | 1.98 |
| Os08g0518900 | C10122 | Chitinase (EC 3.2.1.14) | 8.14 | 0.24 | 0.03 | 0.29 | 1.42 | 0.03 |
| * Os11g0462100 | - | Glycoside hydrolase, family 18 protein | 0.90 | 2.08 | 2.79 | 9.68 | 12.72 | 6.87 |
| * Os11g0700900 | C10923 | Glycoside hydrolase, subgroup, catalytic core domain-containing protein | 1.05 | 0.23 | 0.26 | 0.00 | 18.06 | 0.06 |
| * Os11g0701400 | C10150 | Chitinase (EC 3.2.1.14) III C10150-rice (EC 3.2.1.14) | 0.82 | 0.00 | 0.00 | 0.00 | 4.01 | 0.00 |
| Os11g0701800 | xylanase inhibitor protein, rice xylanase inhibitor | Chitinase (EC 3.2.1.14) III C10701-rice (EC 3.2.1.14) (Class III chitinase homologue) | 29.92 | 2.82 | 2.51 | 3.24 | 8.60 | 0.28 |
| * Os11g0702100 | - | Similar to Class III chitinase homologue | 1.65 | 0.13 | 0.00 | 0.00 | 10.02 | 0.00 |
| Line | Treatment | Panicles Number/Pot | Grain Number /Panicle | Filled Grains Number/Plant | Total Grains Number/Plant | Seed Setting Rate (%) | Relative Seed Setting Rate (%) | 1000-Grain Weight (g) | Yield (g/Pot) |
|---|---|---|---|---|---|---|---|---|---|
| Nipponbare | Normal | 21 ± 2.65 a | 74 ± 4.33 cd | 1293 ± 263.04 a | 1412 ± 108.89 ab | 75.33 ± 2.42 ab | 44.65 ± 5.27 b | 20.97 ± 0.53 c | 21.57 ± 1.80 bc |
| High temp. | 19 ± 0.58 a | 64 ± 2.31 d | 424 ± 35.87 c | 1262 ± 23.23 ac | 31.76 ± 0.16 d | 23.27 ± 0.21 bc | 9.84 ± 0.92 e | ||
| CR-L2 | Normal | 15 ± 2.65 ab | 91 ± 7.48 ad | 1063 ± 89.53 ab | 1354 ± 123.92 a | 78.60 ± 3.79 a | 0.68 ± 0.55 c | 24.60 ± 0.43 ab | 26.15 ± 2.69 a |
| High temp. | 13 ± 2.65 b | 94 ± 6.37 abc | 2 ± 0.50 d | 1406 ± 135.06 ab | 0.51 ± 0.49 e | 10.00 ± 0.60 e | 0.03 ± 0.01 f | ||
| CR-L5 | Normal | 10 ± 1.00 b | 118 ± 12.09 a | 952 ± 67.18 ab | 1183 ± 33.94 c | 68.78 ± 3.17 bc | 58.81 ± 13.94 a | 24.33 ± 0.78 ac | 23.47 ± 0.90 b |
| High temp. | 9 ± 2.08 b | 113 ± 6.70 ab | 467 ± 90.37 c | 1228 ± 251.02 c | 39.95 ± 0.93 d | 24.36 ± 1.37 ac | 11.32 ± 1.84 e | ||
| CR-L6 | Normal | 11 ± 2.12 b | 96 ± 21.33 abc | 709 ± 0.71 bc | 1085 ± 41.01 c | 65.44 ± 2.54 c | 1.22 ± 1.13 c | 22.68 ± 0.61 bc | 16.10 ± 0.45 d |
| High temp. | 11 ± 0.00 b | 98 ± 6.17 abc | 8 ± 7.78 d | 1080 ± 67.88 c | 0.81 ± 0.77 e | 15.60 ± 3.20 d | 0.15 ± 0.15 f | ||
| CR-L7 | Normal | 13 ± 1.73 b | 85 ± 12.94 bd | 799 ± 42.39 b | 1139 ± 79.20 bc | 72.92 ± 2.42 ac | 1.50 ± 2.07 c | 23.49 ± 0.74 ac | 18.76 ± 0.50 cd |
| High temp. | 12 ± 2.65 b | 93 ± 10.43 abc | 2 ± 3.54 d | 1100 ± 137.49 ac | 1.10 ± 1.54 e | 27.29 ± 1.82 a | 0.38 ± 0.54 f |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Y.; Sangarwut, N.; Tongmark, K.; Chakhonkaen, S.; Wang, L.; Muangprom, A. Effects of GS3 Editing in japonica Rice ‘Nipponbare’ on Grain Morphology, Yield Components, and Response to Heat Stress at the Reproductive Stage. Plants 2025, 14, 2897. https://doi.org/10.3390/plants14182897
Qi Y, Sangarwut N, Tongmark K, Chakhonkaen S, Wang L, Muangprom A. Effects of GS3 Editing in japonica Rice ‘Nipponbare’ on Grain Morphology, Yield Components, and Response to Heat Stress at the Reproductive Stage. Plants. 2025; 14(18):2897. https://doi.org/10.3390/plants14182897
Chicago/Turabian StyleQi, Yongbin, Numphet Sangarwut, Keasinee Tongmark, Sriprapai Chakhonkaen, Linyou Wang, and Amorntip Muangprom. 2025. "Effects of GS3 Editing in japonica Rice ‘Nipponbare’ on Grain Morphology, Yield Components, and Response to Heat Stress at the Reproductive Stage" Plants 14, no. 18: 2897. https://doi.org/10.3390/plants14182897
APA StyleQi, Y., Sangarwut, N., Tongmark, K., Chakhonkaen, S., Wang, L., & Muangprom, A. (2025). Effects of GS3 Editing in japonica Rice ‘Nipponbare’ on Grain Morphology, Yield Components, and Response to Heat Stress at the Reproductive Stage. Plants, 14(18), 2897. https://doi.org/10.3390/plants14182897






