Evolution and Functional Dynamics of the BAG Gene Family in Passion Fruit (Passiflora edulis)
Abstract
1. Introduction
2. Methods
2.1. Identification of the Passion Fruit BAG Genes
2.2. Phylogenetic Analysis of the Passion Fruit BAG Genes
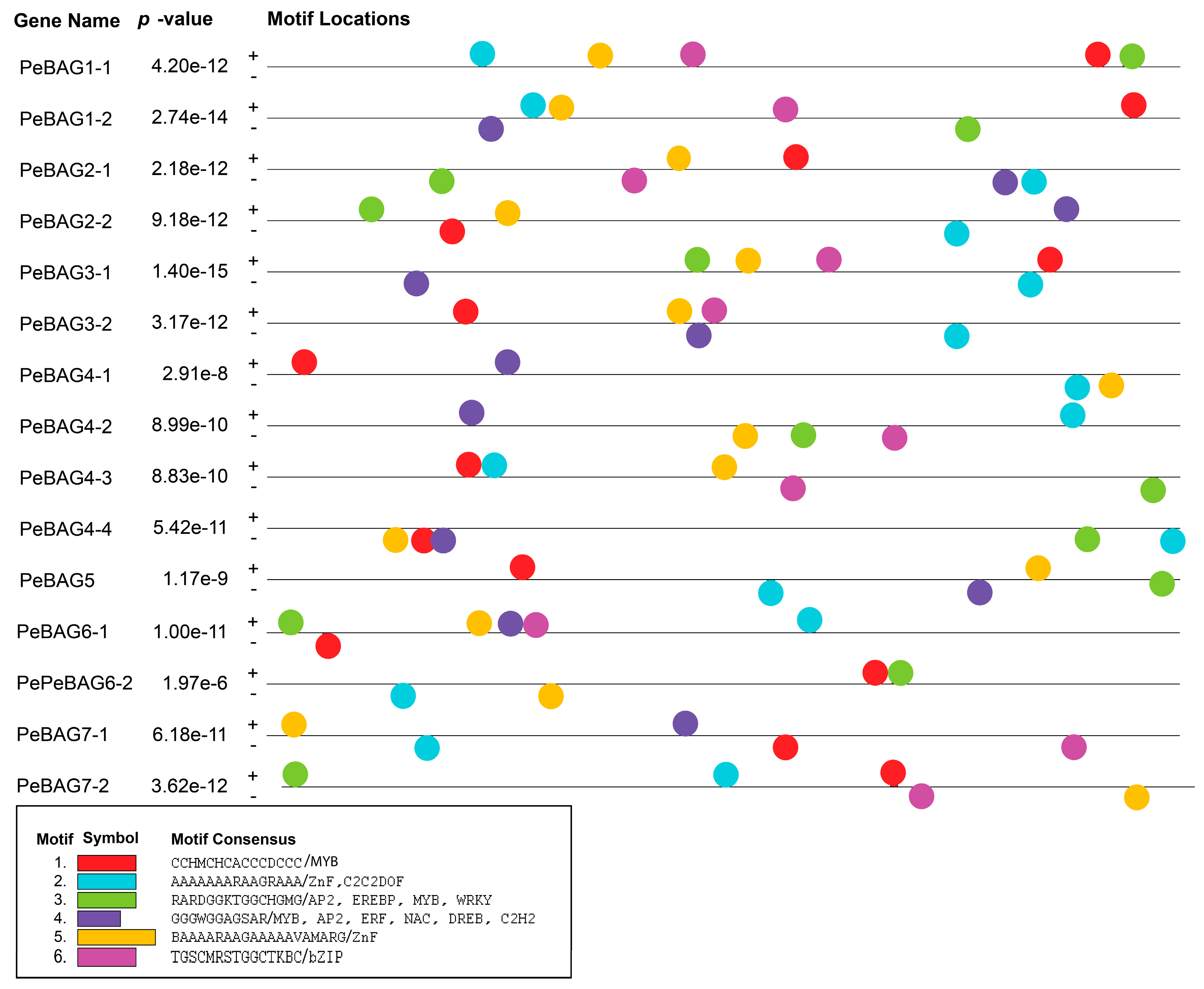
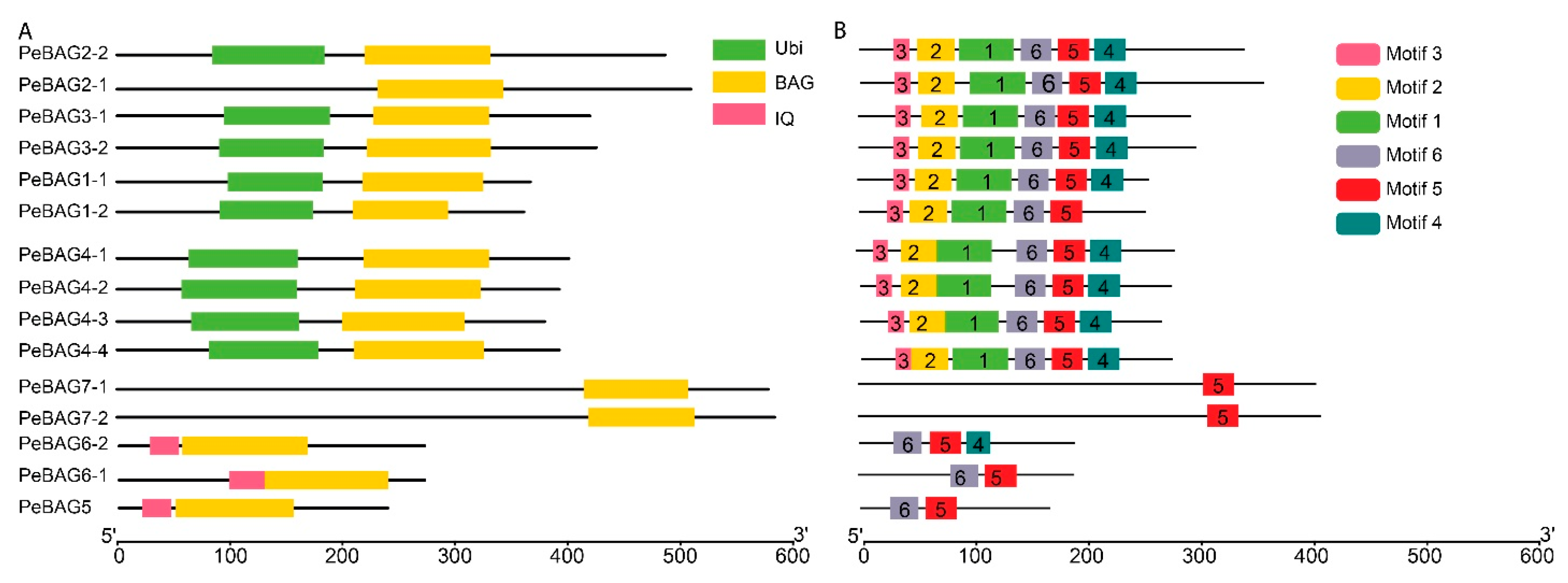
2.3. Conserved Domains and Motif Analysis
2.4. Gene Locations, Duplications, and Synteny Analysis of PeBAGs
2.5. Heatmap Clustering of Expression Data
2.6. Promoter Analysis of PeBAGs and Prediction of Cis-Regulatory Elements
2.7. Heat Stress Treatments, RNA Extraction, and qRT-PCR
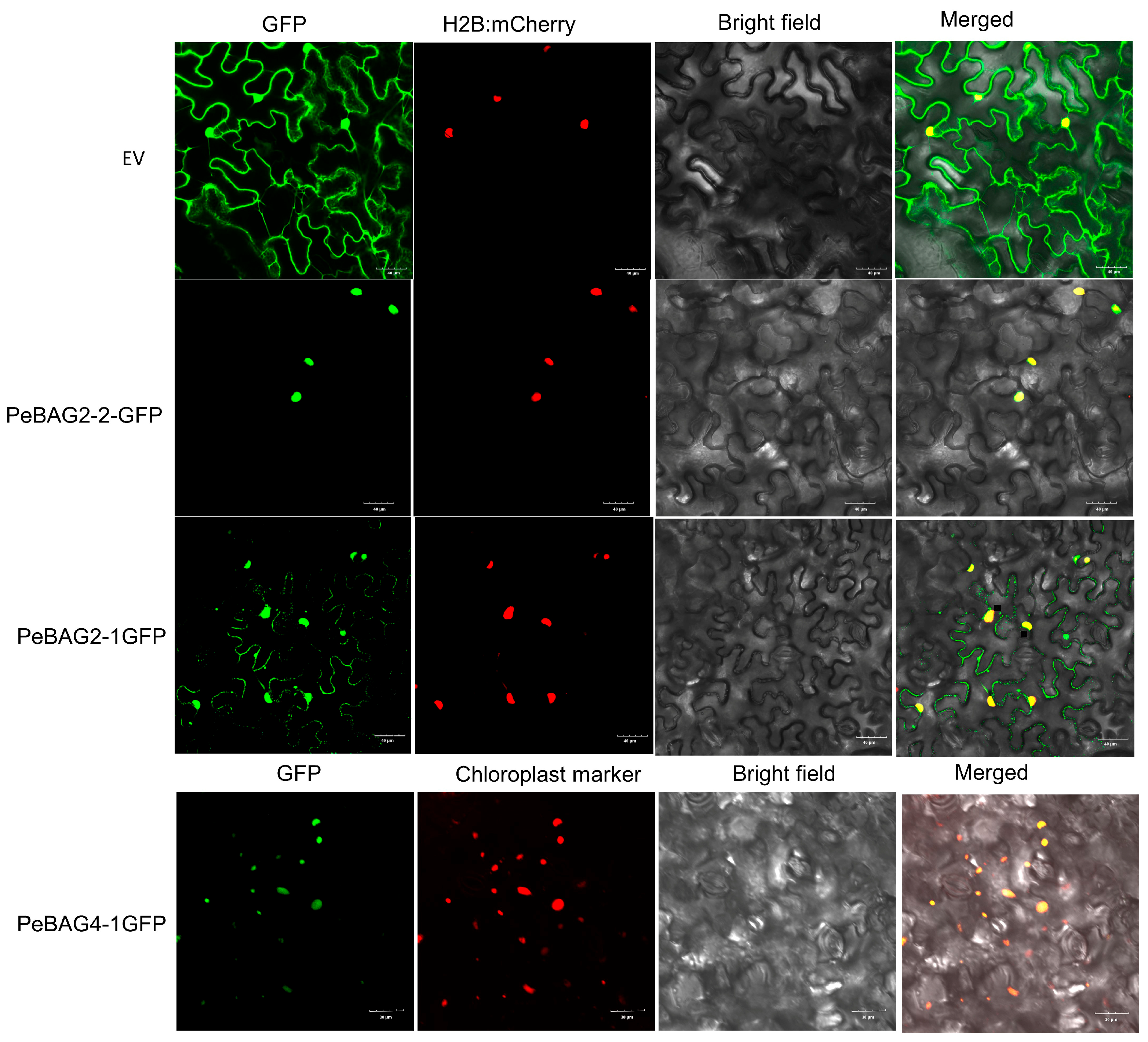
2.8. GFP Subcellular Localization
3. Results
3.1. Identification of the Passion Fruit BAG Gene Family
3.2. Phylogenetic Classification of PeBAGs Genes Encoded Proteins
3.3. Chromosomal Locations, Duplications, and Synteny Analysis of PeBAGs

4. Discussions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ortiz, D.C.; Bohórquez, A.; Duque, M.C.; Tohme, J.; Cuéllar, D.; Mosquera Vásquez, T. Evaluating Purple Passion Fruit (Passiflora edulis Sims f. edulis) Genetic Variability in Individuals from Commercial Plantations in Colombia. Genet. Resour. Crop Evol. 2012, 59, 1089–1099. [Google Scholar] [CrossRef]
- Song, S.; Zhang, D.; Ma, F.; Xing, W.; Huang, D.; Wu, B.; Chen, J.; Chen, D.; Xu, B.; Xu, Y. Genome-Wide Identification and Expression Analyses of the Aquaporin Gene Family in Passion Fruit (Passiflora edulis), Revealing PeTIP3-2 to Be Involved in Drought Stress. Int. J. Mol. Sci. 2022, 23, 5720. [Google Scholar] [CrossRef]
- Siebra, A.L.A.; Oliveira, L.R.; Martins, A.O.B.P.B.; Siebra, D.C.; Albuquerque, R.S.; Lemos, I.C.S.; Delmondes, G.A.; Tintino, S.R.; Figueredo, F.G.; da Costa, J.G.M.; et al. Potentiation of Antibiotic Activity by Passiflora cincinnata Mast. Front of Strains Staphylococcus aureus and Escherichia coli. Saudi J. Biol. Sci. 2018, 25, 37–43. [Google Scholar] [CrossRef]
- Taïwe, G.S.; Kuete, V. Chapter 24—Passiflora edulis. In Medicinal Spices and Vegetables from Africa; Kuete, V., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 513–526. ISBN 978-0-12-809286-6. [Google Scholar]
- He, X.; Luan, F.; Yang, Y.; Wang, Z.; Zhao, Z.; Fang, J.; Wang, M.; Zuo, M.; Li, Y. Passiflora edulis: An Insight Into Current Researches on Phytochemistry and Pharmacology. Front. Pharmacol. 2020, 11, 617. [Google Scholar] [CrossRef]
- Ma, F.; Zhou, H.; Yang, H.; Huang, D.; Xing, W.; Wu, B.; Li, H.; Hu, W.; Song, S.; Xu, Y. WRKY Transcription Factors in Passion Fruit Analysis Reveals Key PeWRKYs Involved in Abiotic Stress and Flavonoid Biosynthesis. Int. J. Biol. Macromol. 2024, 256, 128063. [Google Scholar] [CrossRef]
- Ma, F.; Zhou, H.; Xu, Y.; Huang, D.; Wu, B.; Xing, W.; Chen, D.; Xu, B.; Song, S. Comprehensive Analysis of BZIP Transcription Factors in Passion Fruit. iScience 2023, 26, 106556. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Ma, F.; Huang, D.; Xing, W.; Wu, B.; Sun, P.; Xu, B.; Song, S. Characterization of the NAC Transcription Factor in Passion Fruit (Passiflora edulis) and Functional Identification of PeNAC-19 in Cold Stress. Plants 2023, 12, 1393. [Google Scholar] [CrossRef]
- Arif, M.; Men, S.; Nawaz, A.F.; Li, X.; Xu, L.; Yang, X.; Fahad, S.; Ahmad, P.; Xu, R.; Li, L. Bcl-2-Associated Athanogene (BAG) Co-Chaperones: Key Players in Multiple Abiotic and Biotic Stress Tolerance in Plants. J. Plant Growth Regul. 2024. [Google Scholar] [CrossRef]
- Behl, C. Breaking BAG: The Co-Chaperone BAG3 in Health and Disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef]
- Briknarová, K.; Takayama, S.; Homma, S.; Baker, K.; Cabezas, E.; Hoyt, D.W.; Li, Z.; Satterthwait, A.C.; Ely, K.R. BAG4/SODD Protein Contains a Short BAG Domain. J. Biol. Chem. 2002, 277, 31172–31178. [Google Scholar] [CrossRef]
- Lee, D.W.; Kim, S.J.; Oh, Y.J.; Choi, B.; Lee, J.; Hwang, I. Arabidopsis BAG1 Functions as a Cofactor in Hsc70-Mediated Proteasomal Degradation of Unimported Plastid Proteins. Mol. Plant 2016, 9, 1428–1431. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Koren, S.A.; Cvetojevic, G.; Girardi, P.; Johnson, G.V.W. The Role of BAG3 in Health and Disease: A “Magic BAG of Tricks”. J. Cell. Biochem. 2022, 123, 4–21. [Google Scholar] [CrossRef] [PubMed]
- Brive, L.; Takayama, S.; Briknarová, K.; Homma, S.; Ishida, S.K.; Reed, J.C.; Ely, K.R. The Carboxyl-Terminal Lobe of Hsc70 ATPase Domain Is Sufficient for Binding to BAG1. Biochem. Biophys. Res. Commun. 2001, 289, 1099–1105. [Google Scholar] [CrossRef]
- Cheung, N.S.; Beart, P.M.; Pascoe, C.J.; John, C.A.; Bernard, O. Human Bcl-2 Protects Against AMPA Receptor-Mediated Apoptosis. J. Neurochem. 2000, 74, 1613–1620. [Google Scholar] [CrossRef]
- Arif, M.; Li, Z.; Luo, Q.; Li, L.; Shen, Y.; Men, S. The BAG2 and BAG6 Genes Are Involved in Multiple Abiotic Stress Tolerances in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 5856. [Google Scholar] [CrossRef]
- Farid, B.; Saddique, M.A.B.; Tahir, M.H.N.; Ikram, R.M.; Ali, Z.; Akbar, W. Expression Divergence of BAG Gene Family in Maize under Heat Stress. BMC Plant Biol. 2025, 25, 16. [Google Scholar] [CrossRef] [PubMed]
- Nawkar, G.M.; Maibam, P.; Park, J.H.; Woo, S.G.; Kim, C.Y.; Lee, S.Y.; Kang, C.H. In Silico Study on Arabidopsis BAG Gene Expression in Response to Environmental Stresses. Protoplasma 2017, 254, 409–421. [Google Scholar] [CrossRef]
- Wang, J.; Ao, M.; Ma, A.; Yu, J.; Guo, P.; Huang, S.; Peng, X.; Yun, D.-J.; Xu, Z.-Y. A Mitochondrial Localized Chaperone Regulator OsBAG6 Functions in Saline-Alkaline Stress Tolerance in Rice. Rice 2024, 17, 10. [Google Scholar] [CrossRef]
- Jiang, H.; Ji, Y.; Sheng, J.; Wang, Y.; Liu, X.; Xiao, P.; Ding, H. Genome-Wide Identification of the Bcl-2 Associated Athanogene (BAG) Gene Family in Solanum Lycopersicum and the Functional Role of SlBAG9 in Response to Osmotic Stress. Antioxidants 2022, 11, 598. [Google Scholar] [CrossRef]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.-J.; Hwang, I.; Liu, B.; Xu, Z.-Y. A DNA Methylation Reader-Chaperone Regulator-Transcription Factor Complex Activates OsHKT1;5 Expression during Salinity Stress. Plant Cell 2020, 32, 3535–3558. [Google Scholar] [CrossRef]
- Thanthrige, N.; Jain, S.; Bhowmik, S.D.; Ferguson, B.J.; Kabbage, M.; Mundree, S.; Williams, B. Centrality of BAGs in Plant PCD, Stress Responses, and Host Defense. Trends Plant Sci. 2020, 25, 1131–1140. [Google Scholar] [CrossRef]
- He, M.; Wang, Y.; Jahan, M.S.; Liu, W.; Raziq, A.; Sun, J.; Shu, S.; Guo, S. Characterization of SlBAG Genes from Solanum Lycopersicum and Its Function in Response to Dark-Induced Leaf Senescence. Plants 2021, 10, 947. [Google Scholar] [CrossRef]
- You, Q.; Zhai, K.; Yang, D.; Yang, W.; Wu, J.; Liu, J.; Pan, W.; Wang, J.; Zhu, X.; Jian, Y.; et al. An E3 Ubiquitin Ligase-BAG Protein Module Controls Plant Innate Immunity and Broad-Spectrum Disease Resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef]
- Kandoth, P.K.; Ithal, N.; Recknor, J.; Maier, T.; Nettleton, D.; Baum, T.J.; Mitchum, M.G. The Soybean Rhg1 Locus for Resistance to the Soybean Cyst Nematode Heterodera Glycines Regulates the Expression of a Large Number of Stress- and Defense-Related Genes in Degenerating Feeding Cells. Plant Physiol. 2011, 155, 1960–1975. [Google Scholar] [CrossRef]
- Ghag, S.B.; Shekhawat, U.K.S.; Ganapathi, T.R. Native Cell-Death Genes as Candidates for Developing Wilt Resistance in Transgenic Banana Plants. AoB Plants 2014, 6, plu037. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Takato, H.; Fujita, K.; Suzuki, S. HSG1, a Grape Bcl-2-Associated Athanogene, Promotes Floral Transition by Activating CONSTANS Expression in Transgenic Arabidopsis Plant. Mol. Biol. Rep. 2012, 39, 4367–4374. [Google Scholar] [CrossRef]
- Zheng, Y.-Y.; Chen, L.-H.; Fan, B.-L.; Xu, Z.; Wang, Q.; Zhao, B.-Y.; Gao, M.; Yuan, M.-H.; Tahir ul Qamar, M.; Jiang, Y.; et al. Integrative Multiomics Profiling of Passion Fruit Reveals the Genetic Basis for Fruit Color and Aroma. Plant Physiol. 2024, 194, 2491–2510. [Google Scholar] [CrossRef]
- Xia, Z.; Huang, D.; Zhang, S.; Wang, W.; Ma, F.; Wu, B.; Xu, Y.; Xu, B.; Chen, D.; Zou, M.; et al. Chromosome-Scale Genome Assembly Provides Insights into the Evolution and Flavor Synthesis of Passion Fruit (Passiflora edulis Sims). Hortic. Res. 2021, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Dong, S.; Zhang, S.; Wei, X.; Xie, Q.; Ding, Q.; Xia, R.; Zhang, X. Chromosome-Level Reference Genome Assembly Provides Insights into Aroma Biosynthesis in Passion Fruit (Passiflora edulis). Mol. Ecol. Resour. 2021, 21, 955–968. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Yu, C.; Wang, P.; Zhang, S.; Liu, J.; Cheng, Y.; Zhang, S.; Wu, Z. Passionfruit Genomic Database (PGD): A Comprehensive Resource for Passionfruit Genomics. BMC Genomics 2024, 25, 157. [Google Scholar] [CrossRef]
- Shad, M.A.; Wu, S.; Rao, M.J.; Luo, X.; Huang, X.; Wu, Y.; Zhou, Y.; Wang, L.; Ma, C.; Hu, L. Evolution and Functional Dynamics of TCP Transcription Factor Gene Family in Passion Fruit (Passiflora edulis). Plants 2024, 13, 2568. [Google Scholar] [CrossRef]
- Wu, Y.; Tian, Q.; Huang, W.; Liu, J.; Xia, X.; Yang, X.; Mou, H. Identification and Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis in Passiflora Edulis under Stem Rot Condition. Mol. Biol. Rep. 2020, 47, 2951–2962. [Google Scholar] [CrossRef]
- Huang, X.; Shad, M.A.; Shu, Y.; Nong, S.; Li, X.; Wu, S.; Yang, J.; Rao, M.J.; Aslam, M.Z.; Huang, X.; et al. Genome-Wide Analysis of the Auxin/Indoleacetic Acid (Aux/IAA) Gene Family in Autopolyploid Sugarcane (Saccharum spontaneum). Int. J. Mol. Sci. 2024, 25, 7473. [Google Scholar] [CrossRef]
- Jiang, H.; Liu, X.; Xiao, P.; Wang, Y.; Xie, Q.; Wu, X.; Ding, H. Functional Insights of Plant Bcl-2-Associated Ahanogene (BAG) Proteins: Multi-Taskers in Diverse Cellular Signal Transduction Pathways. Front. Plant Sci. 2023, 14, 1136873. [Google Scholar] [CrossRef]
- Ge, S.; Kang, Z.; Li, Y.; Zhang, F.; Shen, Y.; Ge, R.; Huang, Z. Cloning and Function Analysis of BAG Family Genes in Wheat. Funct. Plant Biol. 2016, 43, 393–402. [Google Scholar] [CrossRef]
- Alekhya, C.; Tejaswi, A.; Harika, G.; Bomma, N.; Gangashetty, P.I.; Tyagi, W.; Yogendra, K. Identification and Evaluation of BAG (B-Cell Lymphoma-2 Associated Athanogene) Family Gene Expression in Pigeonpea (Cajanus cajan) under Terminal Heat Stress. Front. Genet. 2024, 15, 1418380. [Google Scholar] [CrossRef]
- Xin, M.; Li, C.; He, X.; Li, L.; Yi, P.; Tang, Y.; Li, J.; Liu, G.; Sheng, J.; Sun, J. Integrated Metabolomic and Transcriptomic Analyses of Quality Components and Associated Molecular Regulation Mechanisms during Passion Fruit Ripening. Postharvest Biol. Technol. 2021, 180, 111601. [Google Scholar] [CrossRef]
- Pereira, Z.C.; dos Anjos Cruz, J.M.; Corrêa, R.F.; Sanches, E.A.; Campelo, P.H.; de Araújo Bezerra, J. Passion Fruit (Passiflora spp.) Pulp: A Review on Bioactive Properties, Health Benefits and Technological Potential. Food Res. Int. 2023, 166, 112626. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, J.; Lai, M.; Zhang, Y.; Qiu, W.; Li, Y.; Tu, H.; Ling, Q.; Fu, X. Differential Gene Expression Analysis and Physiological Response Characteristics of Passion Fruit (Passiflora edulis) Buds under High-Temperature Stress. PeerJ 2023, 11, e14839. [Google Scholar] [CrossRef] [PubMed]
- Doukhanina, E.V.; Chen, S.; van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and Functional Characterization of the BAG Protein Family in Arabidopsis Thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef]
- Zhou, H.; Li, J.; Liu, X.; Wei, X.; He, Z.; Hu, L.; Wang, J.; Duan, M.; Xie, G.; Wang, J.; et al. The Divergent Roles of the Rice Bcl-2 Associated Athanogene (BAG) Genes in Plant Development and Environmental Responses. Plants 2021, 10, 2169. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Zhou, H.; He, D.; Wang, J.; Yang, J.; Wang, L. Identification and Expression Analysis of BAG Gene Family in Sugarcane Saccharum Spontaneum. Genomics Appl. Biol. 2023, 42, 1364–1378. [Google Scholar] [CrossRef]
- Dash, A.; Ghag, S.B. Genome-Wide in Silico Characterization and Stress Induced Expression Analysis of BcL-2 Associated Athanogene (BAG) Family in Musa spp. Sci. Rep. 2022, 12, 625. [Google Scholar] [CrossRef]
- Li, L.; Xing, Y.; Chang, D.; Fang, S.; Cui, B.; Li, Q.; Wang, X.; Guo, S.; Yang, X.; Men, S.; et al. CaM/BAG5/Hsc70 Signaling Complex Dynamically Regulates Leaf Senescence. Sci. Rep. 2016, 6, 31889. [Google Scholar] [CrossRef]
- Kabbage, M.; Kessens, R.; Bartholomay, L.; Williams, B. The Life and Death of a Plant Cell. Annu. Rev. Plant Biol. 2017, 68, 1–30. [Google Scholar] [CrossRef]
- Kabbage, M.; Dickman, M.B. The BAG Proteins: A Ubiquitous Family of Chaperone Regulators. Cell. Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ma, Z.; Mao, H. Duplicate Genes Contribute to Variability in Abiotic Stress Resistance in Allopolyploid Wheat. Plants 2023, 12, 2465. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Zhao, T.; Holmer, R.; de Bruijn, S.; Angenent, G.C.; van den Burg, H.A.; Schranz, M.E. Phylogenomic Synteny Network Analysis of MADS-Box Transcription Factor Genes Reveals Lineage-Specific Transpositions, Ancient Tandem Duplications, and Deep Positional Conservation. Plant Cell 2017, 29, 1278–1292. [Google Scholar] [CrossRef]
- Altenhoff, A.M.; Boeckmann, B.; Capella-Gutierrez, S.; Dalquen, D.A.; DeLuca, T.; Forslund, K.; Huerta-Cepas, J.; Linard, B.; Pereira, C.; Pryszcz, L.P.; et al. Standardized Benchmarking in the Quest for Orthologs. Nat. Methods 2016, 13, 425–430. [Google Scholar] [CrossRef]
- Williams, B.; Kabbage, M.; Britt, R.; Dickman, M.B. AtBAG7, an Arabidopsis Bcl-2–Associated Athanogene, Resides in the Endoplasmic Reticulum and Is Involved in the Unfolded Protein Response. Proc. Natl. Acad. Sci. USA 2010, 107, 6088–6093. [Google Scholar] [CrossRef]
- Li, Y.; Williams, B.; Dickman, M. Arabidopsis B-Cell Lymphoma2 (Bcl-2)-Associated Athanogene 7 (BAG7)-Mediated Heat Tolerance Requires Translocation, Sumoylation and Binding to WRKY29. New Phytol. 2017, 214, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Locascio, A.; Marqués, M.C.; García-Martínez, G.; Corratgé-Faillie, C.; Andrés-Colás, N.; Rubio, L.; Fernández, J.A.; Véry, A.-A.; Mulet, J.M.; Yenush, L. BCL2-ASSOCIATED ATHANOGENE4 Regulates the KAT1 Potassium Channel and Controls Stomatal Movement. Plant Physiol. 2019, 181, 1277–1294. [Google Scholar] [CrossRef]
- Ding, H.; Qian, L.; Jiang, H.; Ji, Y.; Fang, Y.; Sheng, J.; Xu, X.; Ge, C. Overexpression of a Bcl-2-Associated Athanogene SlBAG9 Negatively Regulates High-Temperature Response in Tomato. Int. J. Biol. Macromol. 2022, 194, 695–705. [Google Scholar] [CrossRef]




| Gene Name | Locus ID | Chromosome | Size (aa) | MW (Da) | pI | A.I. | Stability | GRAVY | Predicted Location |
|---|---|---|---|---|---|---|---|---|---|
| PeBAG1-1 | Pe6g01112 | LG6 | 257 | 29,041.22 | 8.92 | 88.33 | Unstable | −0.445 | Nucleus |
| PeBAG1-2 | Pe9g01431 | LG9 | 251 | 28,073.24 | 6.37 | 88.45 | stable | −0.324 | chloroplast |
| PeBAG2-1 | Pe2g00028 | LG2 | 348 | 38,366.55 | 9.59 | 75.89 | Unstable | −0.643 | Nucleus |
| PeBAG2-2 | Pe2g02075 | LG2 | 343 | 37,833.91 | 9.57 | 72.16 | Unstable | −0.684 | Nucleus |
| PeBAG3-1 | Pe4g00348 | LG4 | 294 | 33,283.48 | 9.42 | 87.79 | Unstable | −0.652 | Cytoplasm |
| PeBAG3-2 | Pe3g01971 | LG3 | 298 | 34,012.46 | 9.82 | 90.23 | Unstable | −0.584 | Cytoplasm |
| PeBAG4-1 | Pe2g01777 | LG2 | 281 | 31,512.85 | 4.82 | 73.06 | Unstable | −0.92 | Nucleus |
| PeBAG4-2 | Pe2g03293 | LG2 | 275 | 30,762.77 | 5.64 | 74.73 | Stable | −0.675 | Cytoplasm |
| PeBAG4-3 | Pe1g00342 | LG1 | 295 | 33,205.96 | 5.88 | 79.59 | Unstable | −0.499 | Nucleus |
| PeBAG4-4 | Pe7g01275 | LG7 | 275 | 30,808.53 | 8.79 | 91.09 | Stable | −0.417 | chloroplast |
| PeBAG5 | LG9 | 167 | 18,975.48 | 5.49 | 81.74 | unstable | −0.535 | chloroplast | |
| PeBAG6-1 | Pe3g01508 | LG3 | 518 | 56,789.93 | 4.69 | 74.46 | Unstable | −0.726 | Nucleus |
| PeBAG6-2 | Pe3g01506 | LG3 | 157 | 17,927.22 | 5.21 | 83.89 | Stable | −0.89 | chloroplast |
| PeBAG7-1 | LG2 | 409 | 47,230.17 | 8.95 | 83.47 | unstable | −0.634 | chloroplast | |
| PeBAG7-2 | LG2 | 405 | 46,606.38 | 9.47 | 78.57 | unstable | −0.65 | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shad, M.A.; Wu, S.; Wu, Y.; Zhang, L.; Zhou, Y.; Wang, J.; Wang, L.; Ma, C.; Hu, L. Evolution and Functional Dynamics of the BAG Gene Family in Passion Fruit (Passiflora edulis). Plants 2025, 14, 2887. https://doi.org/10.3390/plants14182887
Shad MA, Wu S, Wu Y, Zhang L, Zhou Y, Wang J, Wang L, Ma C, Hu L. Evolution and Functional Dynamics of the BAG Gene Family in Passion Fruit (Passiflora edulis). Plants. 2025; 14(18):2887. https://doi.org/10.3390/plants14182887
Chicago/Turabian StyleShad, Munsif Ali, Songguo Wu, Yuxin Wu, Lijie Zhang, Yuhong Zhou, Jingzheng Wang, Lingqiang Wang, Chongjian Ma, and Lihua Hu. 2025. "Evolution and Functional Dynamics of the BAG Gene Family in Passion Fruit (Passiflora edulis)" Plants 14, no. 18: 2887. https://doi.org/10.3390/plants14182887
APA StyleShad, M. A., Wu, S., Wu, Y., Zhang, L., Zhou, Y., Wang, J., Wang, L., Ma, C., & Hu, L. (2025). Evolution and Functional Dynamics of the BAG Gene Family in Passion Fruit (Passiflora edulis). Plants, 14(18), 2887. https://doi.org/10.3390/plants14182887






