Analysis of the Metabolic and Structural Changes in Ulmus pumila ‘Zhonghua Jinye’ Leaf Under Shade Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Treatments
2.2. Pigment and Leaf Colour Determination and Ultra-Structural Analyses
2.3. RNA Extraction, Library Construction and Sequencing
2.4. Illumina Sequencing, Assembly, and Functional Annotation
2.5. The Trend, GO and KEGG Pathway Enrichment of DEG
2.6. Real-Time qPCR Verification
3. Results
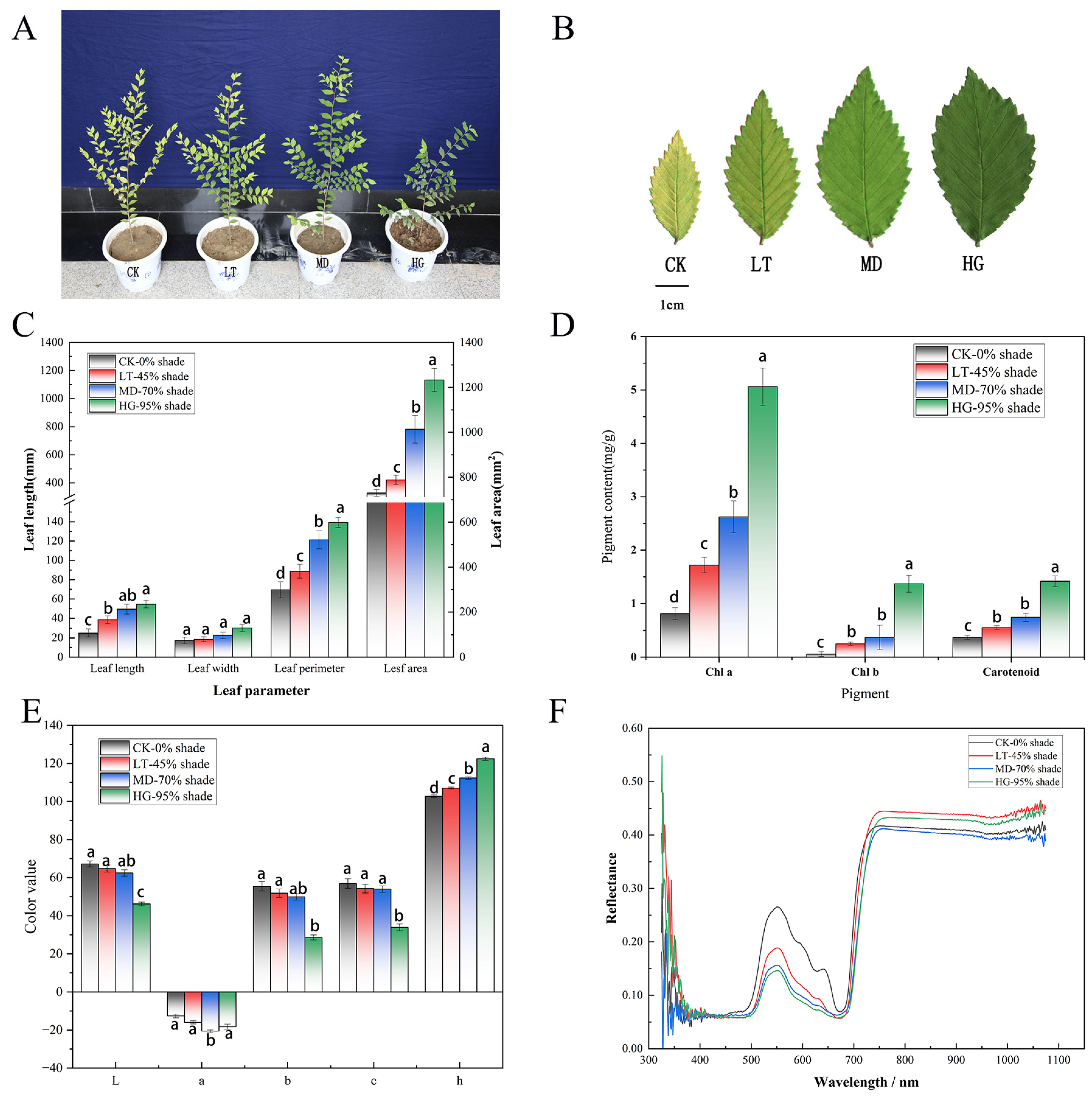
3.1. Effects of Shade Stress on Phenotype and Physiological Indices
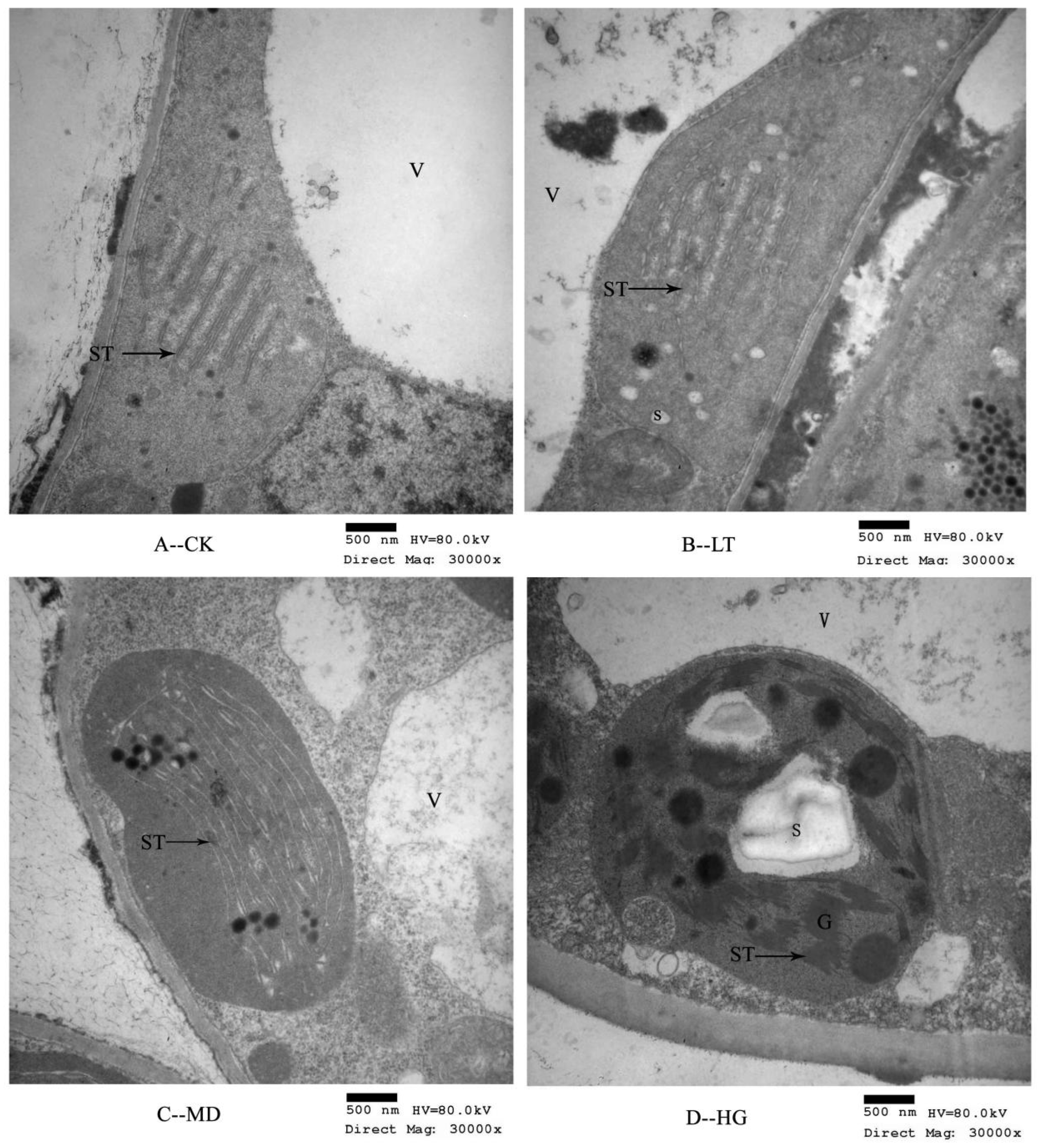
3.2. Chloroplast Thylakoid Structural Changes
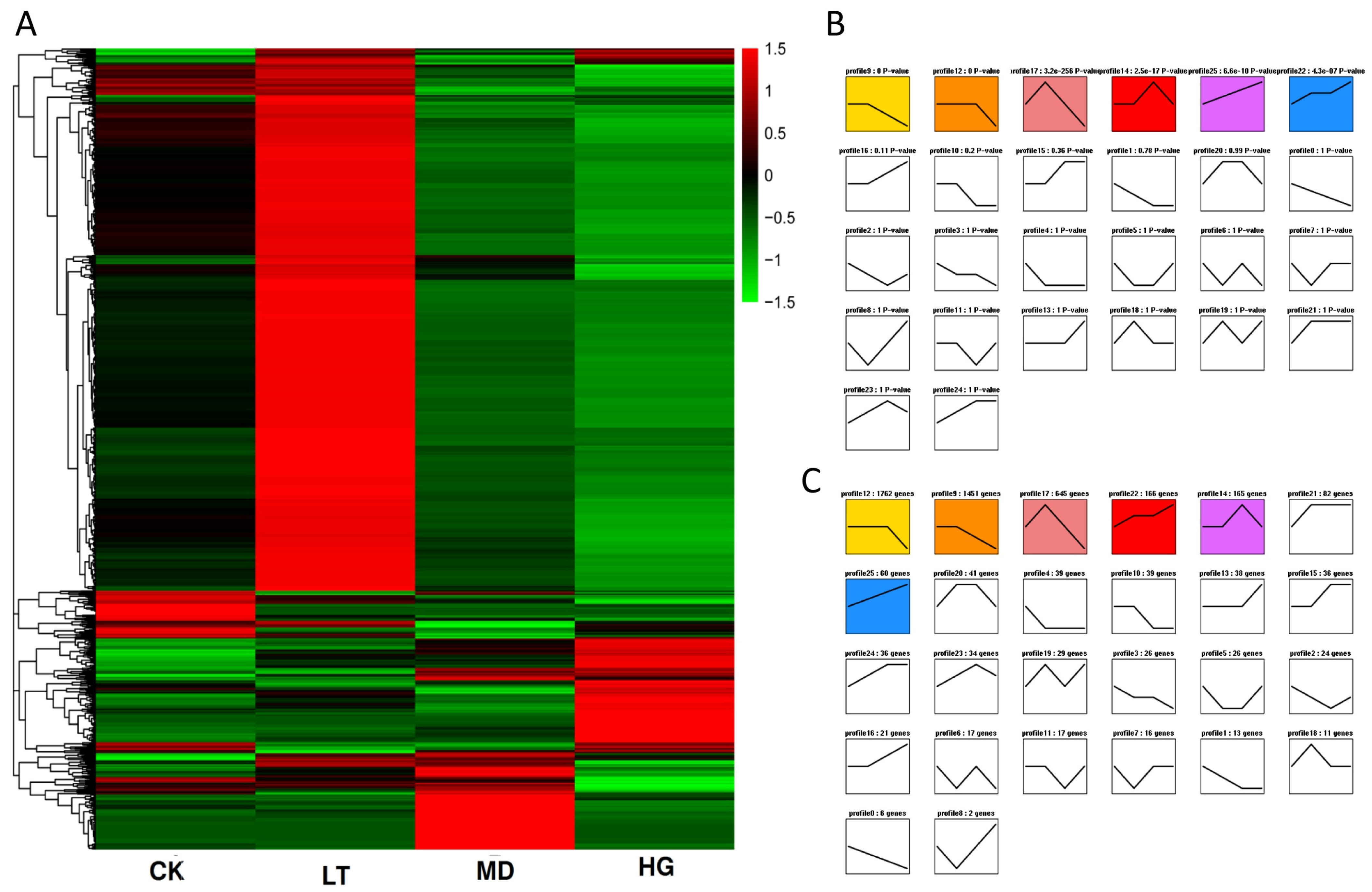
3.3. Transcriptome Analysis
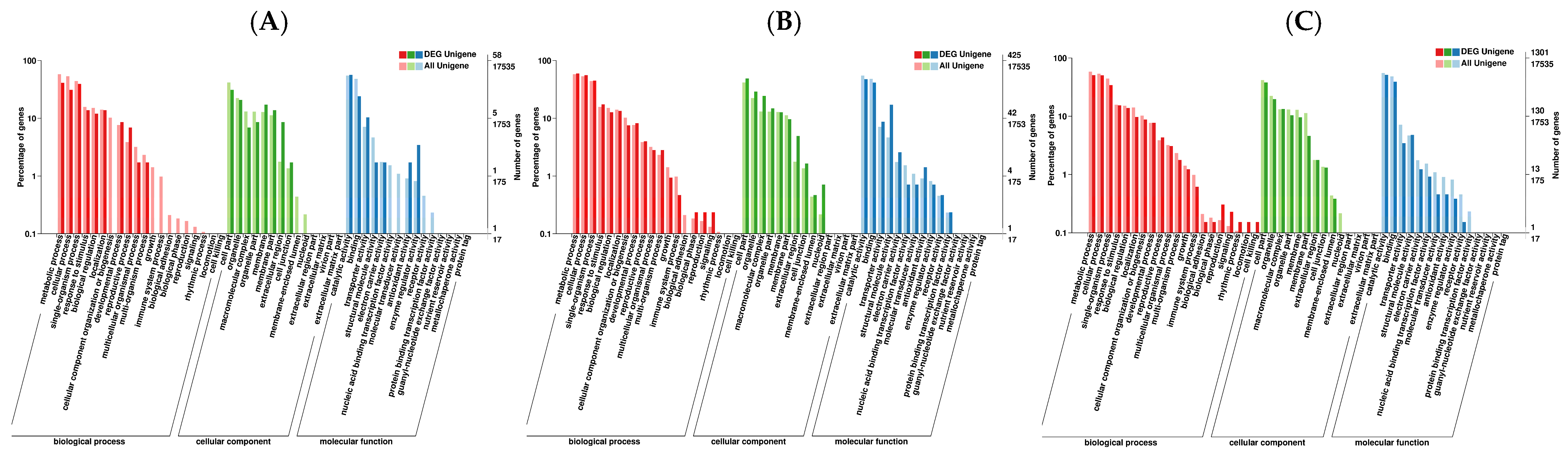
3.4. GO Functional Classification of DEGs
3.5. Trend Analysis of DEGs
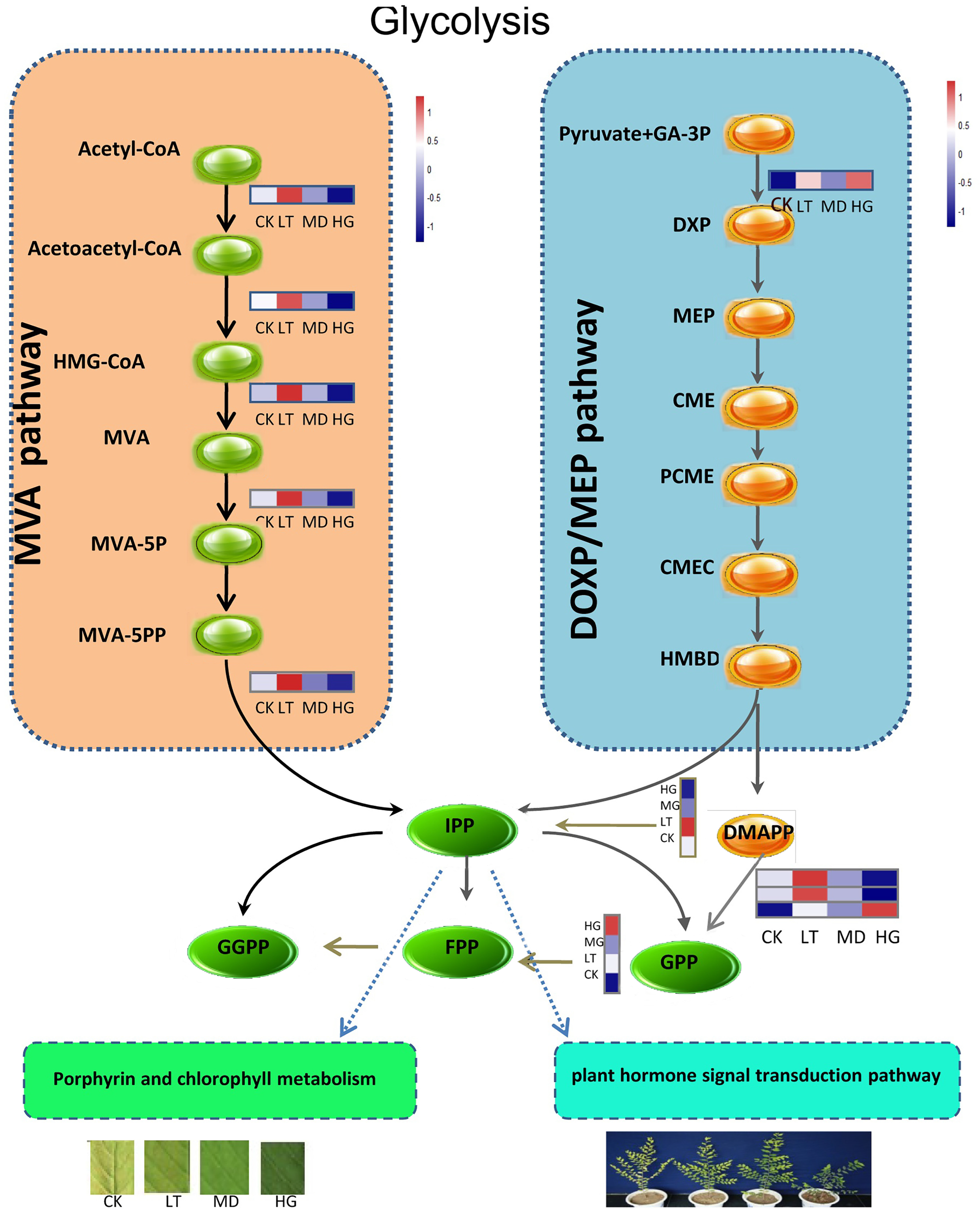
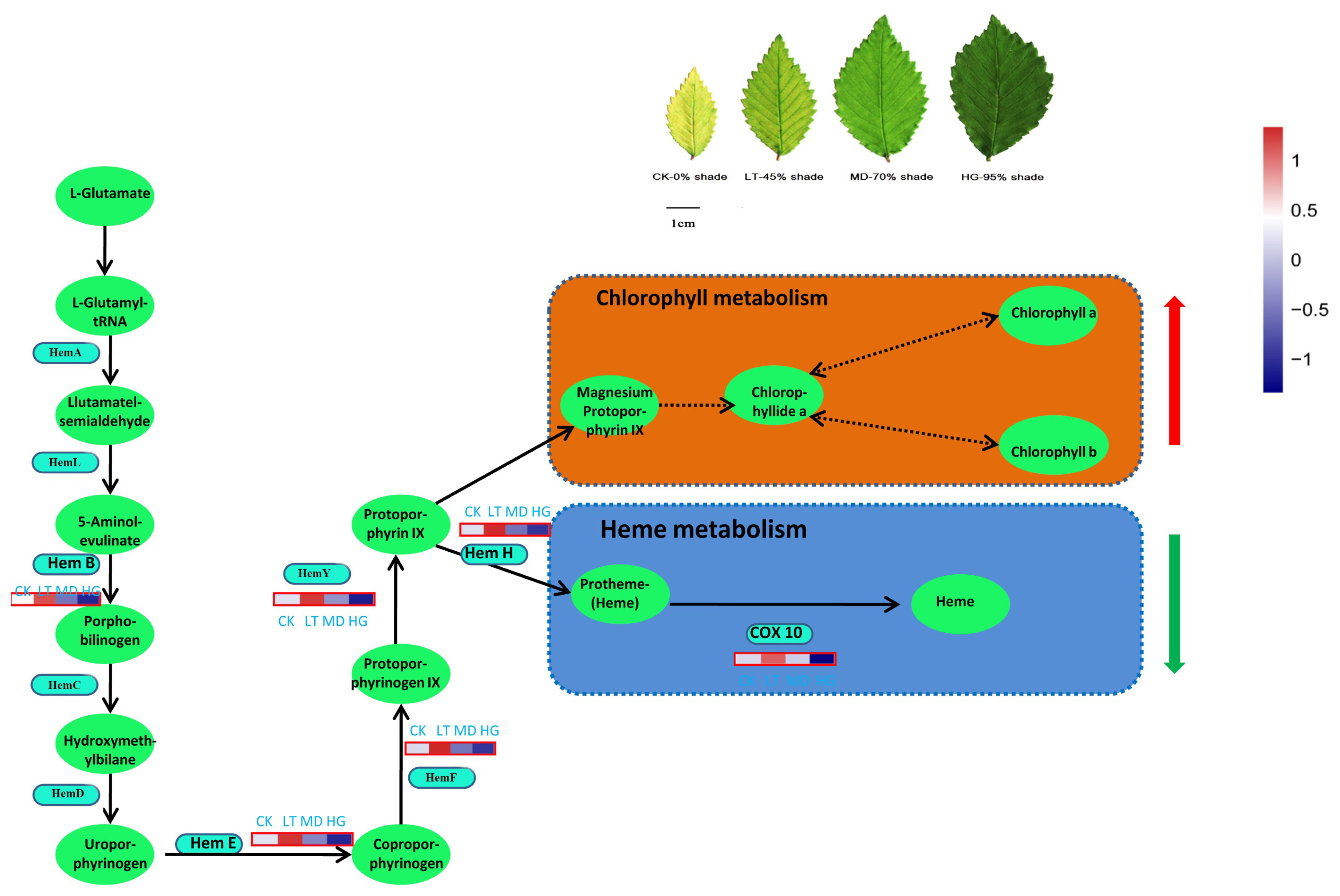
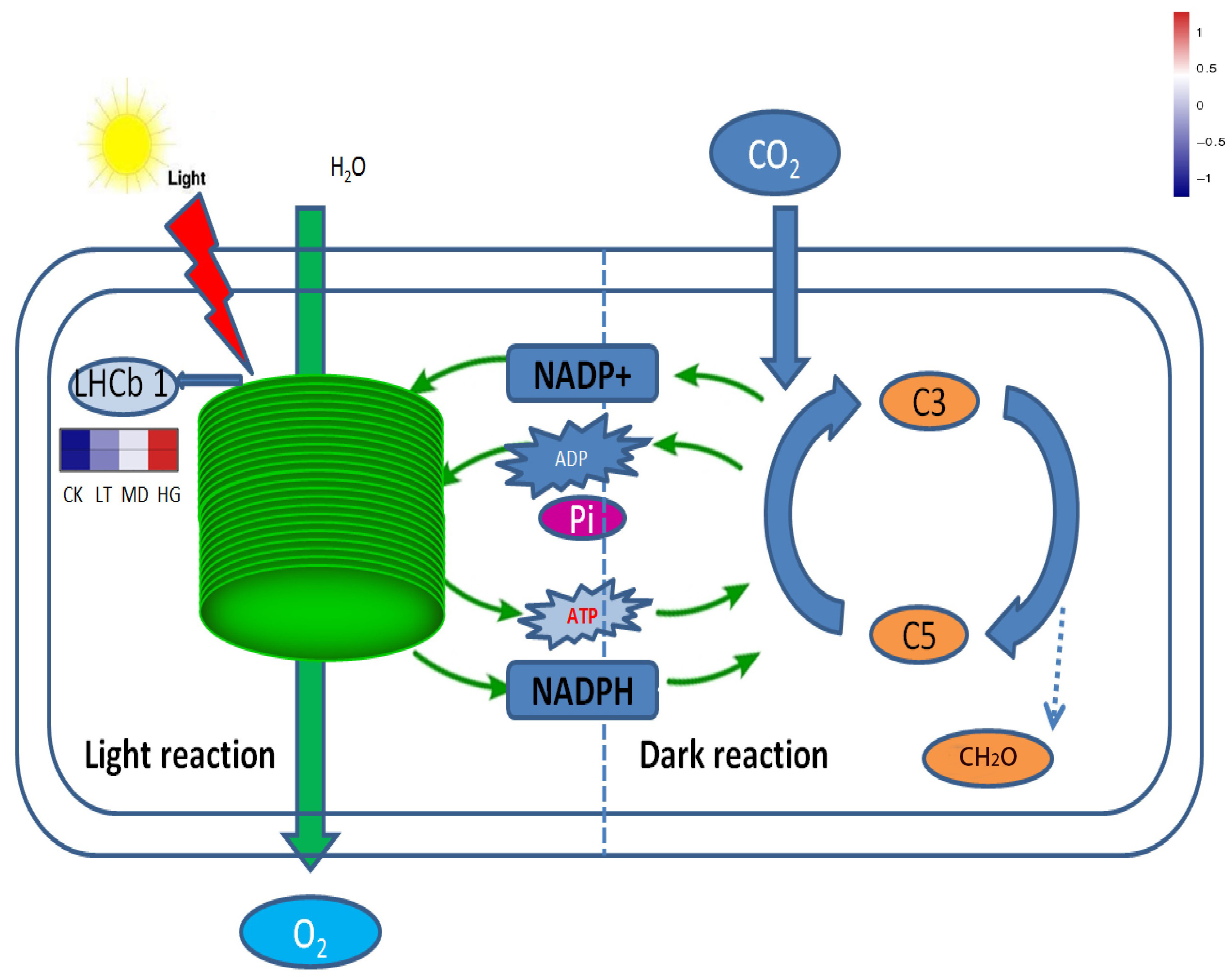
3.6. KEGG Metabolism Pathway Analysis
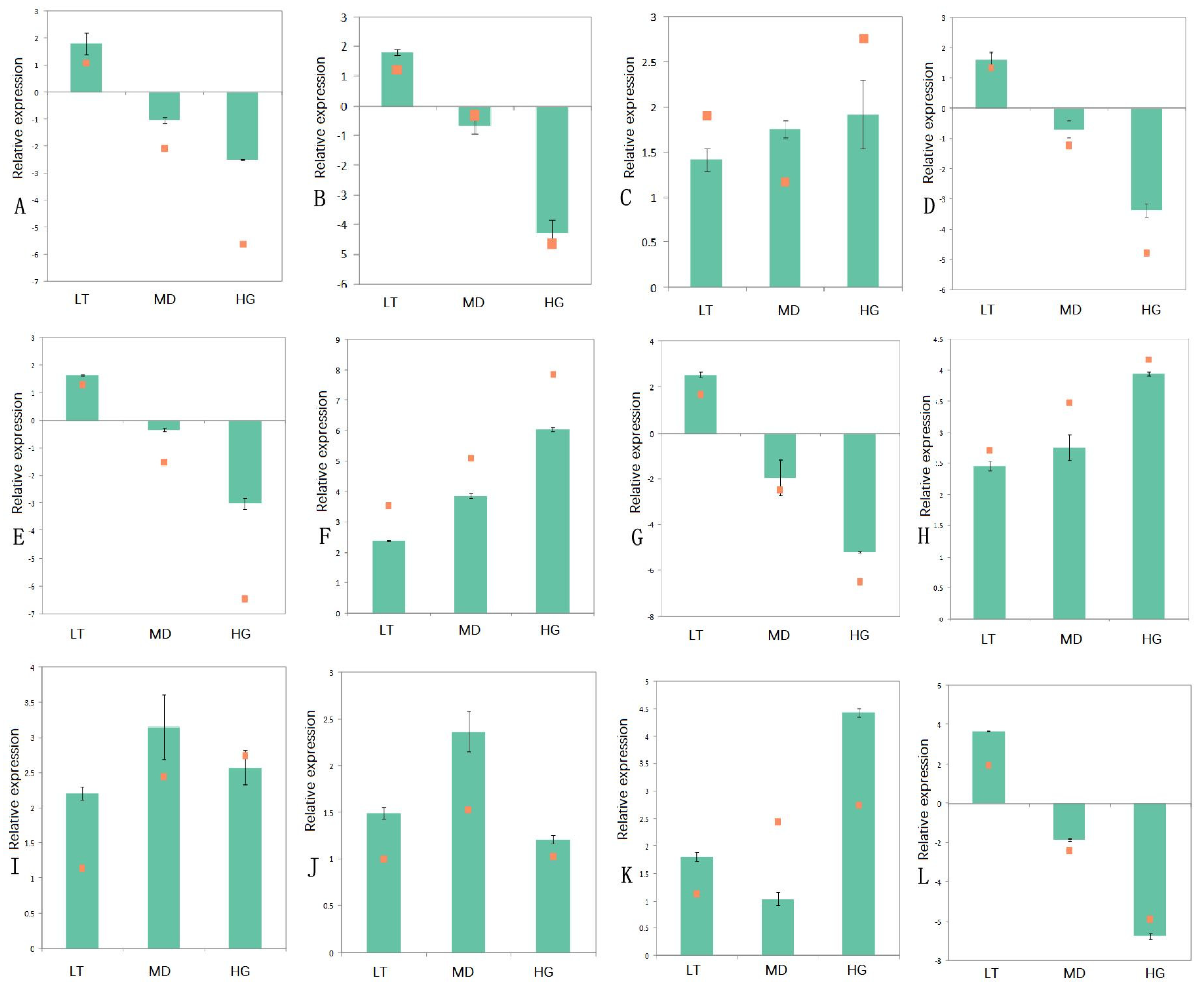
3.7. qPCR Assays
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Conde, P.; Sousa, A.; Costa, A.; Santos, C. A protocol for Ulmus minor Mill. micropropagation and acclimatization. Plant Cell Tissue Organ Cult. 2008, 92, 113–119. [Google Scholar] [CrossRef]
- Dias, M.C.C.; Moutinho-Pereira, J.; Correia, C.; Gonçalves, B.; Santos, C. Cadmium toxicity affects photosynthesis and plant growth at different levels. Acta Physiol. Plant. 2013, 35, 1281–1289. [Google Scholar] [CrossRef]
- Yuan, T.; Su, X.M. Caryopteris × clandonensis ‘worcester gold’ and its domestication, culture. Acta Hortic. Sin. 2004, 31, 112–114. [Google Scholar]
- Jiang, W.B.; Zhuang, M.; Han, H.Z.; Dai, M.S.; Hua, G.P. Progress on color emerging mechanism and photosynthetic characteristics of colored-leaf plants. Acta Hortic. Sin. 2005, 32, 352–358. [Google Scholar]
- Hu, H.Z.; Zhang, R.; Shang, A.Q.; Zhao, L.J.; Lu, Z.M. Response of pigment content of golden-leaf plants to light intensity. Acta Hortic. Sin. 2007, 34, 717–722. [Google Scholar]
- Kang, Y.; Pan, J.; Chen, J.; Liu, Q.; Dong, S. Contents of pigments and anatomical structure of leaves in Acer negundo ‘Aurea’. Bull. Bot. Res. 2023, 43, 591–600. [Google Scholar]
- Zhang, S.; Zuo, L.; Zhang, J.; Chen, P.; Wang, J.; Yang, M. Transcriptome analysis of Ulmus pumila ‘Jinye’ responses to different shading involved in chlorophyll metabolism. Tree Genet. Genomes 2017, 13, 64. [Google Scholar] [CrossRef]
- Zuo, L.; Zhang, S.; Liu, Y.; Huang, Y.; Yang, M.; Wang, J. The reason for growth inhibition of Ulmus pumila ‘Jinye’: Lower resistance and abnormal development of chloroplasts slow down the accumulation of Energy. Int. J. Mol. Sci. 2019, 20, 4227. [Google Scholar] [CrossRef]
- Chen, A. The effects of different light treatments on the leaf color and physiological indicators of Berberis thunbergii var. Atropurpurea. Anhui Agri. Sci. Bull. 2019, 25, 9–10. [Google Scholar]
- Zou, Q. Experimental Instruction of Plant Physiology; China Agricultural Press: Beijing, China, 2000. [Google Scholar]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A. The COG database: A tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Jason, E.; Ziv, B.J. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Zhang, M.; Li, K.; Zhang, C.; Gai, J.; Yu, D. Identification and characterization of class 1 DXS gene encoding 1-deoxy-D-xylulose-5-phosphate synthase, the first committed enzyme of the MEP pathway from soybean. Mol. Biol. Rep. 2009, 36, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Thornber, J.P. Chlorophyll proteins: Light-harvesting and reaction center components of plants. Annu. Rev. Plant Physiol. 1975, 26, 127–158. [Google Scholar] [CrossRef]
- Zhang, Q.D.; Lou, S.Q.; Li, T.Z.; Ma, G.Z.; Zhang, G.Z.; Kuang, T.Y. Structure and function of chloroplast membranes: Effect of potassium and magnesium lons on the strcture and absorption spectrum of two kinds chloroplast membranes. Acta Bot. Sin. 1980, 21, 250–258. [Google Scholar]
- Mishanin, V.I.; Trubitsin, B.V.; Benkov, M.A.; Minin, A.A.; Tikhonov, A.N. Light acclimation of shade-tolerant and light-resistant tradescantia species: Induction of chlorophyll a fluorescence and p700 photooxidation, expression of psbs and lhcb1 proteins. Photosynth. Res. 2016, 130, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Morishige, D.T.; Dreyfuss, B.W. Light-harvesting complexes of higher plants. In Photosynthesis: A Comprehensive Treatise; Raghavendra, A.S., Ed.; Cambridge University Press: Cambridge, UK, 1998; pp. 18–28. [Google Scholar]
- Labate, M.T.; Ko, K.; Ko, Z.W.; Pinto, L.S.; Real, M.J.; Romano, M.R.; Barja, P.R.; Granell, A.; Friso, G.; van Wijk, K.J.; et al. Constitutive expression of pea Lhcb 1-2 in tobacco affects plant development, morphology and photosynthetic capacity. Plant Mol. Biol. 2004, 55, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef]


| Annotation Database | Annotated Number | 300 ≤ Length < 1000 | Length ≥ 1000 | Frequency |
|---|---|---|---|---|
| Nr | 30,802 | 9553 | 15,520 | 48.93% |
| Pfam | 24,272 | 6831 | 14,338 | 38.56% |
| KOG | 20,861 | 6240 | 11,171 | 33.14% |
| Swissprot | 19,220 | 5639 | 11,063 | 30.53% |
| GO | 17,535 | 5134 | 8882 | 27.86% |
| KEGG | 12,983 | 3880 | 7011 | 20.63% |
| COG | 11,990 | 3038 | 6950 | 19.05% |
| All | 33,023 | 10,543 | 16,149 | 52.46% |
| Pathways | Related Unigene | DEG Unigene | Enrichment Factor | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LT | MD | HG | LT | MD | HG | LT | MD | HG | ||
| Calcium signalling pathway | 94 | 2 | 10 | 29 | 3.27 | 2.36 | 0.00 | 0.00 | 0.00 | 3.27 |
| Carbon metabolism | 643 | 2 | 18 | 66 | 5.89 | 0.78 | 0.00 | 0.78 | 1.00 | 5.89 |
| cGMP-PKG signalling pathway | 149 | 2 | 10 | 33 | 3.27 | 1.69 | 0.02 | 0.02 | 0.01 | 3.27 |
| Cutin, suberine and wax biosynthesis | 37 | 3 | 2 | 5 | 0.65 | 1.03 | 0.02 | 0.34 | 0.65 | 0.65 |
| Cyanoamino acid metabolism | 67 | 2 | 3 | 7 | 0.98 | 0.80 | 0.02 | 0.37 | 0.88 | 0.98 |
| Glutathione metabolism | 133 | 2 | 4 | 16 | 1.31 | 0.92 | 0.05 | 0.63 | 0.84 | 1.31 |
| Glycerolipid metabolism | 116 | 2 | 7 | 19 | 2.29 | 1.25 | 0.05 | 0.08 | 0.33 | 2.29 |
| Glycolysis/Gluconeogenesis | 315 | 3 | 12 | 27 | 3.92 | 0.65 | 0.06 | 0.33 | 1.00 | 3.92 |
| Inositol phosphate metabolism | 103 | 2 | 3 | 18 | 0.98 | 1.33 | 0.08 | 0.66 | 0.24 | 0.98 |
| Methane metabolism | 178 | 2 | 6 | 15 | 1.96 | 0.64 | 0.10 | 0.52 | 1.00 | 1.96 |
| Peroxisome | 181 | 4 | 5 | 26 | 1.64 | 1.10 | 0.12 | 0.71 | 0.57 | 1.64 |
| Phenylpropanoid biosynthesis | 171 | 2 | 5 | 11 | 1.64 | 0.49 | 0.13 | 0.66 | 1.00 | 1.64 |
| Pyruvate metabolism | 254 | 2 | 11 | 23 | 3.60 | 0.69 | 0.22 | 0.21 | 1.00 | 3.60 |
| Starch and sucrose metabolism | 265 | 2 | 7 | 21 | 2.29 | 0.61 | 0.24 | 0.77 | 1.00 | 2.29 |
| Tryptophan metabolism | 99 | 2 | 8 | 16 | 2.62 | 1.23 | 0.67 | 0.02 | 0.37 | 2.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Li, Y.; Wang, N.; Zuo, L.; Zhou, Y.; Li, P.; Wang, S.; Feng, S.; Yan, S.; Huang, Y.; et al. Analysis of the Metabolic and Structural Changes in Ulmus pumila ‘Zhonghua Jinye’ Leaf Under Shade Stress. Plants 2025, 14, 2868. https://doi.org/10.3390/plants14182868
Liu Y, Li Y, Wang N, Zuo L, Zhou Y, Li P, Wang S, Feng S, Yan S, Huang Y, et al. Analysis of the Metabolic and Structural Changes in Ulmus pumila ‘Zhonghua Jinye’ Leaf Under Shade Stress. Plants. 2025; 14(18):2868. https://doi.org/10.3390/plants14182868
Chicago/Turabian StyleLiu, Yichao, Yongtan Li, Ning Wang, Lihui Zuo, Yang Zhou, Ping Li, Shijie Wang, Shuxiang Feng, Shufang Yan, Yinran Huang, and et al. 2025. "Analysis of the Metabolic and Structural Changes in Ulmus pumila ‘Zhonghua Jinye’ Leaf Under Shade Stress" Plants 14, no. 18: 2868. https://doi.org/10.3390/plants14182868
APA StyleLiu, Y., Li, Y., Wang, N., Zuo, L., Zhou, Y., Li, P., Wang, S., Feng, S., Yan, S., Huang, Y., & Yang, M. (2025). Analysis of the Metabolic and Structural Changes in Ulmus pumila ‘Zhonghua Jinye’ Leaf Under Shade Stress. Plants, 14(18), 2868. https://doi.org/10.3390/plants14182868







