Lemna gibba Clones Show Differences in Phenotypic Responses to the Light Environment
Abstract
1. Introduction
2. Materials and Methods
2.1. Stock Culturing and Experimental Conditions
2.2. Growth and Morphological Traits
2.3. Chlorophyll Fluorescence
2.3.1. Fast Chlorophyll Fluorescence Induction Parameters
2.3.2. PAM Fluorometry and Chlorophyll Fluorescence Imaging
| Protocol | Parameter | Physiological Interpretation | Reference |
|---|---|---|---|
| Fast chlorophyll fluorescence kinetics | Sm | Normalized area—an assumed proportional to the number of reductions and oxidations of one QA-molecule during the fast OJIP transient, and therefore related to the number of electron carriers per electron transport chain | [39,45] |
| Phi_Pav | Time needed to reach maximal ChlF yield (in ms) | [39,45] | |
| Pi_Abs | Performance index on absorption basis for energy conservation from photons absorbed by PSII antenna to the reduction of QB | [39,45] | |
| ABS/RC | Absorption flux per reaction center | [39,45] | |
| TRo/RC | Maximum trapped exciton flux per active PSII unit; inversely proportional to the number of PSII reaction centers in the sample | [39,45] | |
| ETo/RC | Electron transport flux per reaction center | [39,45] | |
| DIo/RC | Dissipated energy flux per reaction center | [39,45] | |
| Fv/Fm | Maximal (i.e., dark-adapted) photochemical energy conversion rate | [39,45] | |
| Slow chlorophyll fluorescence kinetics | Y(II) | Actual (i.e., adapted to the ambient irradiation) photochemical energy conversion rate | [42] |
| Y(NPQ) | Regulated non-photochemical quenching; proportional to the efficiency of photoprotective mechanisms | [42] | |
| Y(NO) | Non-regulated non-photochemical quenching; proportional to constitutive energy losses in photochemistry | [42] | |
| Rapid light curve | α | Initial slope of the RLC, that is, the maximal light use efficiency; the number of electrons moved through the electron transport chain by 1 absorbed photon (as e− photon−1) | [46] |
| rETRmax | Maximal rate of the electron transport under the applied ambient conditions (as μmol e− m−2 s−1) | [46] | |
| Ek | Onset of light saturation in the course of RLC (as μmol m−2 s−1) | [46] |
2.3.3. Chlorophyll Fluorescence Image Analysis
2.4. Biomass Sampling
2.5. Measurement of Photosynthetic Pigment Contents
2.6. Data Analysis and Statistics
3. Results and Discussion
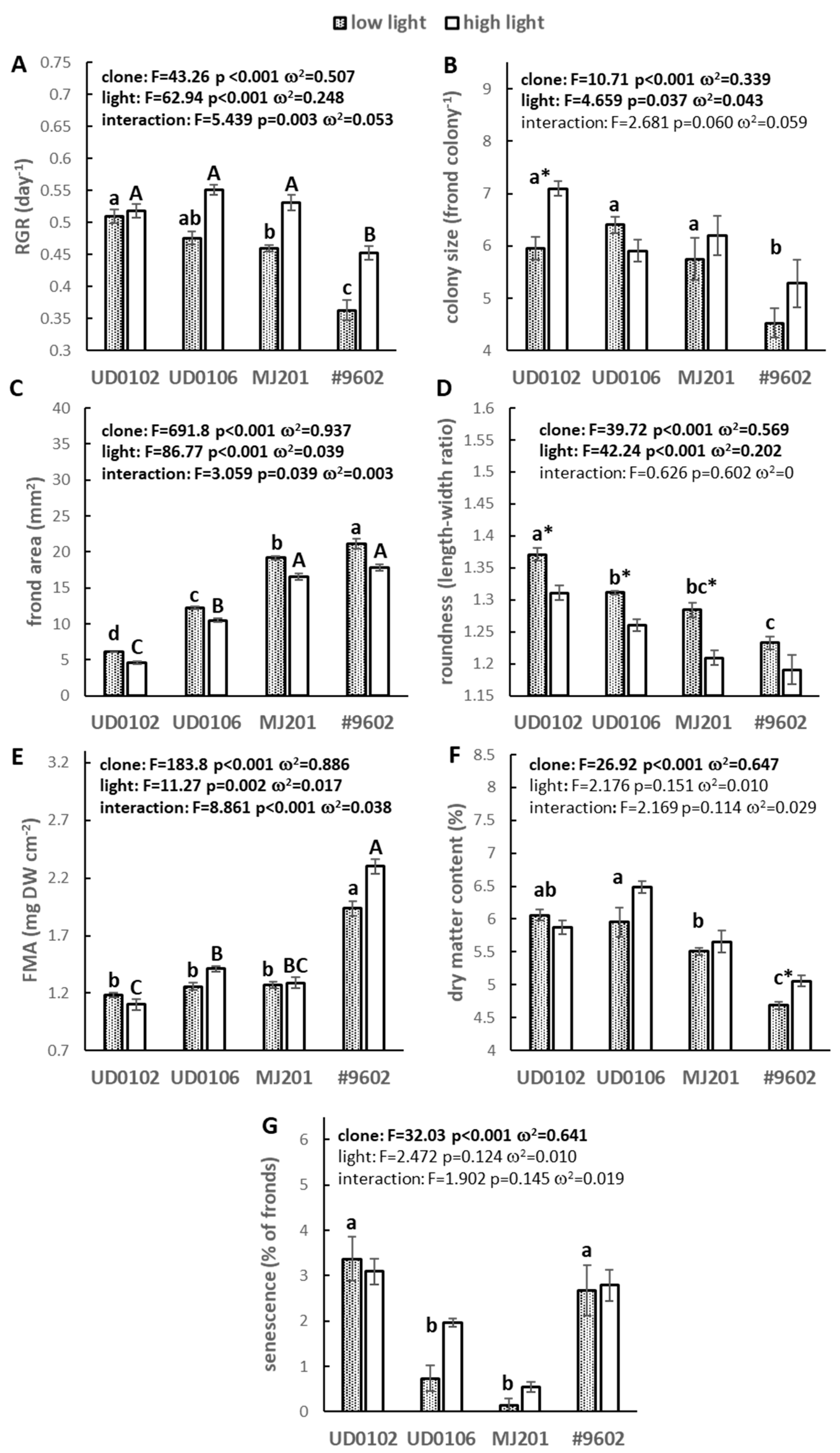
3.1. Growth and Morphology
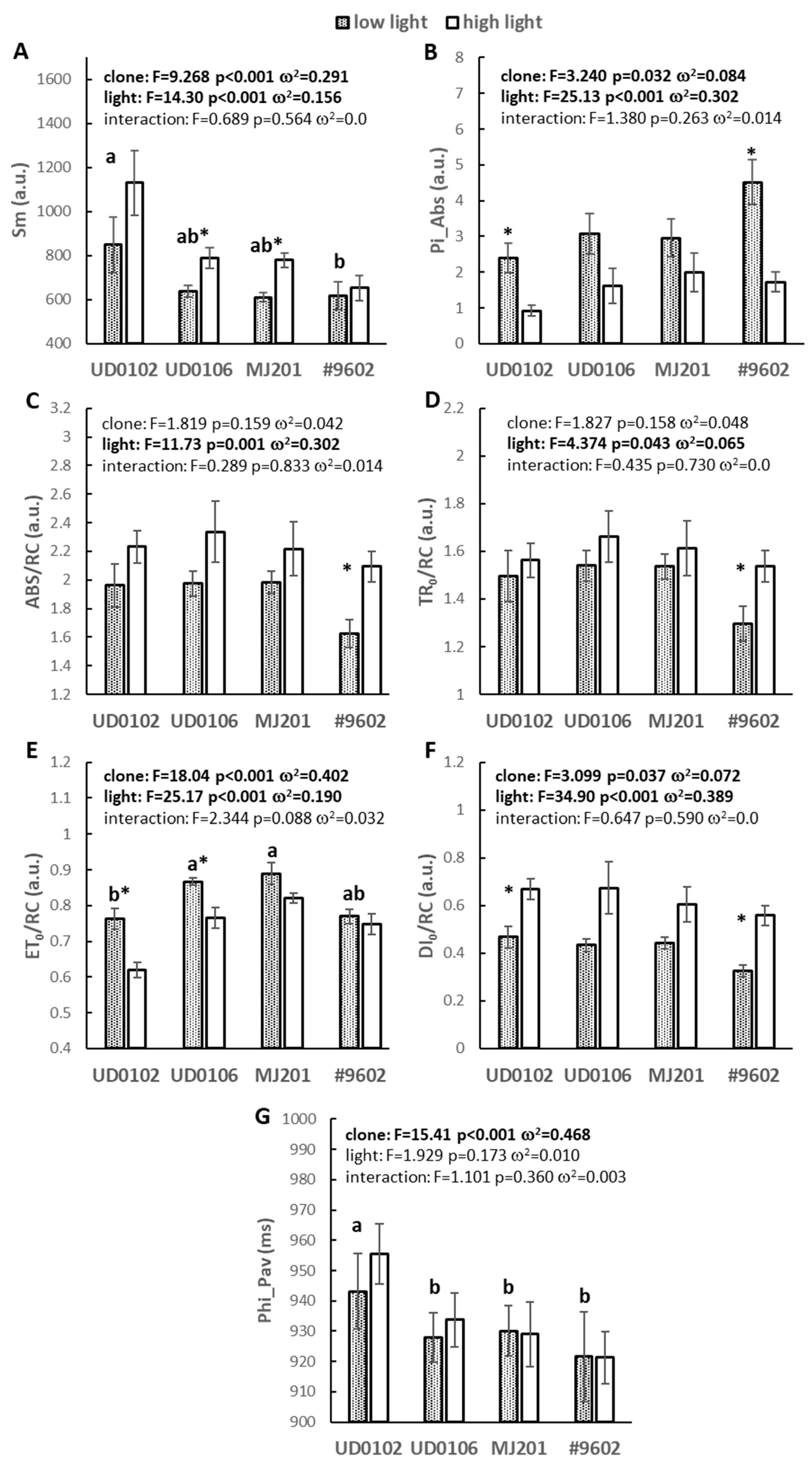
3.2. Fast Chlorophyll Fluorescence Induction
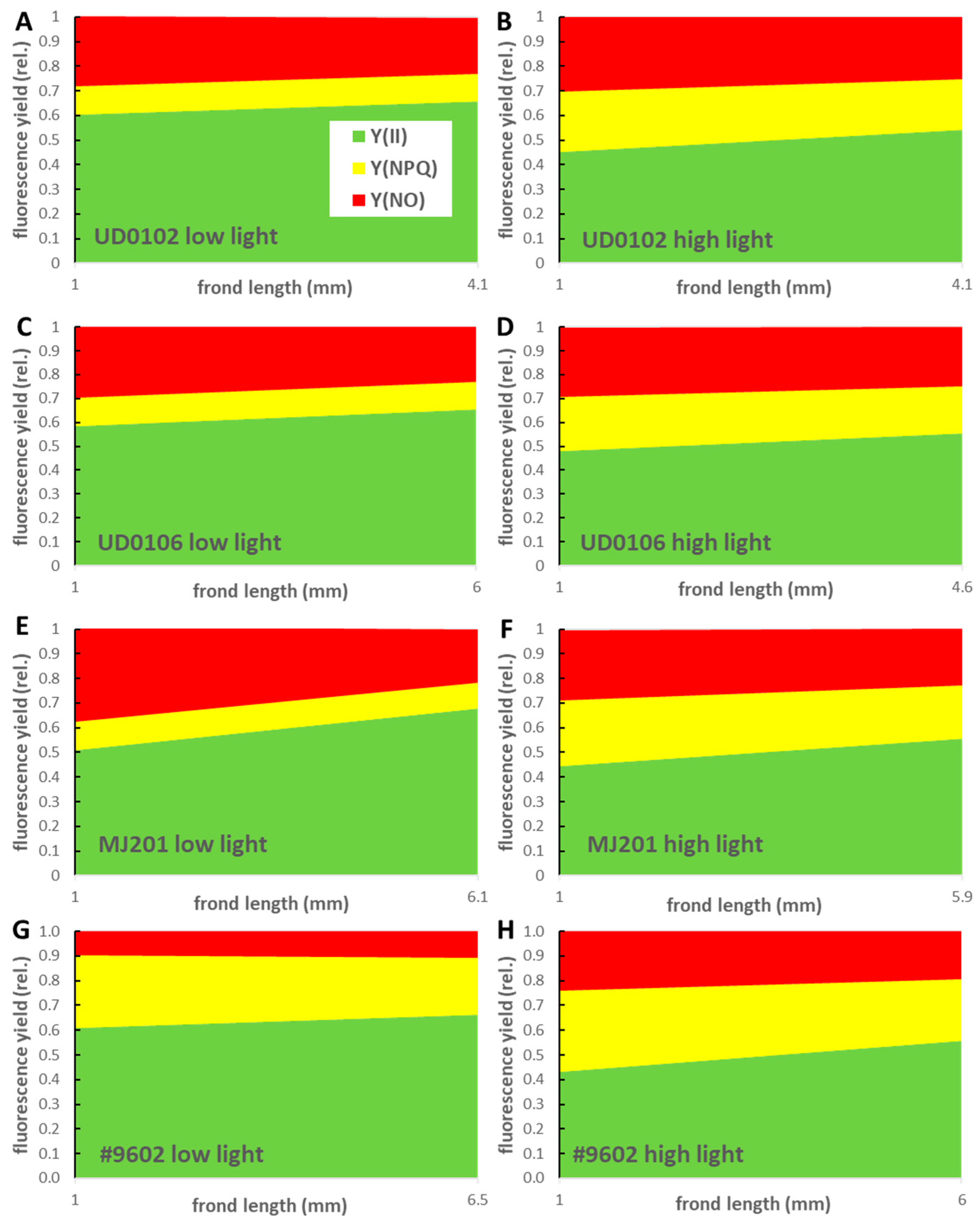
3.3. PAM Fluorometry
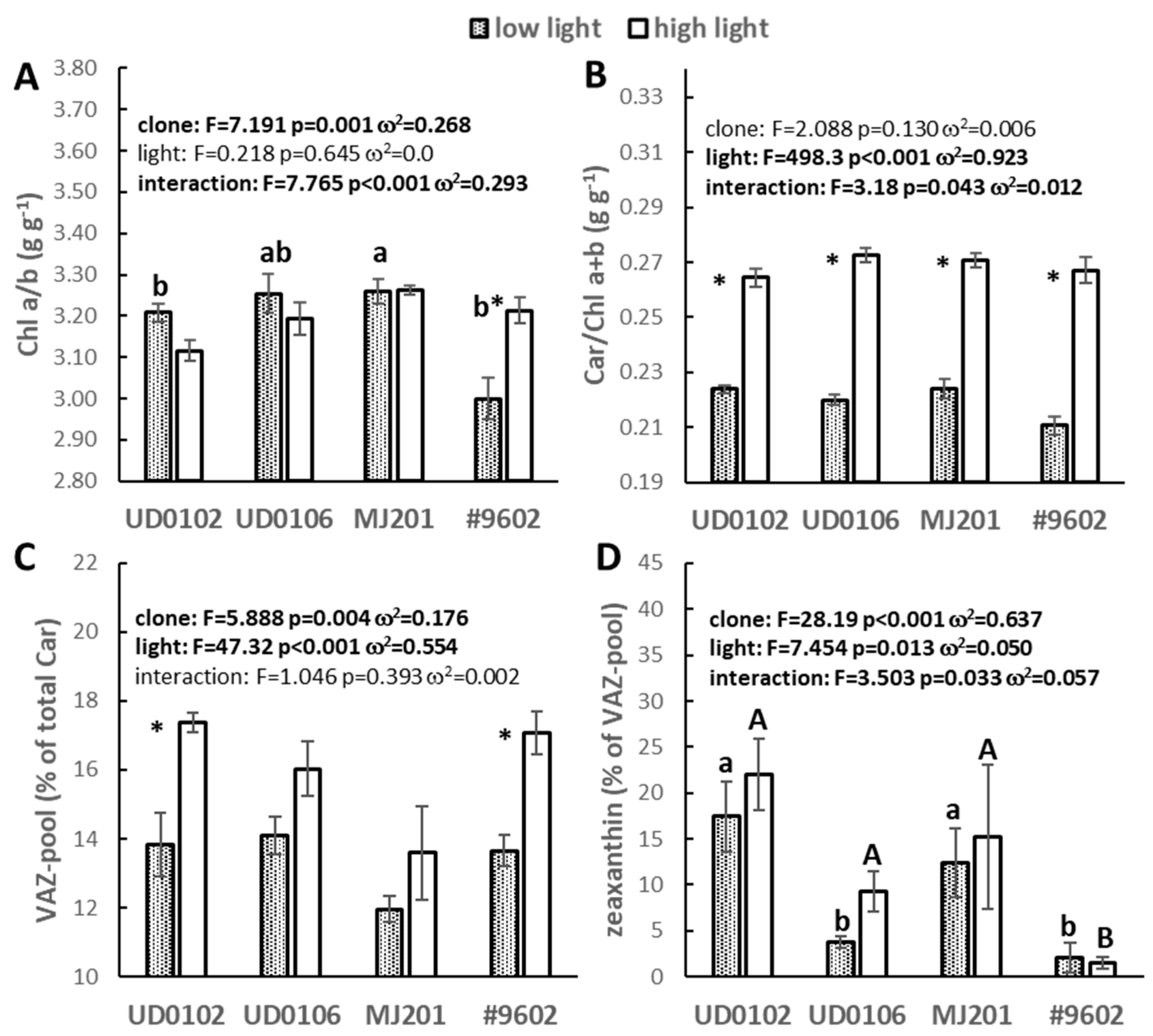
3.4. Photosynthetic Pigment Composition
3.5. Ontogenetic Acclimation of Fronds to Ambient Light
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| α | maximal light use efficiency |
| ABS/RC | absorption flux per reaction center |
| Car | carotenoids |
| Chl-a | chlorophyll-a |
| Chl-b | chlorophyll-b |
| ChlF | chlorophyll fluorescence |
| DIo/RC | dissipated energy flux per reaction center |
| DLI | daily light integral |
| Ek | onset of light saturation in the course of RLC |
| ETo/RC | electron transport flux per reaction center |
| FMA | frond mass-to-area |
| Fv/Fm | maximal (i.e., dark-adapted) photochemical energy conversion rate |
| PAM | pulse amplitude modulated fluorometry |
| Phi_Pav | time needed to reach maximal ChlF yield |
| Pi_Abs | performance index for energy conservation |
| PPFD | photosynthetic photon flux density |
| rETRmax | maximal electron transport rate during an RLC |
| RGR | relative growth rate |
| RLC | rapid light curve |
| Sm | normalized area above the fast chlorophyll fluorescence induction curve |
| TRo/RC | maximum trapped exciton flux per active PSII unit |
| VAZ/Car | relative proportion of the VAZ-pool within the total carotenoids |
| VAZ-pool | the summarized amount of Violaxanthin, Antheraxanthin, and Zeaxanthin |
| Y(II) | actual (i.e., light-adapted) photochemical energy conversion rate |
| Y(NO) | non-regulated non-photochemical quenching |
| Y(NPQ) | regulated non-photochemical quenching |
| Z/VAZ | relative proportion of Zeaxanthin within the VAZ-pool |
References
- Bog, M.; Appenroth, K.J.; Sree, K.S. Key to the Determination of Taxa of Lemnaceae: An Update. Nord. J. Bot. 2020, 38, 1–12. [Google Scholar] [CrossRef]
- Ishizawa, H.; Onoda, Y.; Kitajima, K.; Kuroda, M.; Inoue, D.; Ike, M. Coordination of Leaf Economics Traits within the Family of the World’s Fastest Growing Plants (Lemnaceae). J. Ecol. 2021, 109, 2950–2962. [Google Scholar] [CrossRef]
- Smith, K.E.; Zhou, M.; Flis, P.; Jones, D.H.; Bishopp, A.; Yant, L. The Evolution of the Duckweed Ionome Mirrors Losses in Structural Complexity. Ann. Bot. 2024, 133, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Ware, A.; Jones, D.H.; Flis, P.; Chrysanthou, E.; Smith, K.E.; Kümpers, B.M.C.; Yant, L.; Atkinson, J.A.; Wells, D.M.; Bhosale, R.; et al. Loss of Ancestral Function in Duckweed Roots Is Accompanied by Progressive Anatomical Reduction and a Re-Distribution of Nutrient Transporters. Curr. Biol. 2023, 33, 1795–1802.e4. [Google Scholar] [CrossRef] [PubMed]
- Lemon, G.D.; Posluszny, U.; Husband, B.C. Potential and Realized Rates of Vegetative Reproduction in Spirodela polyrhiza, Lemna minor, and Wolffia borealis. Aquat. Bot. 2001, 70, 79–87. [Google Scholar] [CrossRef]
- Dutt, P.; Laird, R.A. A Demographic Assessment of the Lansing Effect in Duckweed (Lemna turionifera Landolt). Biol. Lett. 2024, 20, 20240271. [Google Scholar] [CrossRef]
- Ziegler, P.; Adelmann, K.; Zimmer, S.; Schmidt, C.; Appenroth, K.-J. Relative In Vitro Growth Rates of Duckweeds (Lemnaceae)—The Most Rapidly Growing Higher Plants. Plant Biol. 2015, 17, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Romano, L.E.; Iovane, M.; Izzo, L.G.; Aronne, G. A Machine-Learning Method to Assess Growth Patterns in Plants of the Family Lemnaceae. Plants 2022, 11, 1910. [Google Scholar] [CrossRef]
- Oláh, V.; Appenroth, K.-J.; Sree, K.S. Duckweed: Research Meets Applications. Plants 2023, 12, 3307. [Google Scholar] [CrossRef]
- Szabó, S.; Zavanyi, G.; Koleszár, G.; Del Castillo, D.; Oláh, V.; Braun, M. Phytoremediation, Recovery and Toxic Effects of Ionic Gadolinium Using the Free-Floating Plant Lemna gibba. J. Hazard. Mater. 2023, 458, 131930. [Google Scholar] [CrossRef]
- Landolt, E. The Family of “Lemnaceae”: A Monographic Study—Biosystematic Investigations in the Family of Duckweeds (‘Lemnaceae’); Geobotanisches Institut der ETH: Zürich, Switzerland, 1986; Volume 2, ISBN 0254-9433. [Google Scholar]
- Tippery, N.P.; Les, D.H. Tiny Plants with Enormous Potential: Phylogeny and Evolution of Duckweeds. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2020; pp. 19–38. ISBN 978-3-030-11044-4. [Google Scholar]
- Davidson, D.; Simon, J.-P. Thermal Adaptation and Acclimation of Ecotypic Populations of Spirodela polyrhiza (L.) Schleid. (Lemnaceae): Morphology and Growth Rates. J. Therm. Biol. 1981, 6, 121–128. [Google Scholar] [CrossRef]
- Kufel, L.; Strzałek, M.; Przetakiewicz, A. Plant Response to Overcrowding—Lemna minor Example. Acta Oecologica 2018, 91, 73–80. [Google Scholar] [CrossRef]
- Jewell, M.D.; Bell, G. Environmental and Genetic Variation in an Asexual Plant. Aquat. Bot. 2023, 188, 103675. [Google Scholar] [CrossRef]
- Zallek, T.A.; Turcotte, M.M. Evolution in Response to Management Increases Invasiveness among Experimental Populations of Duckweed (Lemna minor). Evol. Appl. 2024, 17, e70060. [Google Scholar] [CrossRef] [PubMed]
- Demmig-Adams, B.; López-Pozo, M.; Polutchko, S.K.; Fourounjian, P.; Stewart, J.J.; Zenir, M.C.; Adams, W.W. Growth and Nutritional Quality of Lemnaceae Viewed Comparatively in an Ecological and Evolutionary Context. Plants 2022, 11, 145. [Google Scholar] [CrossRef]
- Pasos-Panqueva, J.; Baker, A.; Camargo-Valero, M.A. Unravelling the Impact of Light, Temperature and Nutrient Dynamics on Duckweed Growth: A Meta-Analysis Study. J. Environ. Manag. 2024, 366, 121721. [Google Scholar] [CrossRef]
- Liebers, M.; Hommel, E.; Grübler, B.; Danehl, J.; Offermann, S.; Pfannschmidt, T. Photosynthesis in the Biomass Model Species Lemna minor Displays Plant-Conserved and Species-Specific Features. Plants 2023, 12, 2442. [Google Scholar] [CrossRef]
- Stewart, J.J.; Adams, W.W.; Escobar, C.M.; López-Pozo, M.; Demmig-Adams, B. Growth and Essential Carotenoid Micronutrients in Lemna gibba as a Function of Growth Light Intensity. Front. Plant Sci. 2020, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.; Adams, W.W.; López-Pozo, M.; Doherty Garcia, N.; McNamara, M.; Escobar, C.M.; Demmig-Adams, B. Features of the Duckweed Lemna That Support Rapid Growth under Extremes of Light Intensity. Cells 2021, 10, 1481. [Google Scholar] [CrossRef]
- Lemon, G.D.; Posluszny, U. Comparative Shoot Development and Evolution in the Lemnaceae. Int. J. Plant Sci. 2000, 161, 733–748. [Google Scholar] [CrossRef]
- Lahive, E.; O’Halloran, J.; Jansen, M.A.K. Frond Development Gradients Are a Determinant of the Impact of Zinc on Photosynthesis in Three Species of Lemnaceae. Aquat. Bot. 2012, 101, 55–63. [Google Scholar] [CrossRef]
- Peršić, V.; Antunović Dunić, J.; Domjan, L.; Zellnig, G.; Cesar, V. Time Course of Age-Linked Changes in Photosynthetic Efficiency of Spirodela polyrhiza Exposed to Cadmium. Front. Plant Sci. 2022, 13, 872793. [Google Scholar] [CrossRef]
- Oláh, V.; Kosztankó, K.; Irfan, M.; Barnáné Szabó, Z.; Jansen, M.A.K.; Szabó, S.; Mészáros, I. Frond-Level Analyses Reveal Functional Heterogeneity within Heavy Metal-Treated Duckweed Colonies. Plant Stress 2024, 11, 100405. [Google Scholar] [CrossRef]
- Peršić, V.; Melnjak, A.; Domjan, L.; Zellnig, G.; Antunović Dunić, J. Age-Specific Physiological Adjustments of Spirodela polyrhiza to Sulfur Deficiency. Plants 2025, 14, 1907. [Google Scholar] [CrossRef] [PubMed]
- Anneberg, T.J.; O’Neill, E.M.; Ashman, T.-L.; Turcotte, M.M. Polyploidy Impacts Population Growth and Competition with Diploids: Multigenerational Experiments Reveal Key Life-History Trade-Offs. New Phytol. 2023, 238, 1294–1304. [Google Scholar] [CrossRef]
- Bafort, Q.; Wu, T.; Natran, A.; De Clerck, O.; Van de Peer, Y. The Immediate Effects of Polyploidization of Spirodela polyrhiza Change in a Strain-Specific Way along Environmental Gradients. Evol. Lett. 2023, 7, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Braglia, L.; Ceschin, S.; Iannelli, M.A.; Bog, M.; Fabriani, M.; Frugis, G.; Gavazzi, F.; Gianì, S.; Mariani, F.; Muzzi, M.; et al. Characterization of the Cryptic Interspecific Hybrid Lemna×mediterranea by an Integrated Approach Provides New Insights into Duckweed Diversity. J. Exp. Bot. 2024, 75, 3092–3110. [Google Scholar] [CrossRef]
- Stepanenko, A.; Braglia, L.; Fuchs, J.; Schubert, V.; Hoang, P.T.N.; Lee, Y.; Chen, G.; Gianì, S.; Morello, L.; Schubert, I. Genome Diversity and Evolution of the Duckweed Section Alatae Comprising Diploids, Polyploids, and Interspecific Hybrids. Plant J. 2025, 122, e70158. [Google Scholar] [CrossRef]
- Sarin, L.P.; Sree, K.S.; Bóka, K.; Keresztes, Á.; Fuchs, J.; Tyagi, A.K.; Khurana, J.P.; Appenroth, K.-J. Characterisation of a Spontaneous Mutant of Lemna gibba G3 (Lemnaceae). Plants 2023, 12, 2525. [Google Scholar] [CrossRef] [PubMed]
- Mortier, F.; Bafort, Q.; Milosavljevic, S.; Kauai, F.; Prost Boxoen, L.; Van de Peer, Y.; Bonte, D. Understanding Polyploid Establishment: Temporary Persistence or Stable Coexistence? Oikos 2024, 2024, e09929. [Google Scholar] [CrossRef]
- Wu, T.; Bafort, Q.; Mortier, F.; Almeida-Silva, F.; Natran, A.; Van de Peer, Y. The Immediate Metabolomic Effects of Whole-Genome Duplication in the Greater Duckweed, Spirodela polyrhiza. Am. J. Bot. 2024, 111, e16383. [Google Scholar] [CrossRef]
- Sree, K.S.; Appenroth, K.-J. Worldwide Genetic Resources of Duckweed: Stock Collections. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2020; pp. 39–46. ISBN 978-3-030-11045-1. [Google Scholar]
- Sree, K.S.; Adelmann, K.; Garcia, C.; Lam, E.; Appenroth, K.-J. Natural Variance in Salt Tolerance and Induction of Starch Accumulation in Duckweeds. Planta 2015, 241, 1395–1404. [Google Scholar] [CrossRef]
- Environment Canada. Biological Test Method—Test for Measuring the Inhibition of Growth Using the Freshwater Macrophyte Lemna Minor; Method Development and Applications Section, Environmental Technology Centre, Environment Canada: Ottawa, ON, Canada, 2007.
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2004, 11, 32–64. [Google Scholar]
- OECD Guidelines for the Testing of Chemicals, Revised Proposal for a New Guideline 221, Lemna sp. Growth Inhibition Test; OECD: Paris, France, 2006.
- Strasser, R.J.; Srivastava, A.; Tsimilli-Michael, M. The Fluorescence Transient as a Tool to Characterize and Screen Photosynthetic Samples. In Probing Photosynthesis: Mechanism, Regulation and Adaptation; Yunus, M., Pathre, U., Mohanty, P., Eds.; Taylor and Francis Inc., CRC Press: London, UK, 2000; pp. 445–483. ISBN 978-0-429-17954-9. [Google Scholar]
- Appenroth, K.-J.; Keresztes, Á.; Sárvári, É.; Jaglarz, A.; Fischer, W. Multiple Effects of Chromate on Spirodela polyrhiza: Electron Microscopy and Biochemical Investigations. Plant Biol. 2003, 5, 315–323. [Google Scholar] [CrossRef]
- Oláh, V.; Hepp, A.; Irfan, M.; Mészáros, I. Chlorophyll Fluorescence Imaging-Based Duckweed Phenotyping to Assess Acute Phytotoxic Effects. Plants 2021, 10, 2763. [Google Scholar] [CrossRef]
- Klughammer, C.; Schreiber, U. Complementary PS II Quantum Yields Calculated from Simple Fluorescence Parameters Measured by PAM Fluorometry and the Saturation Pulse Method. PAM Appl. Notes 2008, 1, 27–35. [Google Scholar]
- Heinz Walz GmbH. Walz GmbH Walz GmbH IMAGING-PAM M-Series Chlorophyll Fluorometer: Instrument Description and Information for Users, 5th ed.; Heinz Walz GmbH: Effeltrich, Germany, 2019. [Google Scholar]
- Oláh, V.; Lakatos, G.; Bertók, C.; Kanalas, P.; Szőllősi, E.; Kis, J.; Mészáros, I. Short-Term Chromium(VI) Stress Induces Different Photosynthetic Responses in Two Duckweed Species, Lemna gibba L. and Lemna minor L. Photosynthetica 2010, 48, 513–520. [Google Scholar] [CrossRef]
- PSI. PAR-FluorPen FP 110 Manual and User Guide; Photon Systems Instruments spol. s.r.o.: Drásov, Czech Republic, 2023. [Google Scholar]
- Saroussi, S.; Beer, S. Alpha and Quantum Yield of Aquatic Plants Derived from PAM Fluorometry: Uses and Misuses. Aquat. Bot. 2007, 86, 89–92. [Google Scholar] [CrossRef]
- Wellburn, A.R. The Spectral Determination of Chlorophylls a and b, as Well as Total Carotenoids, Using Various Solvents with Spectrophotometers of Different Resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Láposi, R.; Veres, S.; Lakatos, G.; Oláh, V.; Fieldsend, A.; Mészáros, I. Responses of Leaf Traits of European Beech (Fagus sylvatica L.) Saplings to Supplemental UV-B Radiation and UV-B Exclusion. Agric. For. Meteorol. 2009, 149, 745–755. [Google Scholar] [CrossRef]
- Oláh, V.; Hepp, A.; Mészáros, I. Temporal Dynamics in Photosynthetic Activity of Spirodela polyrhiza Turions during Dormancy Release and Germination. Environ. Exp. Bot. 2017, 136, 50–58. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- RStudio Team RStudio: Integrated Development for R 2020. Available online: http://www.rstudio.com/ (accessed on 17 January 2025).
- Akima, H.; Gebhardt, A. Akima: Interpolation of Irregularly and Regularly Spaced Data 2022. Available online: https://CRAN.R-project.org/package=akima (accessed on 17 January 2025).
- Nychka, D.; Furrer, R.; Paige, J.; Sain, S. Fields: Tools for Spatial Data 2021. Available online: https://github.com/dnychka/fieldsRPackage (accessed on 17 January 2025).
- Smith, K.E.; Cowan, L.; Taylor, B.; McAusland, L.; Heatley, M.; Yant, L.; Murchie, E.H. Physiological Adaptation to Irradiance in Duckweeds Is Species and Accession Specific and Depends on Light Habitat Niche. J. Exp. Bot. 2024, 75, 2046–2063. [Google Scholar] [CrossRef]
- Landolt, E.; Kandeler, R. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae), Vol. 4: The Family of Lemnaceae—A Monographic Study, Vol. 2 (Phytochemistry, Physiology, Application, Bibliography); Geobotanisches Institut der Eidgenoessischen Technischen Hochschule, Stiftung Ruebel: Zurich, Switzerland, 1987. [Google Scholar]
- Chmilar, S.L.; Luzardo, A.C.; Dutt, P.; Pawluk, A.; Thwaites, V.C.; Laird, R.A. Caloric Restriction Extends Lifespan in a Clonal Plant. Ecol. Lett. 2024, 27, e14444. [Google Scholar] [CrossRef]
- Lepeduš, H.; Vidaković-Cifrek, Ž.; Šebalj, I.; Dunić, J.A.; Cesar, V. Effects of Low and High Irradiation Levels on Growth and PSII Efficiency in Lemna minor L. Acta Bot. Croat. 2020, 79, 185–192. [Google Scholar] [CrossRef]
- Oukarroum, A.; Strasser, R.J. Phenotyping of Dark and Light Adapted Barley Plants by the Fast Chlorophyll a Fluorescence Rise OJIP. S. Afr. J. Bot. 2004, 70, 277–283. [Google Scholar] [CrossRef]
- Khan, N.; Essemine, J.; Hamdani, S.; Qu, M.; Lyu, M.-J.A.; Perveen, S.; Stirbet, A.; Govindjee, G.; Zhu, X.-G. Natural Variation in the Fast Phase of Chlorophyll a Fluorescence Induction Curve (OJIP) in a Global Rice Minicore Panel. Photosynth. Res. 2021, 150, 137–158. [Google Scholar] [CrossRef] [PubMed]
- Stirbet, A.; Lazár, D.; Kromdijk, J. Govindjee Chlorophyll a Fluorescence Induction: Can Just a One-Second Measurement Be Used to Quantify Abiotic Stress Responses? Photosynthetica 2018, 56, 86–104. [Google Scholar] [CrossRef]
- Ralph, P.J.; Gademann, R. Rapid Light Curves: A Powerful Tool to Assess Photosynthetic Activity. Aquat. Bot. 2005, 82, 222–237. [Google Scholar] [CrossRef]
- Murchie, E.H.; Lawson, T. Chlorophyll Fluorescence Analysis: A Guide to Good Practice and Understanding Some New Applications. J. Exp. Bot. 2013, 64, 3983–3998. [Google Scholar] [CrossRef] [PubMed]
- Perkins, R.G.; Mouget, J.-L.; Lefebvre, S.; Lavaud, J. Light Response Curve Methodology and Possible Implications in the Application of Chlorophyll Fluorescence to Benthic Diatoms. Mar. Biol. 2006, 149, 703–712. [Google Scholar] [CrossRef]
- Bielczynski, L.W.; Łącki, M.K.; Hoefnagels, I.; Gambin, A.; Croce, R. Leaf and Plant Age Affects Photosynthetic Performance and Photoprotective Capacity. Plant Physiol. 2017, 175, 1634–1648. [Google Scholar] [CrossRef] [PubMed]
- Arber, A. The Vegetative Morphology of Pistia and the Lemnaceae. Proc. R. Soc. Lond. Ser. B Contain. Pap. Biol. Character 1920, 91, 96–103. [Google Scholar]
- Bai, S.-N. On the Plant Developmental Unit: From Virtual Concept to Visual Plantlet. Plants 2025, 14, 396. [Google Scholar] [CrossRef] [PubMed]
- Ware, A.; Smith, C.; Fairburn, R.; Vermonden, N.; Bishopp, A. Resolving the Duckweed Frond with Comparative Transcriptomics. In Proceedings of the 7th ICDRA 2024 International Conference on Duckweed research and Applications—Book of Abstracts, Bangkok, Thailand, 12–15 November 2024; p. 13. [Google Scholar]
- Ulrich, I.; Ulrich, W. High-Resolution Flow Cytometry of Nuclear DNA in Higher Plants. Protoplasma 1991, 165, 212–215. [Google Scholar] [CrossRef]
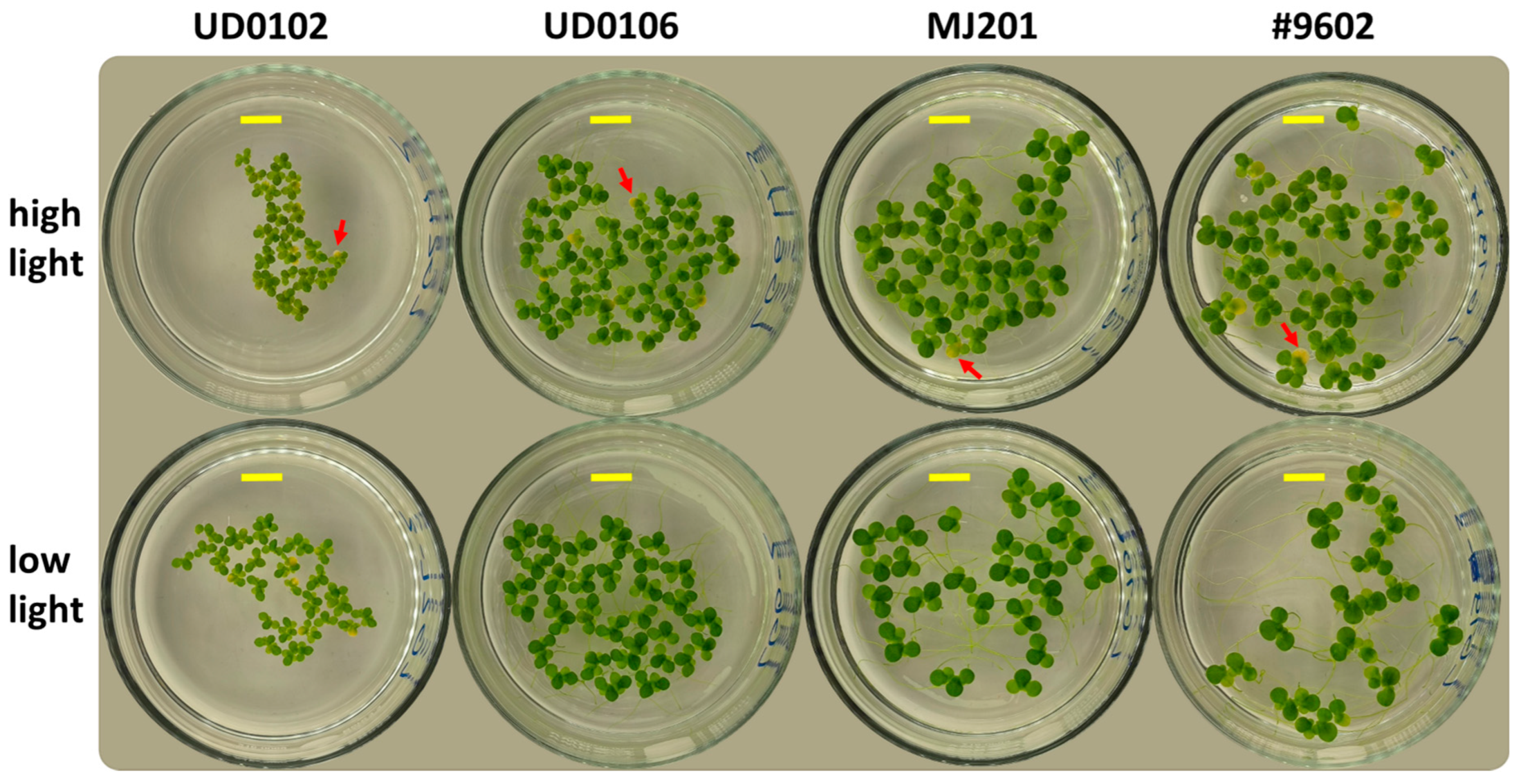



| Clone | Origin | Note |
|---|---|---|
| UD0102 | Poroszló, Hungary (47.646, 20.664) | |
| UD0106 | Hajdúbagos, Hungary (47.386, 21.654) | |
| MJ201 | Mt. Lucas, Ireland (53.275, −7.209) | triploid Lemna × mediterranea |
| #9602 | mutant of 7796 (G3, Sicily, Italy), originally from the Khurana lab (New Delhi, India) | spontaneous tetraploid [31] |
| Chlorophyll a (mg g−1 FW) | Chlorophyll b (mg g−1 FW) | Chlorophyll a + b (mg g−1 FW) | Total Carotenoids (mg g−1 FW) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UD0102 low | 0.723 ± 0.015 a | 0.225 ± 0.005 a * | 0.949 ± 0.020 a * | 0.212 ± 0.003 a | ||||||||
| UD0102 high | 0.588 ± 0.029 A | 0.188 ± 0.008 | 0.776 ± 0.037 | 0.205 ± 0.009 | ||||||||
| UD0106 low | 0.726 ± 0.030 a | 0.223 ± 0.007 a * | 0.949 ± 0.037 a * | 0.209 ± 0.010 a | ||||||||
| UD0106 high | 0.552 ± 0.036 AB | 0.173 ± 0.010 | 0.725 ± 0.047 | 0.198 ± 0.014 | ||||||||
| MJ201 low | 0.555 ± 0.006 b | 0.170 ± 0.003 b * | 0.725 ± 0.009 b * | 0.162 ± 0.003 b | ||||||||
| MJ201 high | 0.423 ± 0.020 BC | 0.130 ± 0.006 | 0.552 ± 0.026 | 0.149 ± 0.007 | ||||||||
| #9602 low | 0.365 ± 0.019 c | 0.122 ± 0.005 c | 0.486 ± 0.024 c | 0.102 ± 0.004 c | ||||||||
| #9602 high | 0.349 ± 0.025 C | 0.108 ± 0.007 | 0.457 ± 0.032 | 0.122 ± 0.010 | ||||||||
| two-way ANOVA | ||||||||||||
| F | p | ω2 | F | p | ω2 | F | p | ω2 | F | p | ω2 | |
| clone | 65.99 | <0.001 | 0.706 | 77.96 | <0.001 | 0.729 | 68.35 | <0.001 | 0.748 | 64.06 | <0.001 | 0.845 |
| light | 40.28 | <0.001 | 0.142 | 47.93 | <0.001 | 0.148 | 33.38 | <0.001 | 0.120 | 0.011 | 0.918 | 0.0 |
| interaction | 4.012 | 0.020 | 0.033 | 2.75 | 0.066 | 0.017 | 2.114 | 0.126 | 0.012 | 2.548 | 0.081 | 0.021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xuan, P.T.H.; Amri, R.; Bach, N.P.; Irfan, M.; Bog, M.; Appenroth, K.J.; Sree, K.S.; Jansen, M.A.K.; Szabó, S.; Mészáros, I.; et al. Lemna gibba Clones Show Differences in Phenotypic Responses to the Light Environment. Plants 2025, 14, 2840. https://doi.org/10.3390/plants14182840
Xuan PTH, Amri R, Bach NP, Irfan M, Bog M, Appenroth KJ, Sree KS, Jansen MAK, Szabó S, Mészáros I, et al. Lemna gibba Clones Show Differences in Phenotypic Responses to the Light Environment. Plants. 2025; 14(18):2840. https://doi.org/10.3390/plants14182840
Chicago/Turabian StyleXuan, Pham Thi Hong, Raja Amri, Nguyen Phuong Bach, Muhammad Irfan, Manuela Bog, Klaus J. Appenroth, K. Sowjanya Sree, Marcel A. K. Jansen, Sándor Szabó, Ilona Mészáros, and et al. 2025. "Lemna gibba Clones Show Differences in Phenotypic Responses to the Light Environment" Plants 14, no. 18: 2840. https://doi.org/10.3390/plants14182840
APA StyleXuan, P. T. H., Amri, R., Bach, N. P., Irfan, M., Bog, M., Appenroth, K. J., Sree, K. S., Jansen, M. A. K., Szabó, S., Mészáros, I., & Oláh, V. (2025). Lemna gibba Clones Show Differences in Phenotypic Responses to the Light Environment. Plants, 14(18), 2840. https://doi.org/10.3390/plants14182840









