Identification of the Chenopodium quinoa HSP90 Gene Family and Functional Analysis of CqHSP90.1c and CqHSP90.6a Under High-Temperature Stress in Transgenic Arabidopsis thaliana
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Properties of the CqHSP90 Gene Family
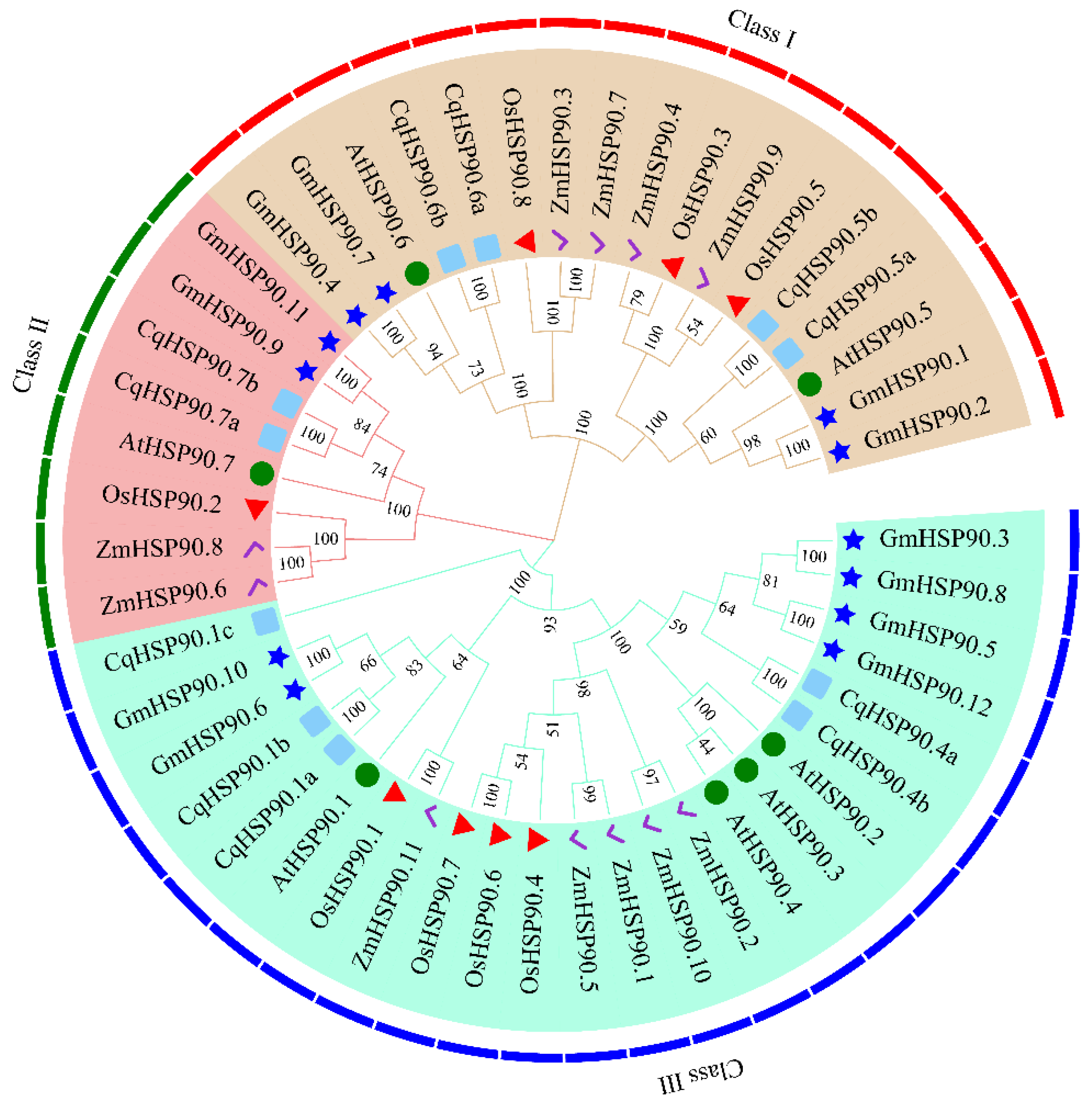
2.2. Phylogenetic Analysis of the HSP90 Gene Family
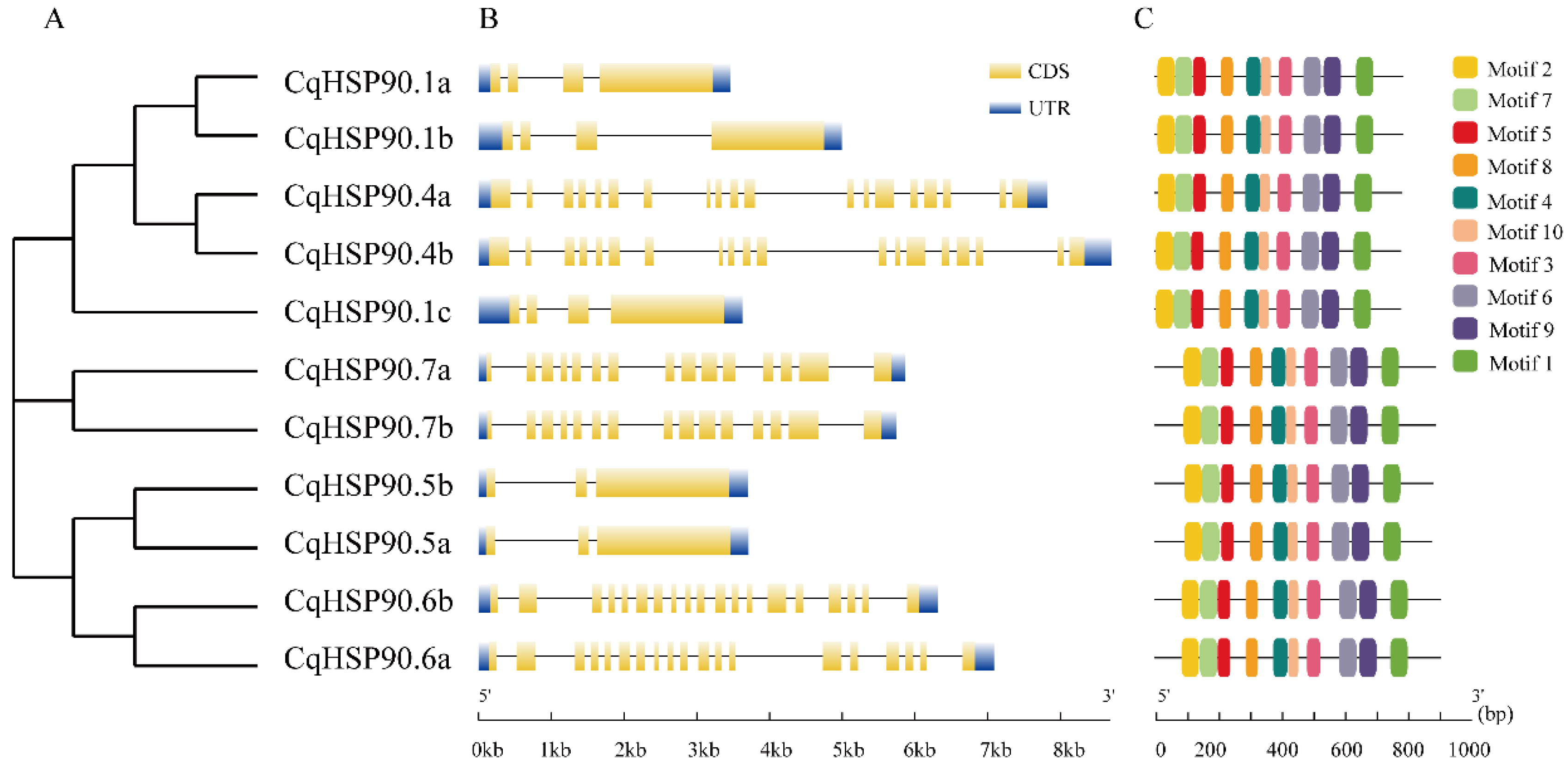
2.3. Motif and Genetic Structure Analysis of the HSP90 Gene Family
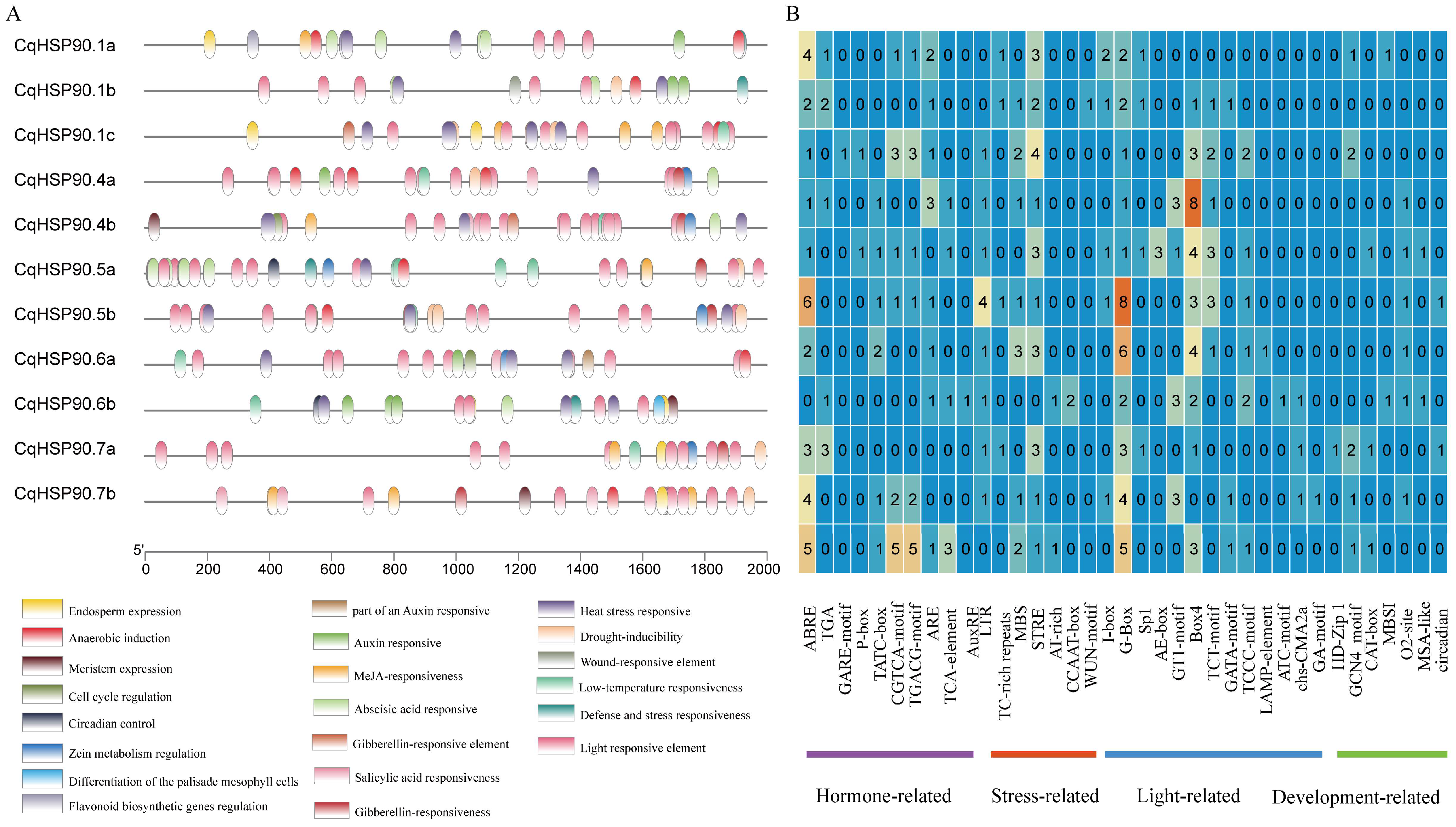
2.4. Analysis of Cis-Acting Elements of the CqHSP90 Gene Family
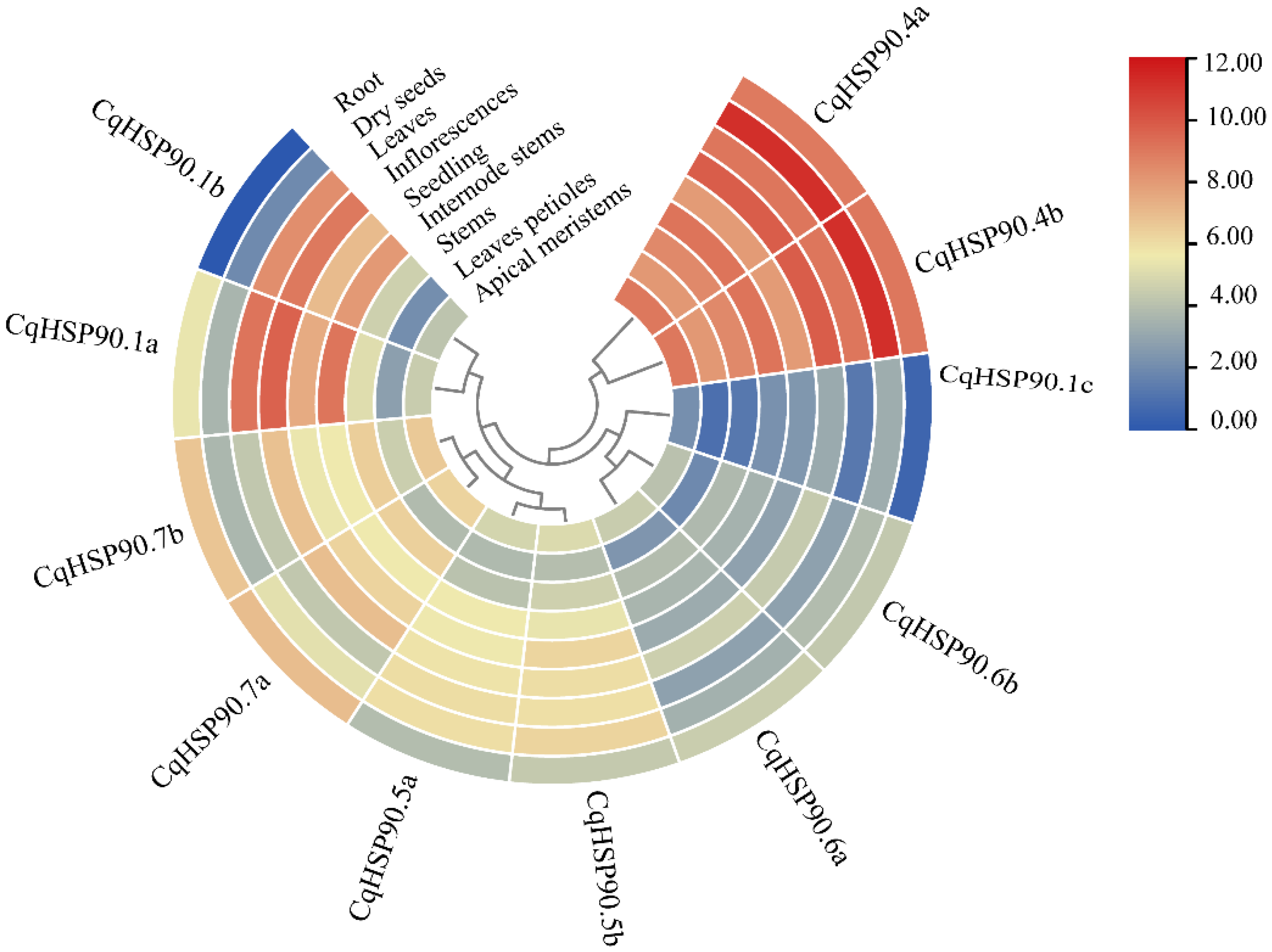
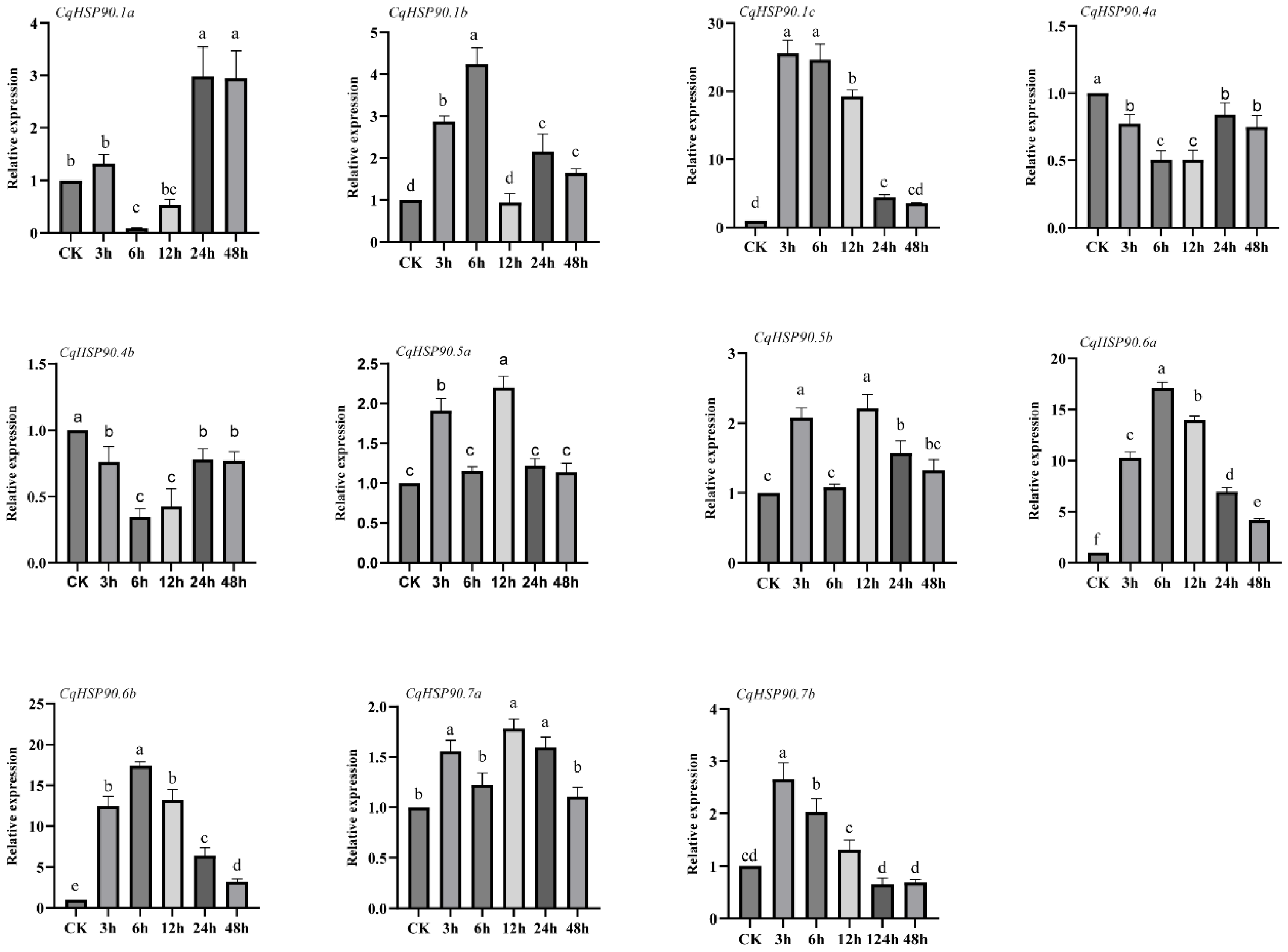
2.5. Heat Stress and Tissue Expression Pattern Analysis of HSP90
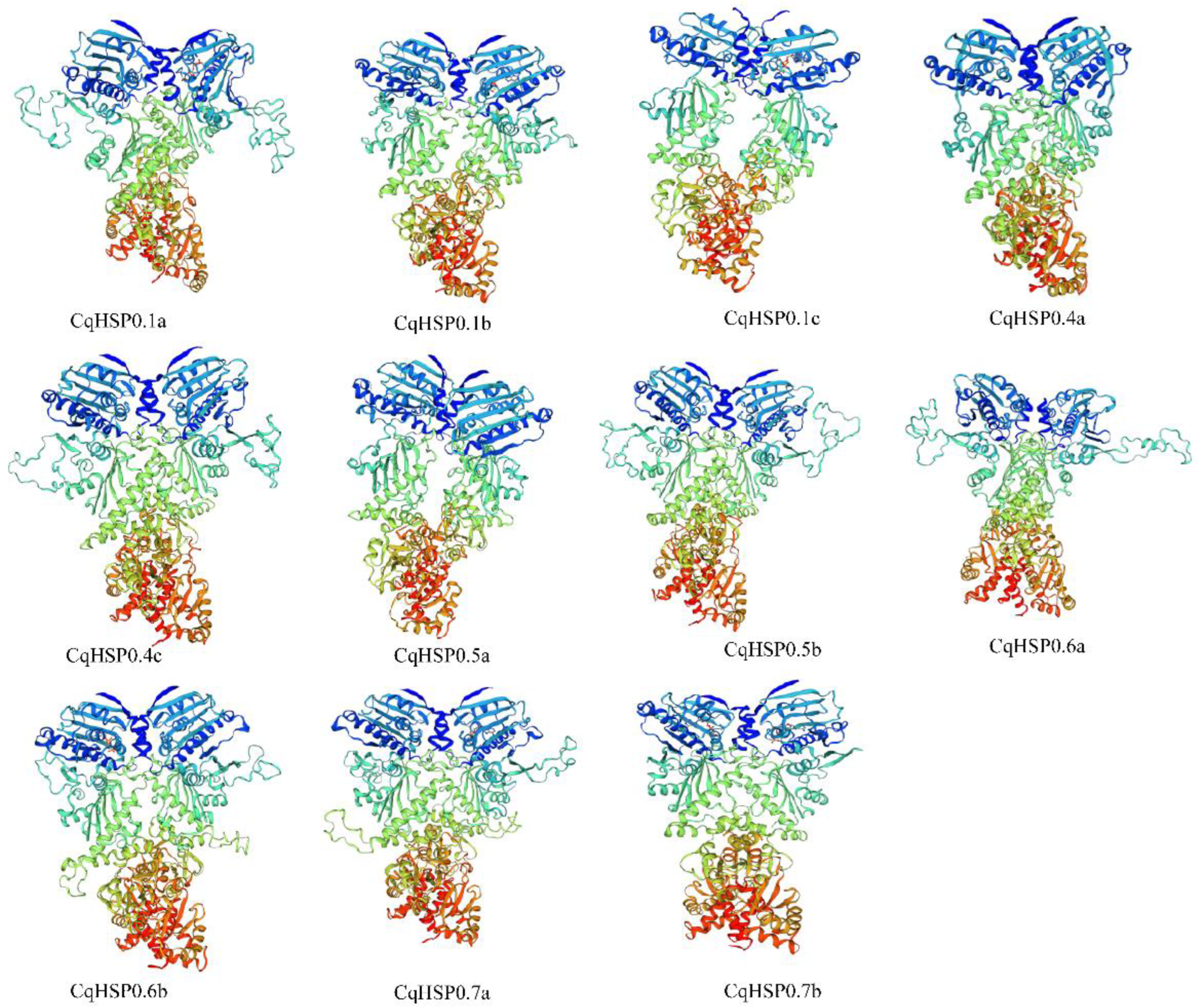
2.6. Protein 3D Structure Prediction
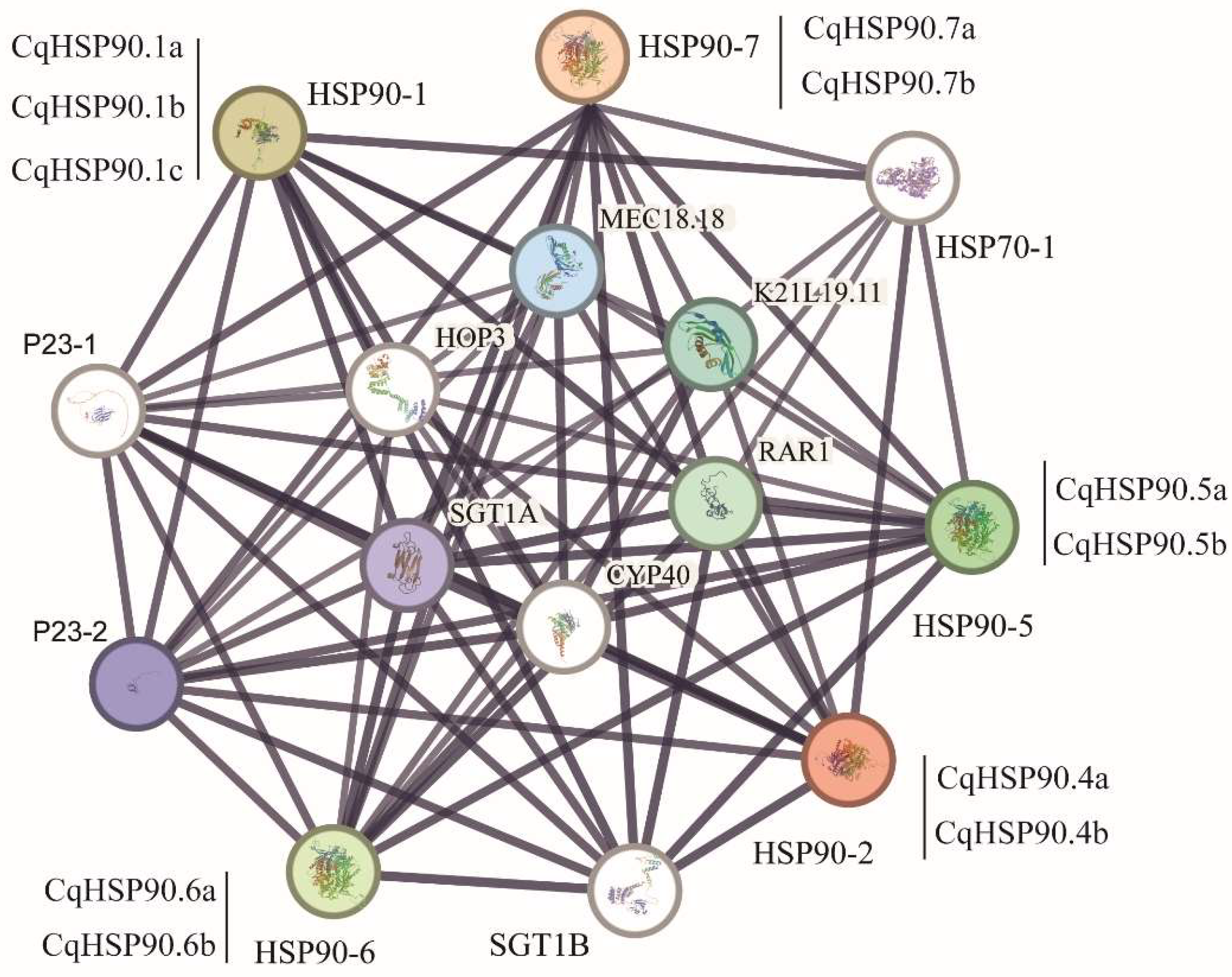
2.7. Analysis of Protein–Protein Interaction Network
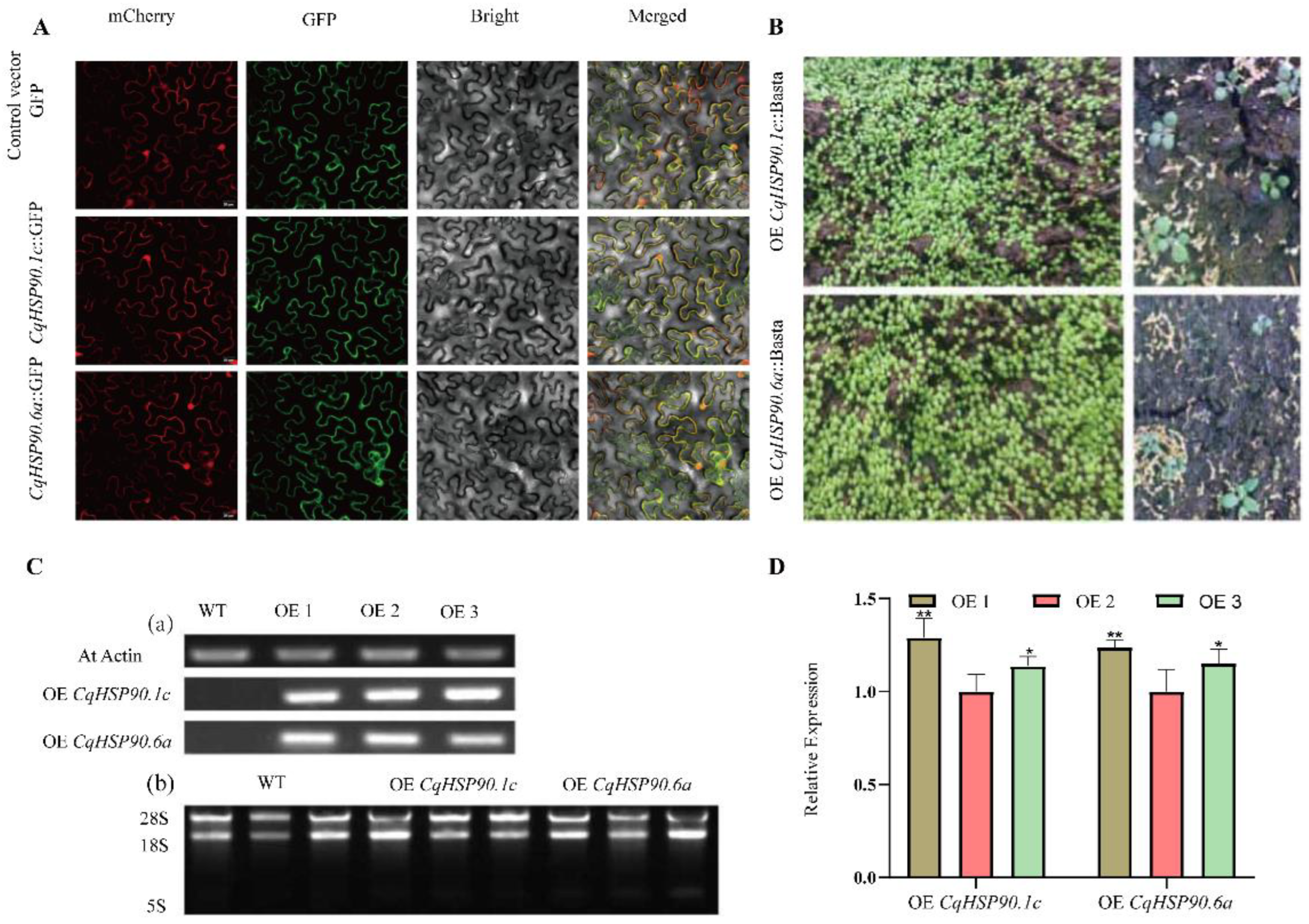
2.8. Subcellular Localization and Acquisition of Transgenic Material
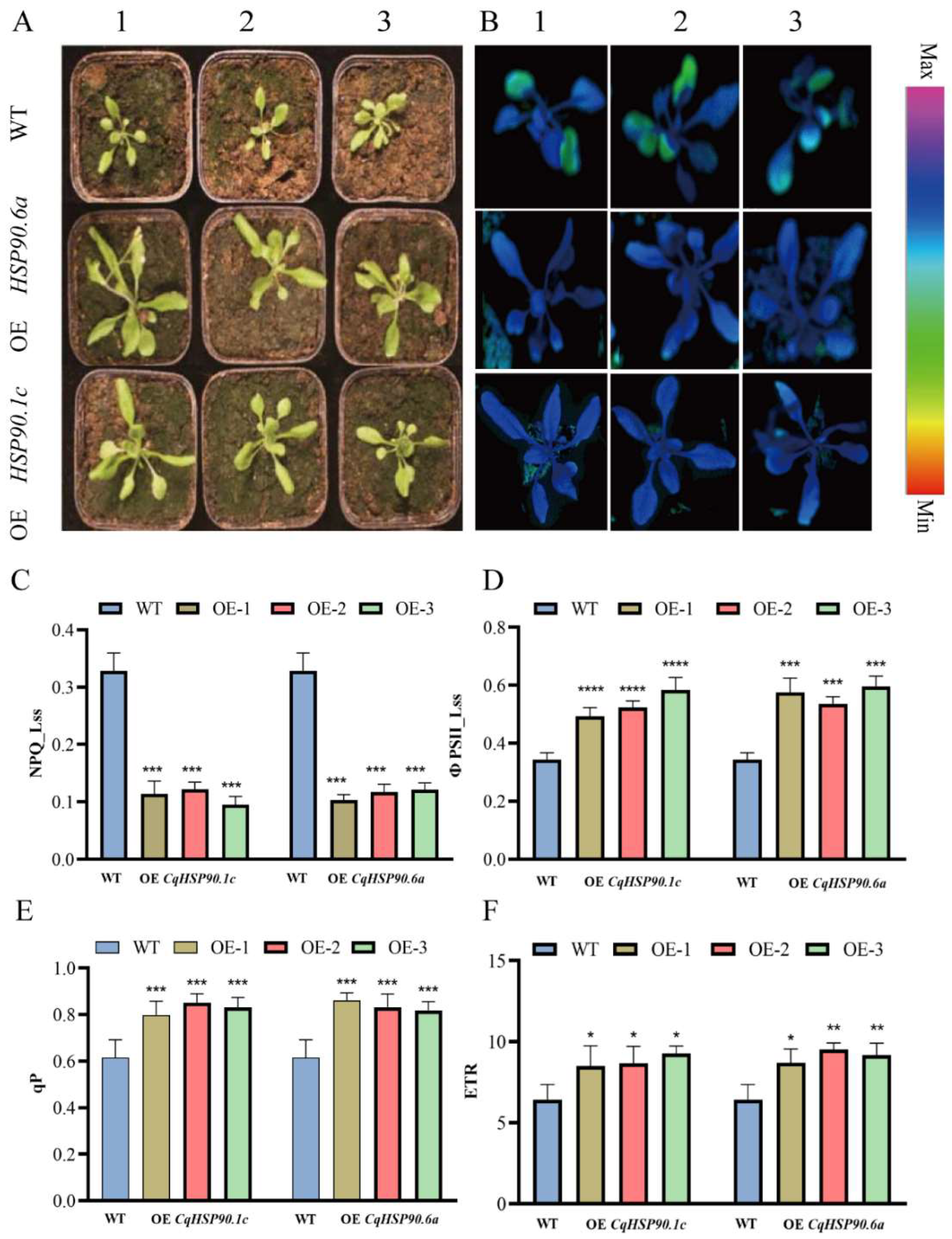
2.9. Overexpression of CqHSP90 Enhances High-Temperature Tolerance in Arabidopsis thaliana
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of Quinoa HSP90
4.2. Subcellular Localization and Physical and Chemical Properties Analysis
4.3. Phylogenetic Analysis
4.4. Conserved Motifs and Gene Structures of the HSP90 Gene Family
4.5. Analysis of Cis-Acting Elements in the HSP90 Gene Family
4.6. Protein 3D Structure Analysis and Protein–Protein Interaction Network
4.7. Plant Treatment and Expression Pattern Analysis
4.8. Subcellular Localization of CqHSP90.1c and HSP90.6a
4.9. Plasmid Construction and Acquisition of Transgenic Plant Materials
4.10. DNA Molecular Level Verification and Quantitative Expression Analysis of Transgenic Lines
4.11. Determination of Physiological Indexes of Wild-Type and Transgenic Arabidopsis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Herman, D.J.; Knowles, L.O.; Knowles, N.R. Heat stress affects carbohydrate metabolism during cold-induced sweetening of potato (Solanum tuberosum L.). Planta 2017, 245, 563–582. [Google Scholar] [CrossRef]
- Gupta, S.C.; Sharma, A.; Mishra, M.; Mishra, R.K.; Chowdhuri, D.K. Heat shock proteins in toxicology: How close and how far? Life Sci. 2010, 86, 377–384. [Google Scholar] [CrossRef]
- Yadav, R.; Juneja, S.; Kumar, S. Cross priming with drought improves heat-tolerance in chickpea (Cicer arietinum L.) by stimulating small heat shock proteins and antioxidative defense. Environ. Sustain. 2021, 4, 171–182. [Google Scholar] [CrossRef]
- Levitt, M.; Gerstein, M.; Huang, E.; Subbiah, S.; Tsai, J. Protein folding: The endgame. Annu. Rev. Biochem. 1997, 66, 549–579. [Google Scholar] [CrossRef]
- Picard, D. Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. CMLS 2002, 59, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.C.; Gorman, K.F.; Rutherford, S. Modularity and intrinsic evolvability of Hsp90-buffered change. PLoS ONE 2006, 1, e76. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.; Inouye, M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000, 25, 24–28. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef]
- Al-Whaibi, M.H. Plant heat-shock proteins: A mini review. J. King Saud Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Fu, X. Chaperone function and mechanism of small heat-shock proteins. Acta Biochim. Biophys. Sin. 2014, 46, 347–356. [Google Scholar] [CrossRef]
- Niforou, K.; Cheimonidou, C.; Trougakos, I.P. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014, 2, 323–332. [Google Scholar] [CrossRef]
- Huang, S.; Monaghan, J.; Zhong, X.; Lin, L.; Sun, T.; Dong, O.X.; Li, X. HSP90s are required for NLR immune receptor accumulation in Arabidopsis. Plant J. Cell Mol. Biol. 2014, 79, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; von Löffelholz, O.; Abell, B.M. Chaperone receptors: Guiding proteins to intracellular compartments. Protoplasma 2012, 249, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Lachowiec, J.; Lemus, T.; Borenstein, E.; Queitsch, C. Hsp90 promotes kinase evolution. Mol. Biol. Evol. 2015, 32, 91–99. [Google Scholar] [CrossRef]
- Pennisi, R.; Ascenzi, P.; di Masi, A. Hsp90: A New Player in DNA Repair? Biomolecules 2015, 5, 2589–2618. [Google Scholar] [CrossRef]
- Zuehlke, A.; Johnson, J.L. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers 2010, 93, 211–217. [Google Scholar] [CrossRef]
- Tapia, H.; Morano, K.A. Hsp90 nuclear accumulation in quiescence is linked to chaperone function and spore development in yeast. Mol. Biol. Cell 2010, 21, 63–72. [Google Scholar] [CrossRef]
- Hao, P.; Zhu, J.; Gu, A.; Lv, D.; Ge, P.; Chen, G.; Li, X.; Yan, Y. An integrative proteome analysis of different seedling organs in tolerant and sensitive wheat cultivars under drought stress and recovery. Proteomics 2015, 15, 1544–1563. [Google Scholar] [CrossRef]
- Lv, D.W.; Subburaj, S.; Cao, M.; Yan, X.; Li, X.; Appels, R.; Sun, D.F.; Ma, W.; Yan, Y.M. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell. Proteom. MCP 2014, 13, 632–652. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, C.Y.; Lv, D.W.; Zhen, S.M.; Li, X.H.; Yan, Y.M. Comparative phosphoproteome analysis of the developing grains in bread wheat (Triticum aestivum L.) under well-watered and water-deficit conditions. J. Proteome Res. 2014, 13, 4281–4297. [Google Scholar] [CrossRef]
- Krishna, P.; Gloor, G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 238–246. [Google Scholar] [CrossRef]
- Lindquist, S. The heat-shock response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Fan, P.; Jiang, P.; Lv, S.; Chen, X.; Li, Y. Chloroplast-targeted Hsp90 plays essential roles in plastid development and embryogenesis in Arabidopsis possibly linking with VIPP1. Physiol. Plant. 2014, 150, 292–307. [Google Scholar] [CrossRef]
- Oh, S.E.; Yeung, C.; Babaei-Rad, R.; Zhao, R. Cosuppression of the chloroplast localized molecular chaperone HSP90.5 impairs plant development and chloroplast biogenesis in Arabidopsis. BMC Res. Notes 2014, 7, 643. [Google Scholar] [CrossRef] [PubMed]
- Sable, A.; Rai, K.M.; Choudhary, A.; Yadav, V.K.; Agarwal, S.K.; Sawant, S.V. Inhibition of Heat Shock proteins HSP90 and HSP70 induce oxidative stress, suppressing cotton fiber development. Sci. Rep. 2018, 8, 3620. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Burch-Smith, T.; Schiff, M.; Feng, S.; Dinesh-Kumar, S.P. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 2004, 279, 2101–2108. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Cha, J.Y.; Ahn, G.; Kim, J.Y.; Kang, S.B.; Kim, M.R.; Su’udi, M.; Kim, W.Y.; Son, D. Structural and functional differences of cytosolic 90-kDa heat-shock proteins (Hsp90s) in Arabidopsis thaliana. Plant Physiol. Biochem. PPB 2013, 70, 368–373. [Google Scholar] [CrossRef]
- Han, Y.; Chen, Y.; Yin, S.; Zhang, M.; Wang, W. Over-expression of TaEXPB23, a wheat expansin gene, improves oxidative stress tolerance in transgenic tobacco plants. J. Plant Physiol. 2015, 173, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Mach, J. Alternative Splicing Produces a JAZ Protein That Is Not Broken Down in Response to Jasmonic Acid. Plant Cell 2009, 21, 14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jackson, S.E.; Queitsch, C.; Toft, D. Hsp90: From structure to phenotype. Nat. Struct. Mol. Biol. 2004, 11, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Pan, F.; Yang, C.; Jia, H.; Jiang, H.; He, F.; Li, N.; Lu, X.; Zhang, H. Genome-wide identification and expression analysis of HSP90 gene family in Nicotiana tabacum. BMC Genet. 2019, 20, 35. [Google Scholar] [CrossRef]
- Song, H.; Zhao, R.; Fan, P.; Wang, X.; Chen, X.; Li, Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 2009, 229, 955–964. [Google Scholar] [CrossRef]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef]
- Azevedo, C.; Betsuyaku, S.; Peart, J.; Takahashi, A.; Noël, L.; Sadanandom, A.; Casais, C.; Parker, J.; Shirasu, K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006, 25, 2007–2016. [Google Scholar] [CrossRef]
- Zhang, M.; Botër, M.; Li, K.; Kadota, Y.; Panaretou, B.; Prodromou, C.; Shirasu, K.; Pearl, L.H. Structural and functional coupling of Hsp90- and Sgt1-centred multi-protein complexes. EMBO J. 2008, 27, 2789–2798. [Google Scholar] [CrossRef]
- LaPointe, P.; Mercier, R.; Wolmarans, A. Aha-type co-chaperones: The alpha or the omega of the Hsp90 ATPase cycle? Biol. Chem. 2020, 401, 423–434. [Google Scholar] [CrossRef]
- Scofield, S.R.; Huang, L.; Brandt, A.S.; Gill, B.S. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005, 138, 2165–2173. [Google Scholar] [CrossRef]
- Takahashi, A.; Casais, C.; Ichimura, K.; Shirasu, K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 11777–11782. [Google Scholar] [CrossRef]
- Iki, T.; Yoshikawa, M.; Meshi, T.; Ishikawa, M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2012, 31, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Duan, M.; Zhang, L.; Li, J.; Shan, L.; Zheng, L.; Liu, J. HOP1 and HOP2 are involved in salt tolerance by facilitating the brassinosteroid-related nucleo-cytoplasmic partitioning of the HSP90-BIN2 complex. Plant Cell Environ. 2022, 45, 3551–3565. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S.H.; Sobott, F.; Yao, Z.P.; Zhang, W.; Nielsen, P.R.; Grossmann, J.G.; Laue, E.D.; Robinson, C.V.; Jackson, S.E. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 2006, 356, 746–758. [Google Scholar] [CrossRef]
- Chong, L.P.; Wang, Y.; Gad, N.; Anderson, N.; Shah, B.; Zhao, R. A highly charged region in the middle domain of plant endoplasmic reticulum (ER)-localized heat-shock protein 90 is required for resistance to tunicamycin or high calcium-induced ER stresses. J. Exp. Bot. 2015, 66, 113–124. [Google Scholar] [CrossRef]
- Phillips, J.J.; Yao, Z.P.; Zhang, W.; McLaughlin, S.; Laue, E.D.; Robinson, C.V.; Jackson, S.E. Conformational dynamics of the molecular chaperone Hsp90 in complexes with a co-chaperone and anticancer drugs. J. Mol. Biol. 2007, 372, 1189–1203. [Google Scholar] [CrossRef]
- Bo, C.; Liu, D.; Yang, J.; Ji, M.; Li, Z.; Zhu, Y.; Duan, Y.; Xue, J.; Xue, T. Comprehensive in silico characterization of NAC transcription factor family of Pinellia ternata and functional analysis of PtNAC66 under high-temperature tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. PPB 2024, 208, 108539. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Rathor, P.; Borza, T.; Stone, S.; Tonon, T.; Yurgel, S.; Potin, P.; Prithiviraj, B. A Novel Protein from Ectocarpus sp. Improves Salinity and High Temperature Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 1971. [Google Scholar] [CrossRef]
| Gene ID | Gene Name | Number of Amino Acids | Molecular Mass (kDa) | pI | Subcellular Location | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|---|
| LOC110729587 | CqHSP90.1a | 703 | 80.9 | 5 | Nucl | 42.65 | 82.11 | −0.62 |
| LOC110722595 | CqHSP90.1b | 703 | 80.9 | 5 | Cyto | 41.65 | 81.96 | −0.631 |
| LOC110693874 | CqHSP90.1c | 700 | 81.1 | 5.07 | E.R. | 42.42 | 84.64 | −0.629 |
| LOC110686052 | CqHSP90.4a | 697 | 79.9 | 4.99 | Cyto | 37.5 | 83.07 | −0.586 |
| LOC110721231 | CqHSP90.4b | 697 | 79.9 | 4.99 | Cyto | 37.38 | 83.34 | −0.586 |
| LOC110711021 | CqHSP90.5a | 795 | 90.2 | 4.95 | Chlo | 41.44 | 78.35 | −0.554 |
| LOC110707029 | CqHSP90.5b | 795 | 90.4 | 4.93 | Chlo | 44.03 | 78.35 | −0.562 |
| LOC110691828 | CqHSP90.6a | 788 | 88.9 | 5.3 | E.R. | 40.8 | 82.04 | −0.532 |
| LOC110701871 | CqHSP90.6b | 784 | 88.4 | 5.24 | Mito | 41 | 82.69 | −0.508 |
| LOC110721133 | CqHSP90.7a | 810 | 93.1 | 4.83 | E.R. | 37.01 | 79.31 | −0.753 |
| LOC110720567 | CqHSP90.7b | 810 | 93.1 | 4.83 | E.R. | 37.43 | 79.19 | −0.758 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Wang, W.; Liu, W.; Song, J.; Chen, S.; An, Y.; Yin, H.; Guo, S. Identification of the Chenopodium quinoa HSP90 Gene Family and Functional Analysis of CqHSP90.1c and CqHSP90.6a Under High-Temperature Stress in Transgenic Arabidopsis thaliana. Plants 2025, 14, 2770. https://doi.org/10.3390/plants14172770
Chen F, Wang W, Liu W, Song J, Chen S, An Y, Yin H, Guo S. Identification of the Chenopodium quinoa HSP90 Gene Family and Functional Analysis of CqHSP90.1c and CqHSP90.6a Under High-Temperature Stress in Transgenic Arabidopsis thaliana. Plants. 2025; 14(17):2770. https://doi.org/10.3390/plants14172770
Chicago/Turabian StyleChen, Fangjun, Wei Wang, Wenli Liu, Jiancheng Song, Shihua Chen, Yibo An, Haibo Yin, and Shanli Guo. 2025. "Identification of the Chenopodium quinoa HSP90 Gene Family and Functional Analysis of CqHSP90.1c and CqHSP90.6a Under High-Temperature Stress in Transgenic Arabidopsis thaliana" Plants 14, no. 17: 2770. https://doi.org/10.3390/plants14172770
APA StyleChen, F., Wang, W., Liu, W., Song, J., Chen, S., An, Y., Yin, H., & Guo, S. (2025). Identification of the Chenopodium quinoa HSP90 Gene Family and Functional Analysis of CqHSP90.1c and CqHSP90.6a Under High-Temperature Stress in Transgenic Arabidopsis thaliana. Plants, 14(17), 2770. https://doi.org/10.3390/plants14172770







