Bioinformatics Analysis Reveals PPR Genes Modulation by Ahyp-miR0005 Under Abiotic Stress Across Diverse Plant Species
Abstract
1. Introduction
2. Results
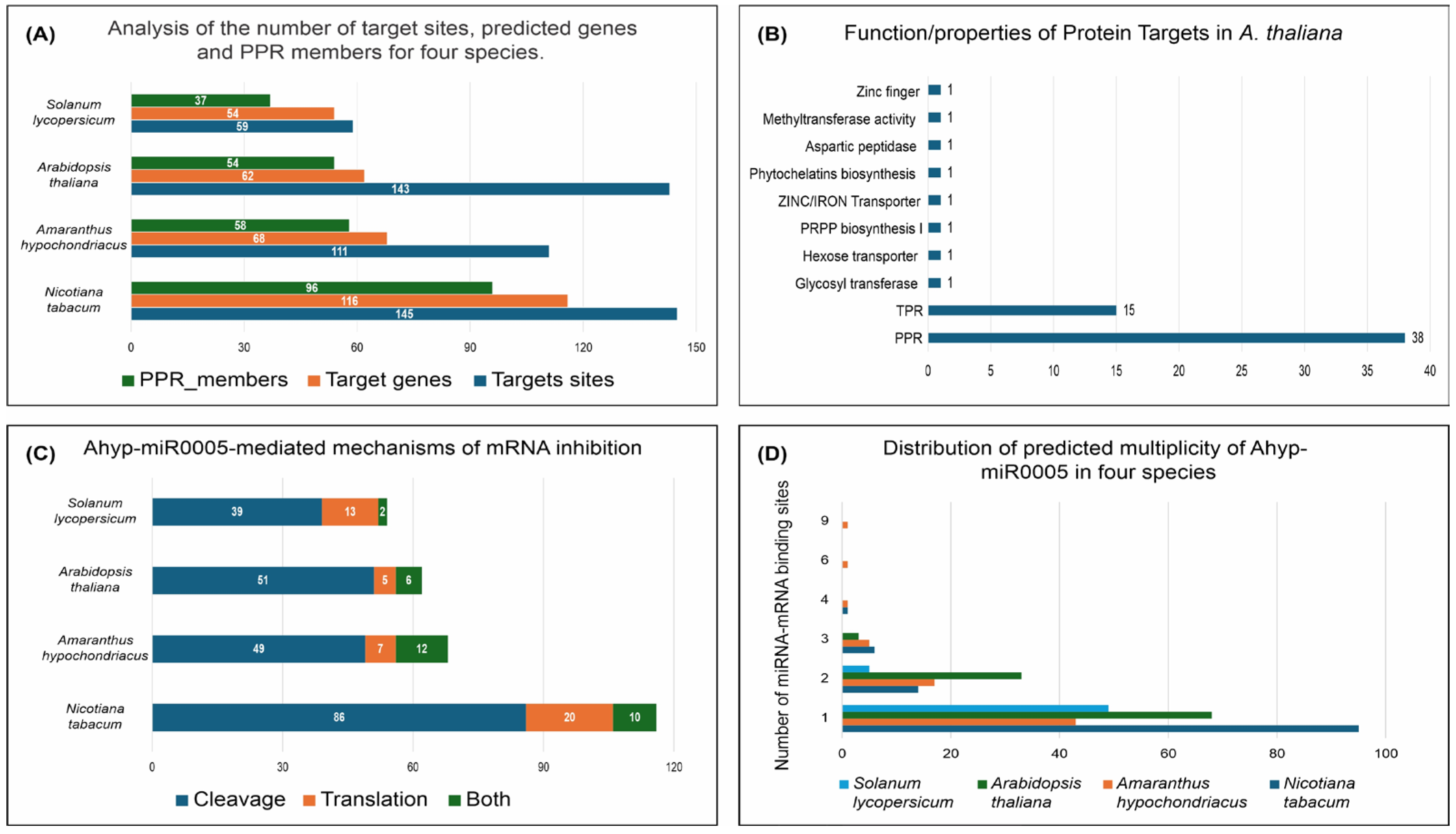
2.1. Identification of miRNA Ahyp-miR0005 Target Transcripts
2.2. Prediction of Cleavage Inhibition and Multi-Site Regulation Analysis of miRNA Ahyp-miR0005
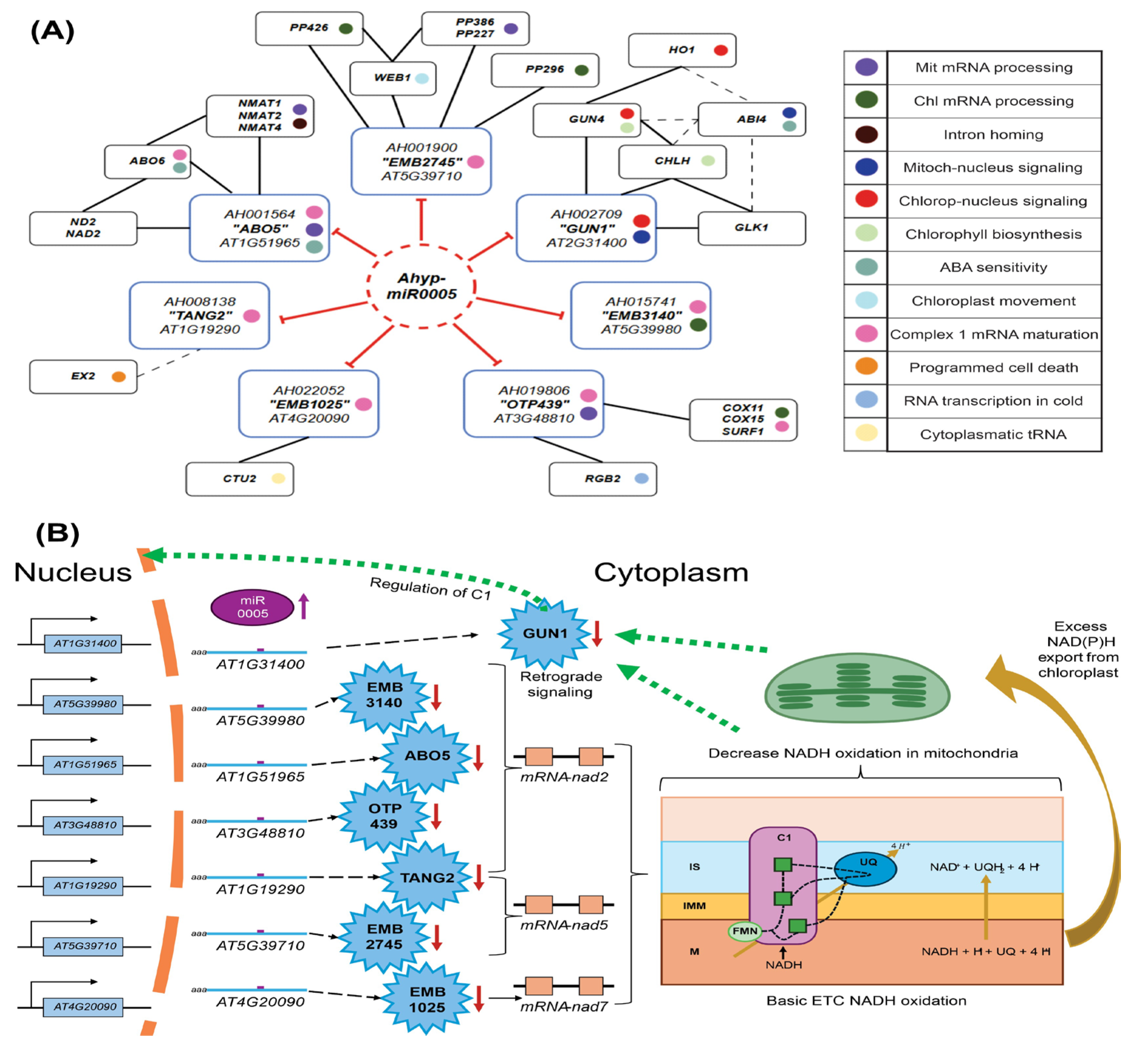
2.3. Functional Analysis of Ahyp-miR0005 Target Genes
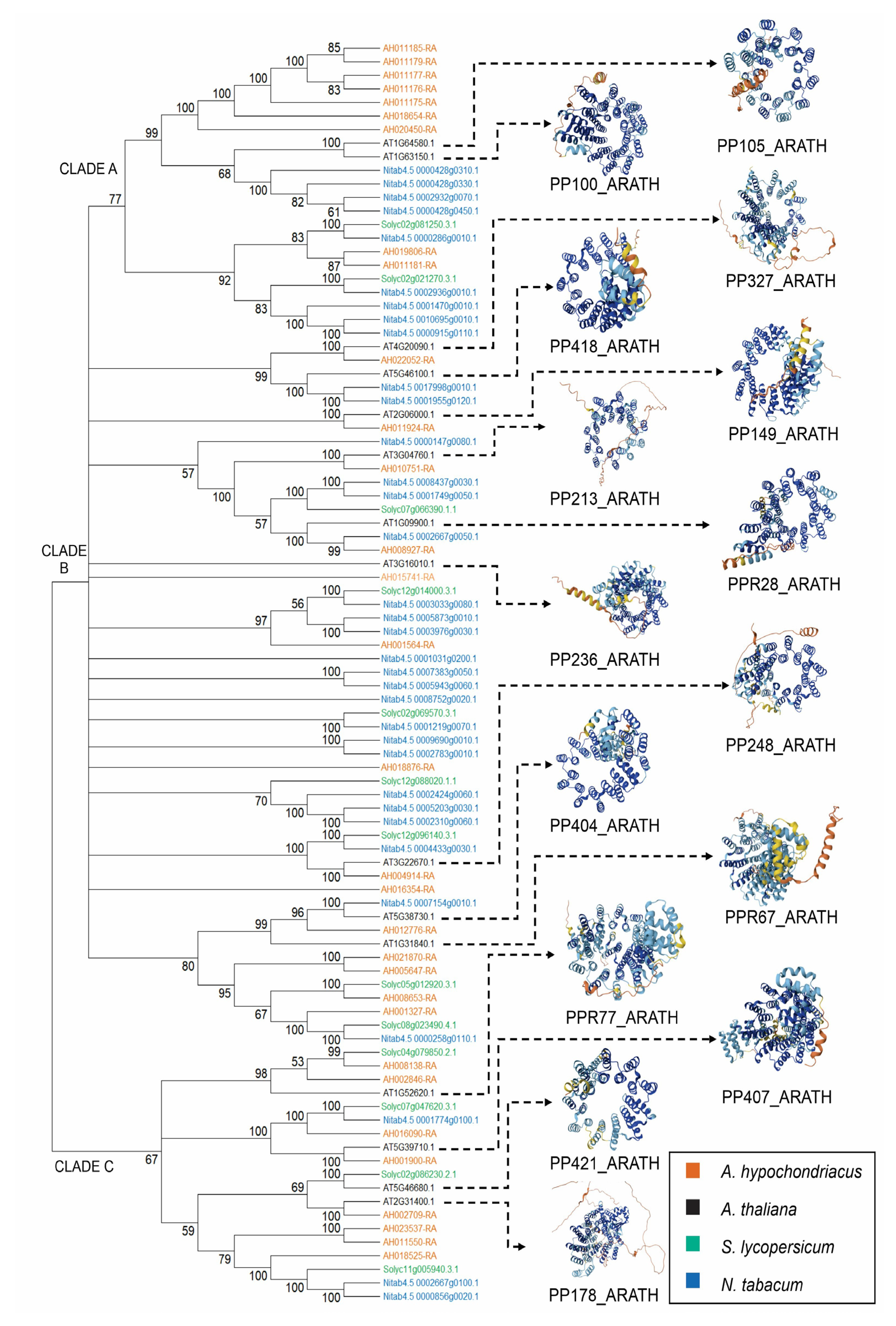
2.4. Distribution and Phylogenetic Analysis of Ahyp-miR0005 Target Genes
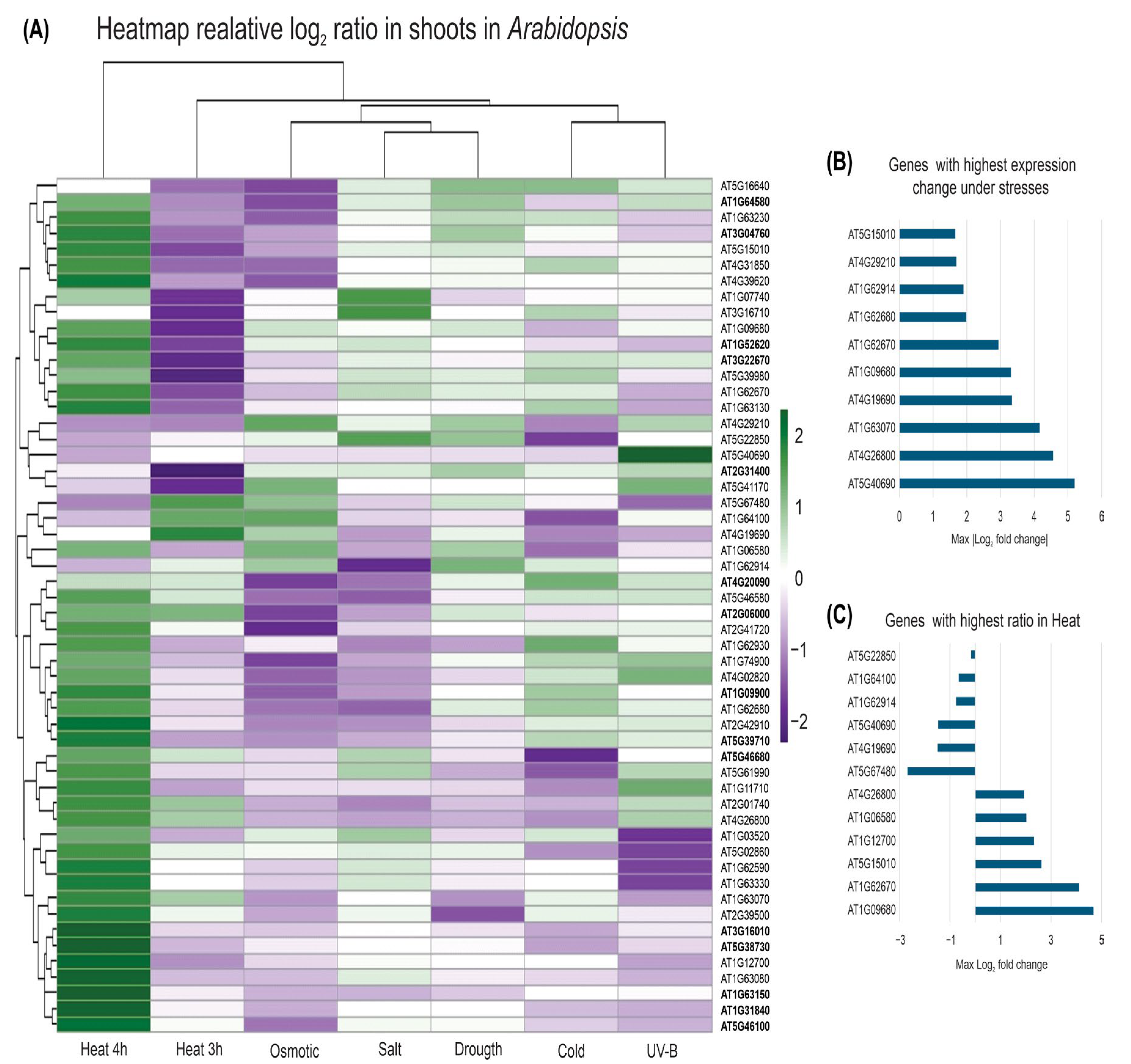
2.5. Gene Expression of Ahyp-miR0005 Targets in Arabidopsis Thaliana Under Abiotic Stress
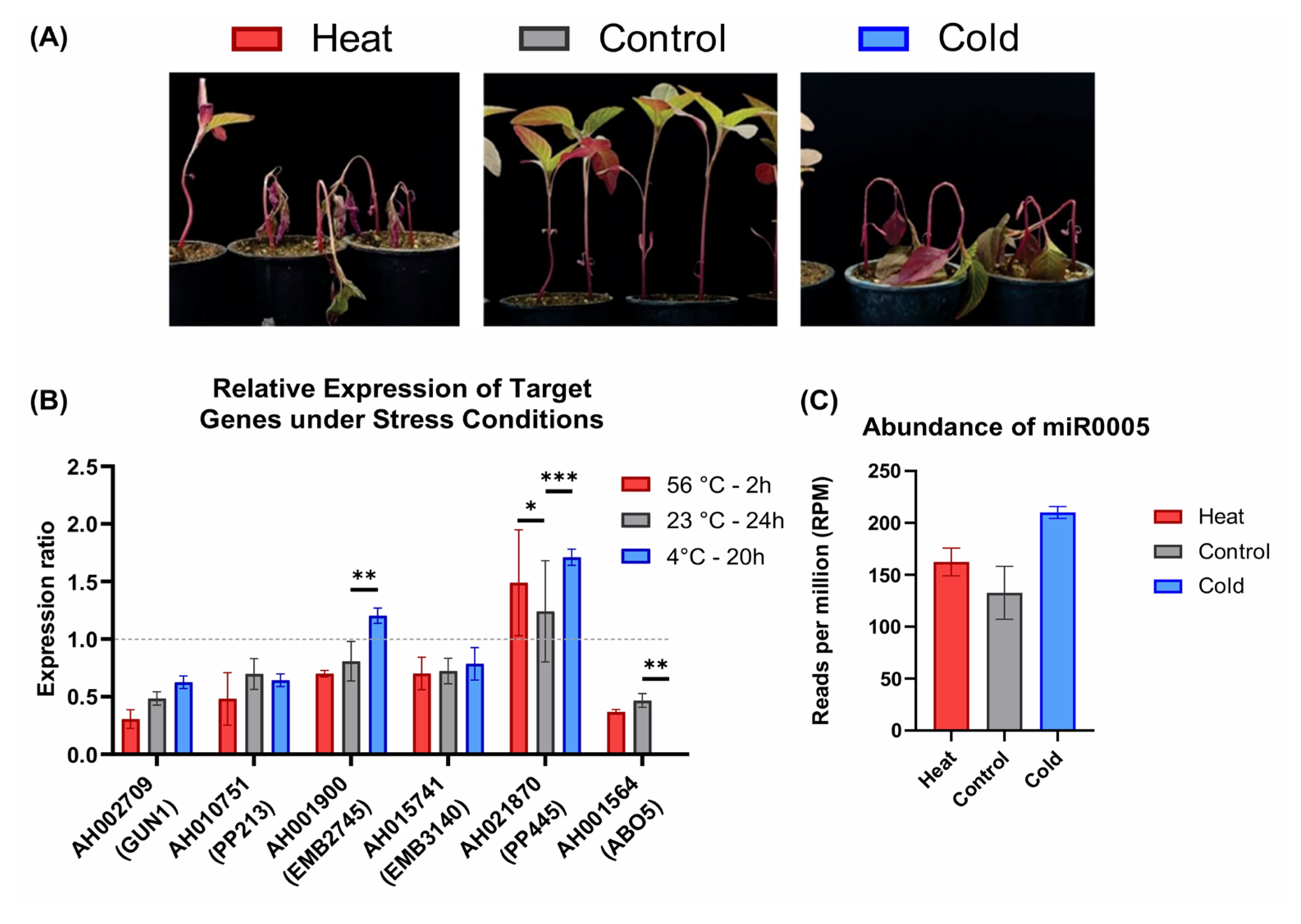
2.6. Gene Expression Analysis of Ahyp-miR0005 Targets Under Abiotic Stress in Amaranth
3. Discussion
3.1. Ahyp-miR0005 Targets PPR Genes Across Different Plant Species
3.2. Ahyp-miR0005-PPR Interactions with Multiple Binding Sites as Combinatorial MECHANISMS of Gene Regulation
3.3. Ahyp-miR0005 Could Regulate Conserved PPR Genes Between Species
3.4. Modulation of PPRs and Its Implications in the Abiotic Stress Response
3.5. Functional Implications of Ahyp-miR0005-PPR Genes and Their Significance in Organelle Biogenesis
4. Materials and Methods
4.1. Prediction of Ahyp-miR0005 Target Genes and Their Action Mechanisms
4.2. Analysis of Conserved Domains in High-Multiplicity Target Genes
4.3. Gene Ontology (GO) Enrichment Analysis and Interaction of Target Genes
4.4. Co-Expression and Functional Interaction Analysis of Homologous Target Genes in Arabidopsis Thaliana
4.5. Gene Expression Analysis Under Abiotic Stress Conditions Using Arabidopsis eFP Browser
4.6. Phylogenetic Analysis of Ahyp-miR0005 Target Genes in Four Species
4.7. Plant Growth Conditions
4.8. Total RNA Extraction
4.9. Quantification of Target Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| miRNAs | MicroRNAs |
| PPR | Pentatricopeptide repeat |
| RSBPN | Retrograde signaling between the plastid and nucleus |
| PPR/TPR | Pentatricopeptide/tetratricopeptide repeat |
| ROS | Reactive oxygen species |
| GUN 1 | GENOMES UNCOUPLED 1 |
| ABO5 | ABA overly-sensitive 5 |
| MORF1 | Multiple organellar RNA editing factor |
| BASS | Bile acid sodium symporter |
| GO | Gene ontology |
| ceRNAs | Competitive endogenous RNAs |
| CDS | Coding sequences |
| CDD | Conserved Domain Database |
| PSSMs | Position-specific scoring matrices |
| MPK6 | Mitogen-Activated Protein Kinase 6 |
| ABA | Abscisic acid |
| SEA | Singular Enrichment Analysis |
References
- Nawaz, M.; Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A Review of Plants Strategies to Resist Biotic and Abiotic Environmental Stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, S.; Parihar, P.; Singh, V.P.; Prasad, S.M. Retrograde Signaling between Plastid and Nucleus: A Review. J. Plant Physiol. 2015, 181, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, M.; Liu, S.; Teng, Q.; Li, S.; Jiang, Y. Functions of PPR Proteins in Plant Growth and Development. Int. J. Mol. Sci. 2021, 22, 11274. [Google Scholar] [CrossRef] [PubMed]
- Jonietz, C.; Forner, J.; Hölzle, A.; Thuss, S.; Binder, S. RNA PROCESSING FACTOR2 Is Required for 5′ End Processing of Nad9 and Cox3 MRNAs in Mitochondria of Arabidopsis thaliana. Plant Cell 2010, 22, 443–453. [Google Scholar] [CrossRef]
- Rovira, A.G.; Smith, A.G. PPR Proteins—Orchestrators of Organelle RNA Metabolism. Physiol. Plant 2019, 166, 451–459. [Google Scholar] [CrossRef]
- Li, K.; Liu, Z.; Xing, L.; Wei, Y.; Mao, J.; Meng, Y.; Bao, L.; Han, M.; Zhao, C.; Zhang, D. MiRNAs Associated with Auxin Signaling, Stress Response, and Cellular Activities Mediate Adventitious Root Formation in Apple Rootstocks. Plant Physiol. Biochem. 2019, 139, 66–81. [Google Scholar] [CrossRef]
- Yagi, Y.; Tachikawa, M.; Noguchi, H.; Satoh, S.; Obokata, J.; Nakamura, T. Pentatricopeptide Repeat Proteins Involved in Plant Organellar RNA Editing. RNA Biol. 2013, 10, 1419–1425. [Google Scholar] [CrossRef]
- Lurin, C.; Andrés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-Wide Analysis of Arabidopsis Pentatricopeptide Repeat Proteins Reveals Their Essential Role in Organelle Biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A Complete Reannotation of the Arabidopsis thaliana Reference Genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Lightfoot, D.J.; Jarvis, D.E.; Ramaraj, T.; Lee, R.; Jellen, E.N.; Maughan, P.J. Single-Molecule Sequencing and Hi-C-Based Proximity-Guided Assembly of Amaranth (Amaranthus hypochondriacus) Chromosomes Provide Insights into Genome Evolution. BMC Biol. 2017, 15, 74. [Google Scholar] [CrossRef]
- Jain, R.; Jenkins, J.; Shu, S.; Chern, M.; Martin, J.A.; Copetti, D.; Duong, P.Q.; Pham, N.T.; Kudrna, D.A.; Talag, J.; et al. Genome Sequence of the Model Rice Variety KitaakeX. BMC Genom. 2019, 20, 905. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An Improved de Novo Assembly and Annotation of the Tomato Reference Genome Using Single-Molecule Sequencing, Hi-C Proximity Ligation and Optical Maps. bioRxiv 2019. [Google Scholar] [CrossRef]
- Laluk, K.; Abuqamar, S.; Mengiste, T. The Arabidopsis Mitochondria-Localized Pentatricopeptide Repeat Protein PGN Functions in Defense against Necrotrophic Fungi and Abiotic Stress Tolerance. Plant Physiol. 2011, 156, 2053–2068. [Google Scholar] [CrossRef]
- Liu, Y.; He, J.; Chen, Z.; Ren, X.; Hong, X.; Gong, Z. ABA Overly-Sensitive 5 (ABO5), Encoding a Pentatricopeptide Repeat Protein Required for Cis-Splicing of Mitochondrial Nad2 Intron 3, Is Involved in the Abscisic Acid Response in Arabidopsis. Plant J. 2010, 63, 749–765. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, S.; Bi, C.; Mei, C.; Jiang, S.C.; Wang, X.F.; Lu, Z.J.; Zhang, D.P. Arabidopsis Exoribonuclease USB1 Interacts with the PPR-Domain Protein SOAR1 to Negatively Regulate Abscisic Acid Signaling. J. Exp. Bot. 2020, 71, 5837–5851. [Google Scholar] [CrossRef] [PubMed]
- Mousa, N.A.; Siaguro, P.; Wiryowidagdo, S.; Wagih, M.E. Evaluation and Selection of Elite Clonal Genotypes of the Sweet Crop Licorice (Glycyrrhiza glabra) in a New Environment. Sugar Tech 2007, 9, 83–94. [Google Scholar] [CrossRef]
- Murayama, M.; Hayashi, S.; Nishimura, N.; Ishide, M.; Kobayashi, K.; Yagi, Y.; Asami, T.; Nakamura, T.; Shinozaki, K.; Hirayama, T. Isolation of Arabidopsis Ahg11, a Weak ABA Hypersensitive Mutant Defective in Nad4 RNA Editing. J. Exp. Bot. 2012, 63, 5301–5310. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, D. Functional Disruption of the Pentatricopeptide Protein SLG1 Affects Mitochondrial RNA Editing, Plant Development, and Responses to Abiotic Stresses in Arabidopsis. Plant J. 2012, 70, 432–444. [Google Scholar] [CrossRef]
- Zsigmond, L.; Rigó, G.; Szarka, A.; Székely, G.; Ötvös, K.; Darula, Z.; Medzihradszky, K.F.; Koncz, C.; Koncz, Z.; Szabados, L. Arabidopsis PPR40 Connects Abiotic Stress Responses to Mitochondrial Electron Transport. Plant Physiol. 2008, 146, 1721–1737. [Google Scholar] [CrossRef]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Takenaka, M.; Kühn, K.; Craddock, C.; Smalle, J.; Karampelias, M.; Denecke, J.; Peters, J.; et al. SLO2, a Mitochondrial Pentatricopeptide Repeat Protein Affecting Several RNA Editing Sites, Is Required for Energy Metabolism. Plant J. 2012, 71, 836–849. [Google Scholar] [CrossRef]
- Kobayashi, K.; Suzuki, M.; Tang, J.; Nagata, N.; Ohyama, K.; Seki, H.; Kiuchi, R.; Kaneko, Y.; Nakazawa, M.; Matsui, M.; et al. Lovastatin Insensitive 1, a Novel Pentatricopeptide Repeat Protein, Is a Potential Regulatory Factor of Isoprenoid Biosynthesis in Arabidopsis. Plant Cell Physiol. 2007, 48, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, X.; Hu, Z.; Wu, C.; Shen, Z. The Pentatricopeptide Repeat Protein GEND1 Is Required for Root Development and High Temperature Tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2021, 578, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Lasorella, C.; Fortunato, S.; Dipierro, N.; Jeran, N.; Tadini, L.; Vita, F.; Pesaresi, P.; de Pinto, M.C. Chloroplast-Localized GUN1 Contributes to the Acquisition of Basal Thermotolerance in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1058831. [Google Scholar] [CrossRef] [PubMed]
- Sechet, J.; Roux, C.; Plessis, A.; Effroy, D.; Frey, A.; Perreau, F.; Biniek, C.; Krieger-Liszkay, A.; Macherel, D.; North, H.M.; et al. The ABA-Deficiency Suppressor Locus HAS2 Encodes the PPR Protein LOI1/MEF11 Involved in Mitochondrial RNA Editing. Mol. Plant 2015, 8, 644–656. [Google Scholar] [CrossRef]
- Liu, J.M.; Zhao, J.Y.; Lu, P.P.; Chen, M.; Guo, C.H.; Xu, Z.S.; Ma, Y.Z. The E-Subgroup Pentatricopeptide Repeat Protein Family in Arabidopsis thaliana and Confirmation of the Responsiveness PPR96 to Abiotic Stresses. Front. Plant Sci. 2016, 7, 1825. [Google Scholar] [CrossRef]
- Xie, T.; Chen, D.; Wu, J.; Huang, X.; Wang, Y.; Tang, K.; Li, J.; Sun, M.; Peng, X. Growing Slowly 1 Locus Encodes a PLS-Type PPR Protein Required for RNA Editing and Plant Development in Arabidopsis. J. Exp. Bot. 2016, 67, 5687–5698. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Zhu, X.; Ren, Y.; Dong, H.; Duan, E.; Teng, X.; Zhao, H.; Chen, R.; Chen, X.; et al. Tetrapyrrole Biosynthesis Pathway Regulates Plastid-to-Nucleus Signaling by Controlling Plastid Gene Expression in Plants. Plant Commun. 2023, 4, 100411. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A Diverse and Evolutionarily Fluid Set of MicroRNAs in Arabidopsis thaliana. Genes. Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Y.H.; Chiang, V.L. Stress-Responsive MicroRNAs in Populus. Plant J. 2008, 55, 131–151. [Google Scholar] [CrossRef]
- Li, Y.F.; Zheng, Y.; Addo-Quaye, C.; Zhang, L.; Saini, A.; Jagadeeswaran, G.; Axtell, M.J.; Zhang, W.; Sunkar, R. Transcriptome-Wide Identification of MicroRNA Targets in Rice. Plant J. 2010, 62, 742–759. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Jiang, J.; Hu, Z.; Lyu, T.; Yang, Y.; Jiang, J.; Cao, J. Over-Expression of MiR158 Causes Pollen Abortion in Brassica campestris Ssp. Chinensis. Plant Mol. Biol. 2017, 93, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Li, P.; Teng, C.; Meyers, B.C. Secondary SiRNAs from Medicago NB-LRRs Modulated via MiRNA-Target Interactions and Their Abundances. Plant J. 2015, 83, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Liang, C.; Wang, S.; Hou, Y.; Gao, L.; Liu, L.; He, W.; Ma, W.; Mo, B.; Chen, X. The Disease Resistance Protein SNC1 Represses the Biogenesis of MicroRNAs and Phased SiRNAs. Nat. Commun. 2018, 9, 5080. [Google Scholar] [CrossRef]
- Lin, P.C.; Lu, C.W.; Shen, B.N.; Lee, G.Z.; Bowman, J.L.; Arteaga-Vazquez, M.A.; Daisy Liu, L.Y.; Hong, S.F.; Lo, C.F.; Su, G.M.; et al. Identification of MiRNAs and Their Targets in the Liverwort Marchantia polymorpha by Integrating RNA-Seq and Degradome Analyses. Plant Cell Physiol. 2016, 57, 339–358. [Google Scholar] [CrossRef]
- Martínez Núñez, M.; Ruíz Rivas, M.; Gregorio Jorge, J.; Hernández, P.F.V.; Luna Suárez, S.; de Folter, S.; Chávez Montes, R.A.; Rosas Cárdenas, F.d.F. Identification of Genuine and Novel MiRNAs in Amaranthus hypochondriacus from High-Throughput Sequencing Data. Genomics 2021, 113, 88–103. [Google Scholar] [CrossRef]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-Wide Analysis of the Rice PPR Gene Family and Their Expression Profiles under Different Stress Treatments. BMC Genom. 2018, 19, 720. [Google Scholar] [CrossRef]
- Ding, X.; Guo, J.; Zhang, Q.; Yu, L.; Zhao, T.; Yang, S. Heat-Responsive Mirnas Participate in the Regulation of Male Fertility Stability in Soybean Cms-Based F1 under High Temperature Stress. Int. J. Mol. Sci. 2021, 22, 2446. [Google Scholar] [CrossRef]
- Téllez Valerio, C.E.; Gregorio Jorge, J.; Luna Suárez, S.; Maldonado Mendoza, I.E.; Rosas Cárdenas, F.d.F. MiR6024 Overexpression Increases the Susceptibility of Nicotiana tabacum to Sclerotinia sclerotiorum. Eur. J. Plant Pathol. 2023, 165, 97–113. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Ruwe, H.; Wang, G.; Gusewski, S.; Schmitz-Linneweber, C. Systematic Analysis of Plant Mitochondrial and Chloroplast Small RNAs Suggests Organelle-Specific MRNA Stabilization Mechanisms. Nucleic Acids Res. 2016, 44, 7406–7417. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Bhattacharya, A.; Bhardwaj, V.; Jha, V.; Mandal, A.K.; Mukerji, M. Alu-MiRNA Interactions Modulate Transcript Isoform Diversity in Stress Response and Reveal Signatures of Positive Selection. Sci. Rep. 2016, 6, 32348. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. The MiRNA–Target Interactions: An Underestimated Intricacy. Nucleic Acids Res. 2024, 52, 1544–1557. [Google Scholar] [CrossRef]
- Wilczynska, A.; Bushell, M. The Complexity of MiRNA-Mediated Repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of MicroRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory Network of MiRNA on Its Target: Coordination between Transcriptional and Post-Transcriptional Regulation of Gene Expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef]
- Filipovska, A.; Rackham, O. Modular Recognition of Nucleic Acids by PUF, TALE and PPR Proteins. Mol. Biosyst. 2012, 8, 699–708. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. MicroRNA Sponges: Progress and Possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef]
- Farberov, L.; Ionescu, A.; Zoabi, Y.; Shapira, G.; Ibraheem, A.; Azan, Y.; Perlson, E.; Shomron, N. Multiple Copies of MicroRNA Binding Sites in Long 3′UTR Variants Regulate Axonal Translation. Cells 2023, 12, 233. [Google Scholar] [CrossRef]
- Naeli, P.; Winter, T.; Hackett, A.P.; Alboushi, L.; Jafarnejad, S.M. The Intricate Balance between MicroRNA-Induced MRNA Decay and Translational Repression. FEBS J. 2023, 290, 2508–2524. [Google Scholar] [CrossRef]
- Kang, W.; Bang-Berthelsen, C.H.; Holm, A.; Houben, A.J.S.; Müller, A.H.; Thymann, T.; Pociot, F.; Estivill, X.; Friedländer, M.R. Survey of 800+ Data Sets from Human Tissue and Body Fluid Reveals XenomiRs Are Likely Artifacts. RNA 2017, 23, 433–445. [Google Scholar] [CrossRef]
- Tillich, M.; Beick, S.; Schmitz-Linneweber, C. Chloroplast RNA-Binding Proteins: Repair and Regulation of Chloroplast Transcripts. RNA Biol. 2010, 7, 172–178. [Google Scholar] [CrossRef]
- Arae, T.; Isai, S.; Sakai, A.; Mineta, K.; Yokota Hirai, M.; Suzuki, Y.; Kanaya, S.; Yamaguchi, J.; Naito, S.; Chiba, Y. Co-Ordinated Regulations of MRNA Synthesis and Decay during Cold Acclimation in Arabidopsis Cells. Plant Cell Physiol. 2017, 58, 1090–1102. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S.; Barron, C.; Brady, S.M.; Schiefelbein, J.; Benfey, P.N. Cell Identity Mediates the Response of Arabidopsis Roots to Abiotic Stress. Science 2008, 320, 942–945. [Google Scholar] [CrossRef]
- Okuda, K.; Chateigner-Boutin, A.L.; Nakamura, T.; Delannoy, E.; Sugita, M.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Small, I.; Shikanai, T. Pentatricopeptide Repeat Proteins with the DYW Motif Have Distinct Molecular Functions in RNA Editing and RNA Cleavage in Arabidopsis Chloroplasts. Plant Cell 2009, 21, 146–156. [Google Scholar] [CrossRef]
- Liu, X.; Yu, F.; Rodermel, S. An Arabidopsis Pentatricopeptide Repeat Protein, SUPPRESSOR OF VARIEGATION7, Is Required for FtsH-Mediated Chloroplast Biogenesis. Plant Physiol. 2010, 154, 1588–1601. [Google Scholar] [CrossRef]
- Chang, R.; Jang, C.J.H.; Branco-Price, C.; Nghiem, P.; Bailey-Serres, J. Transient MPK6 Activation in Response to Oxygen Deprivation and Reoxygenation Is Mediated by Mitochondria and Aids Seedling Survival in Arabidopsis. Plant Mol. Biol. 2012, 78, 109–122. [Google Scholar] [CrossRef]
- Yang, X.; Jia, Z.; Pu, Q.; Tian, Y.; Zhu, F.; Liu, Y. ABA Mediates Plant Development and Abiotic Stress via Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 3796. [Google Scholar] [CrossRef] [PubMed]
- Tadini, L.; Jeran, N.; Pesaresi, P. GUN1 and Plastid RNA Metabolism: Learning from Genetics. Cells 2020, 9, 2307. [Google Scholar] [CrossRef] [PubMed]
- Kempken, F. (Ed.) Plant Mitochondria; Springer: New York, NY, USA, 2010; Volume 1, ISBN 978-0-387-89781-3. [Google Scholar]
- Dai, X.; Zhuang, Z.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A Reference Genome for Nicotiana Tabacum Enables Map-Based Cloning of Homeologous Loci Implicated in Nitrogen Utilization Efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)-from Genotype to Phenotype to Breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO Analysis Toolkit for the Agricultural Community, 2017 Update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress Global Stress Expression Data Set: Protocols, Evaluation and Model Data Analysis of UV-B Light, Drought and Cold Stress Responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; Cukura, A.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Vennapusa, A.R.; Somayanda, I.M.; Doherty, C.J.; Jagadish, S.V.K. A Universal Method for High-Quality RNA Extraction from Plant Tissues Rich in Starch, Proteins and Fiber. Sci. Rep. 2020, 10, 16887. [Google Scholar] [CrossRef]
- Vera Hernández, F.P.; Martínez Núñez, M.; Ruiz Rivas, M.; Vázquez Portillo, R.E.; Bibbins Martínez, M.D.; Luna Suárez, S.; Rosas Cárdenas, F.d.F. Reference Genes for RT-QPCR Normalisation in Different Tissues, Developmental Stages and Stress Conditions of Amaranth. Plant Biol. 2018, 20, 713–721. [Google Scholar] [CrossRef]

| Amaranth | Name | Ath_Name UniProt | Arabidopsis | Tomato | Tobacco | Function |
|---|---|---|---|---|---|---|
| AH001327 | - | F4I4T7_ARATH | AT1G30290 | Solyc08g023490 | Nitab4.5_0002071g0050.1 Nitab4.5_0000315g0050.1 | - |
| AH001564 | ABO5 | PPR76_ARATH | AT1G51965 | Solyc03g121110 | Nitab4.5_0005085g0030.1 Nitab4.5_0004969g0060.1 | Embryogenesis, metabolism |
| AH001900 | EMB2745 | PP407_ARATH | AT5G39710 | Solyc01g108410 | Nitab4.5_0004334g0100.1 Nitab4.5_0005583g0010.1 | Essential in embryo |
| AH002709 | GUN1 | PP178_ARATH | AT2G31400 | Solyc06g009520.3.1 | Nitab4.5_0000363g0190.1 Nitab4.5_0007407g0040.1 | Stress response |
| AH002755 | - | - | - | Solyc03g114000 Solyc06g071310 | Nitab4.5_0001422g0090.1 Nitab4.5_0009569g0010.1 Nitab4.5_0002314g0060.1 Nitab4.5_0004495g0010.1 | - |
| AH002846 | - | PP440_ARATH | AT5G61400 | Solyc06g069700 | Nitab4.5_0003021g0070.1 Nitab4.5_0004286g0010.1 | RNA processing |
| AH004914 | - | PP211_ARATH | AT3G04130 | Solyc05g014490 | Nitab4.5_0001898g0020.1 | RNA processing |
| AH005647 | - | PP445_ARATH | AT5G65560 | Solyc10g081880 | Nitab4.5_0002165g0010.1 Nitab4.5_0005257g0020.1 Nitab4.5_0002364g0030.1 Nitab4.5_0000574g0030.1 | RNA processing |
| AH008138 | TANG2 | PPR50_ARATH | AT1G19290 | Solyc04g079850.2.1 | Nitab4.5_0006518g0030.1 Nitab4.5_0000110g0140.1 | RNA processing |
| AH008653 | - | PP388_ARATH | AT5G16420 | Solyc01g096210 | Nitab4.5_0002527g0080.1 Nitab4.5_0002667g0050.1 | RNA processing |
| AH008927 | MEE40 | PP281_ARATH | AT1G09900 | - | Nitab4.5_0002667g0060.1 Nitab4.5_0002667g0050.1 | RNA processing |
| AH010751 | - | PP213_ARATH | AT3G04760 | - | Nitab4.5_0003780g0130.1 | RNA processing |
| AH011550 | ABO8 | PP306_ARATH | AT4G11690 | - | Nitab4.5_0007891g0010.1 Nitab4.5_0001714g0210.1 | Response to abscisic acid |
| AH011924 | - | PP149_ARATH | AT2G06000 | Solyc01g104630 | Nitab4.5_0009570g0010.1 Nitab4.5_0000249g0340.1 | RNA processing |
| AH012776 | - | PP180_ARATH | AT2G32630 | Solyc10g084080 | Nitab4.5_0005518g0020.1 Nitab4.5_0001297g0100.1 | RNA processing |
| AH015741 | PDM3/EMB3140 | PP408_ARATH | AT5G39980 | Solyc02g087560.1 | Nitab4.5_0002265g0130.1 Nitab4.5_0000564g0400.1 | -Essential in Embryo -Chloroplast development |
| AH016090 | - | PP338_ARATH | AT4G26680 | Solyc07g047620 | Nitab4.5_0001774g0100.1 Nitab4.5_0001701g0020.1 | RNA processing |
| AH016354 | EMB1444 | PPR15_ARATH | AT1G06143 | - | Nitab4.5_0000128g0270.1 | Essential in embryo |
| AH018525 | - | PP325_ARATH | AT4G19440 | Solyc11g005940 | Nitab4.5_0000856g0020.1 Nitab4.5_0002667g0100.1 | RNA processing |
| AH018654 | RPF1 PPR3 - RFL9/RPF4 RPF6 RPF8 - RFL2 - - RPF2 - - PPR-AC RPF3 - | PPR38_ARATH PP247_ARATH PP100_ARATH PPR94_ARATH PPR99_ARATH PPR37_ARATH PPR98_ARATH PPR36_ARATH PP102_ARATH PPR97_ARATH PPR91_ARATH PPR39_ARATH PP101_ARATH PPR90_ARATH PPR96_ARATH PP103_ARATH | AT1G12700 AT3G22470 AT1G63150 AT1G62910 AT1G63130 AT1G12620 AT1G63080 AT1G12300 AT1G63400 AT1G63070 AT1G62670 AT1G12775 AT1G63330 AT1G62590 AT1G62930 AT1G64100 | Solyc06g007740.1 Solyc06g007850 Solyc04g080120.1 Solyc06g007300 Solyc05g009253.1 Solyc06g005220.2 | - | -RNA processing factor -mRNA modification |
| AH019806 | OTP439 | PP270_ARATH | AT3G48810 | Solyc01g058205.1 | Nitab4.5_0000225g0060.1 Nitab4.5_0004303g0030.1 | RNA processing |
| AH021870 | - | PP445_ARATH | AT5G65560 | Solyc07g047820 | Nitab4.5_0000441g0020.1 Nitab4.5_0011940g0010.1 | RNA processing |
| AH022052 | EMB1025 | PP327_ARATH | AT4G20090 | Solyc02g081860 | Nitab4.5_0000844g0300.1 Nitab4.5_0010610g0010.1 | Essential in embryo |
| AH023537 | - | PP156_ARATH | AT2G16880 | Solyc01g111470.3.1 | Nitab4.5_0000061g0180.1 | RNA processing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores Benavides, V.; Montes, R.A.C.; Rosas Cárdenas, F.d.F. Bioinformatics Analysis Reveals PPR Genes Modulation by Ahyp-miR0005 Under Abiotic Stress Across Diverse Plant Species. Plants 2025, 14, 2757. https://doi.org/10.3390/plants14172757
Flores Benavides V, Montes RAC, Rosas Cárdenas FdF. Bioinformatics Analysis Reveals PPR Genes Modulation by Ahyp-miR0005 Under Abiotic Stress Across Diverse Plant Species. Plants. 2025; 14(17):2757. https://doi.org/10.3390/plants14172757
Chicago/Turabian StyleFlores Benavides, Vladimir, Ricardo A. Chávez Montes, and Flor de Fátima Rosas Cárdenas. 2025. "Bioinformatics Analysis Reveals PPR Genes Modulation by Ahyp-miR0005 Under Abiotic Stress Across Diverse Plant Species" Plants 14, no. 17: 2757. https://doi.org/10.3390/plants14172757
APA StyleFlores Benavides, V., Montes, R. A. C., & Rosas Cárdenas, F. d. F. (2025). Bioinformatics Analysis Reveals PPR Genes Modulation by Ahyp-miR0005 Under Abiotic Stress Across Diverse Plant Species. Plants, 14(17), 2757. https://doi.org/10.3390/plants14172757






