Genome-Wide Identification of the Glycosyl Hydrolase Family 1 Genes in Brassica napus L. and Functional Characterization of BnBGLU77
Abstract
1. Introduction
2. Results
2.1. Identification of BGLU Family Genes in B. napus
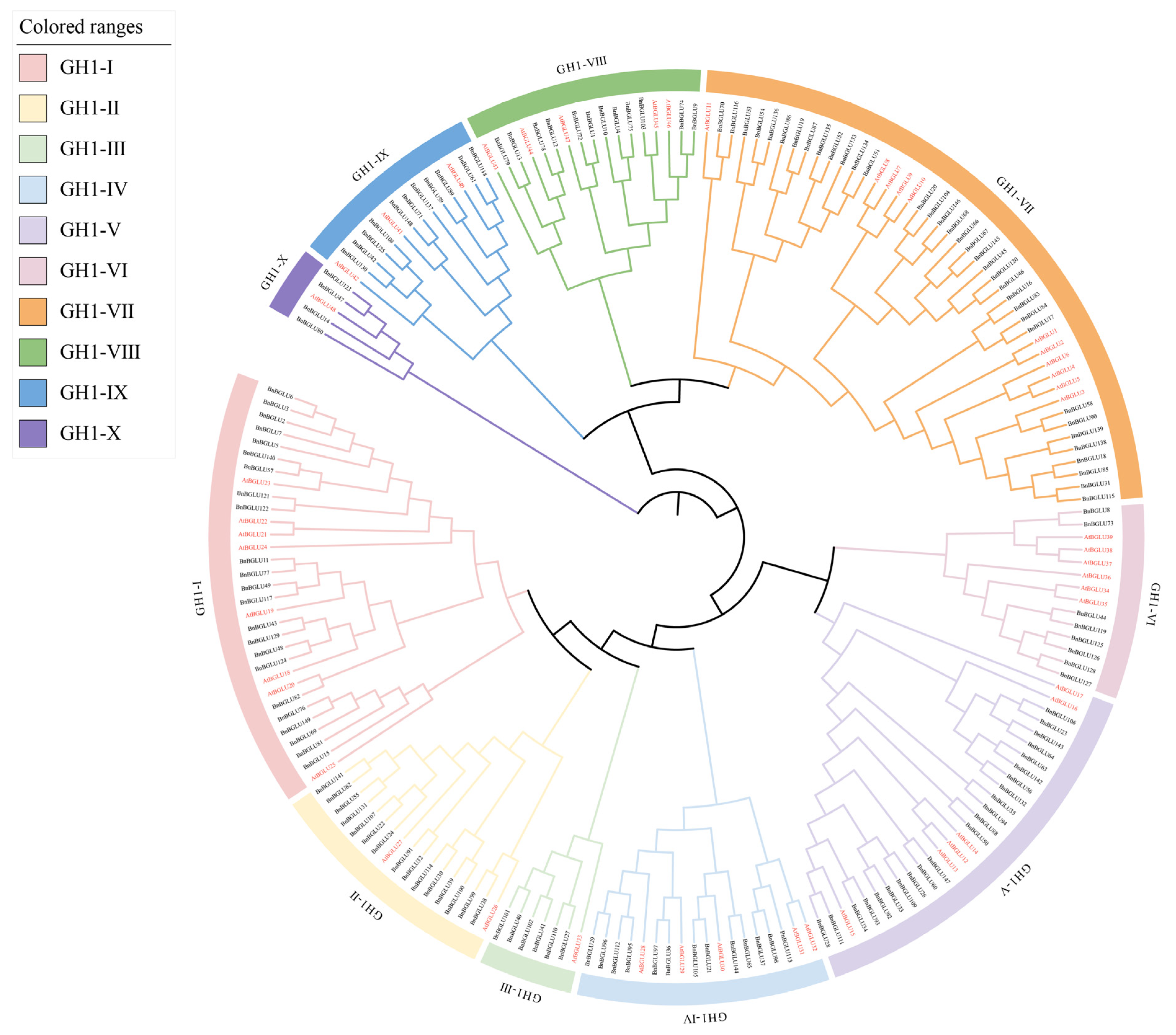
2.2. Phylogenetic Analysis of BGLU Proteins in A. thaliana and B. napus
2.3. Analysis of Conserved Motifs and Gene Structure of the BnBGLU Family
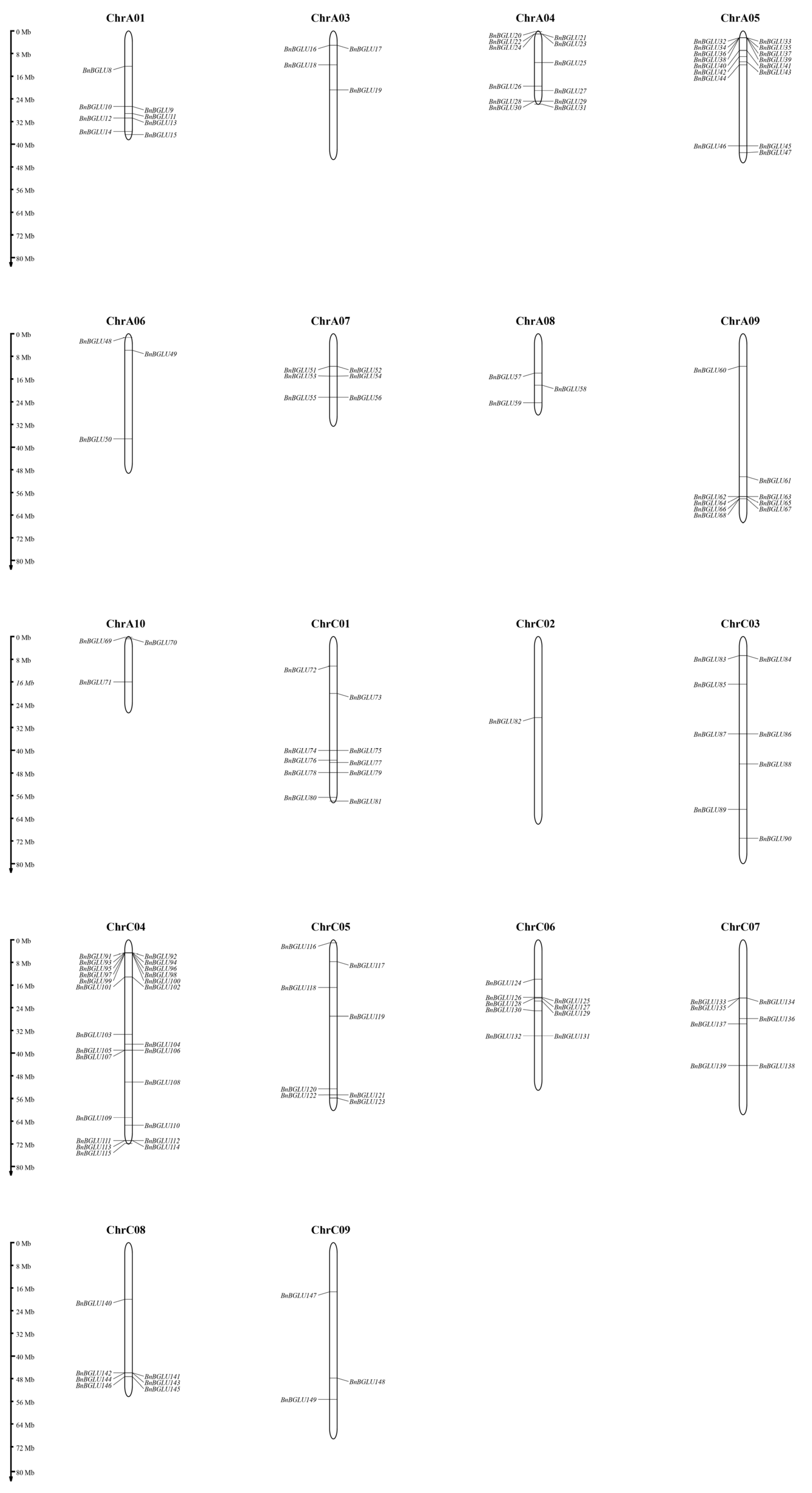
2.4. Chromosome Mapping Analysis of BnBGLU Genes
2.5. Collinearity and Selection Pressure Analysis of BGLU Genes
2.6. Prediction of Cis-Acting Elements in the BnBGLU Promoter Regions
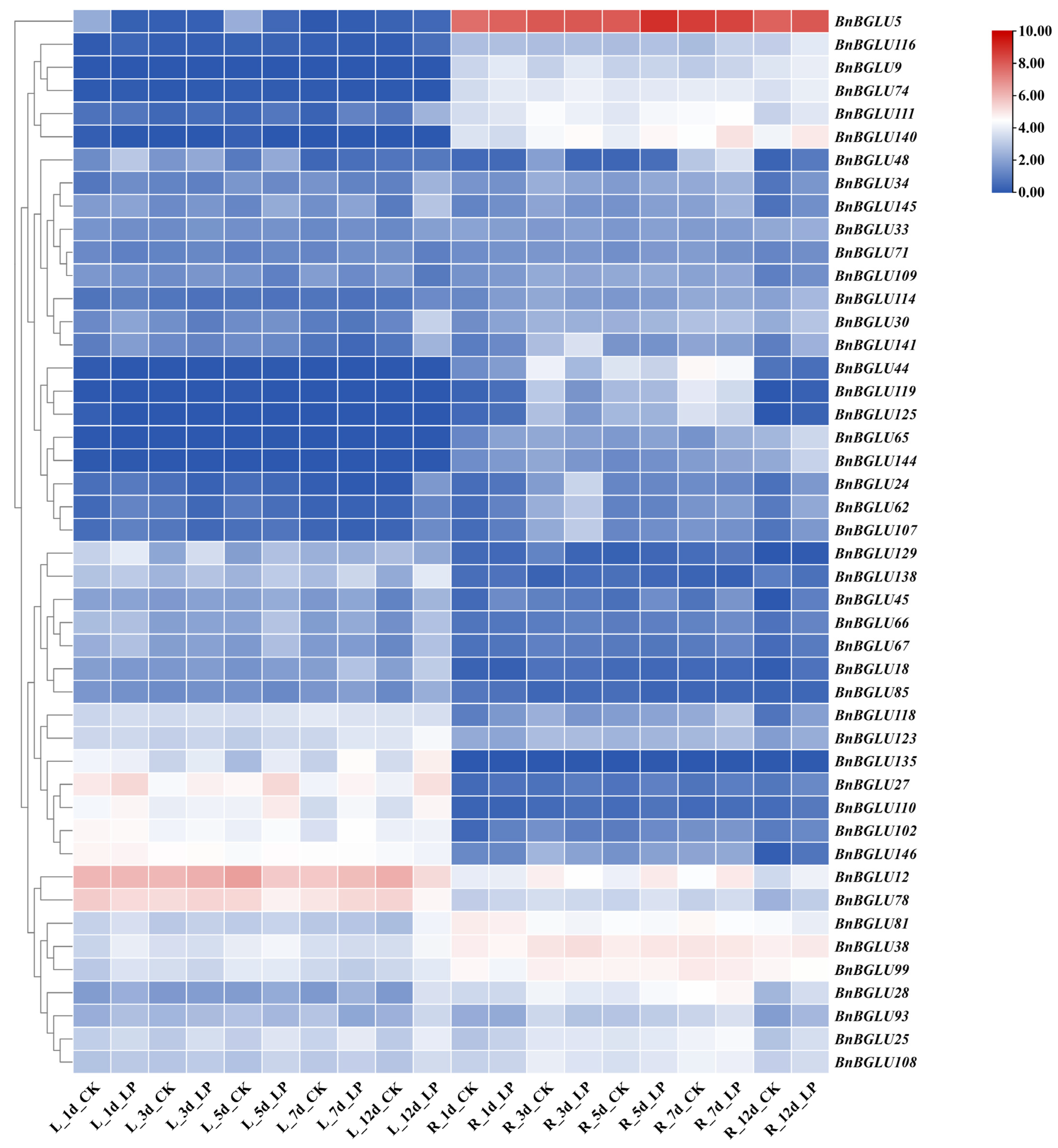
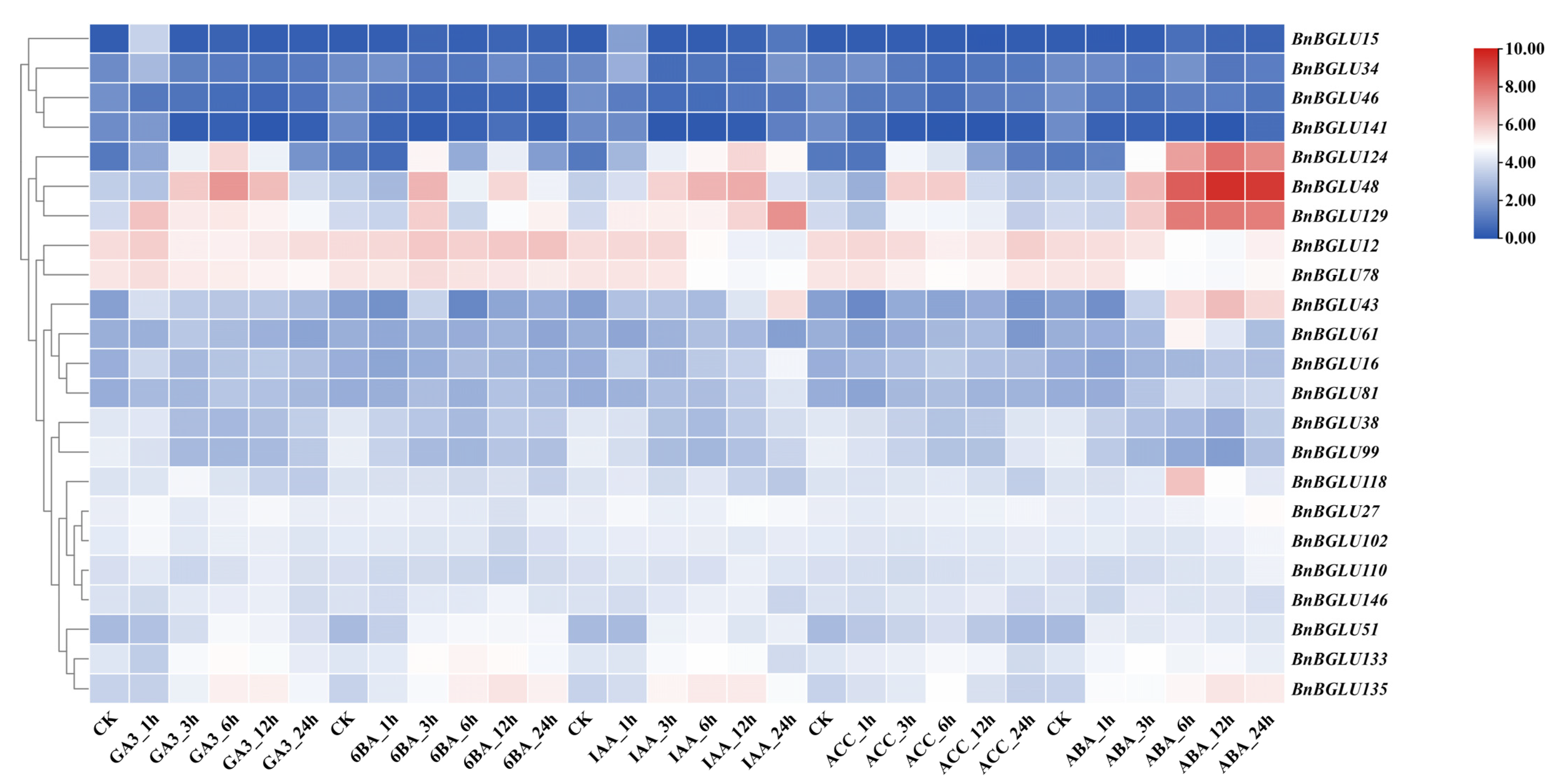
2.7. Expression Analysis of BnBGLU Genes in B. napus Under Phosphorus and Phytohormone Treatment
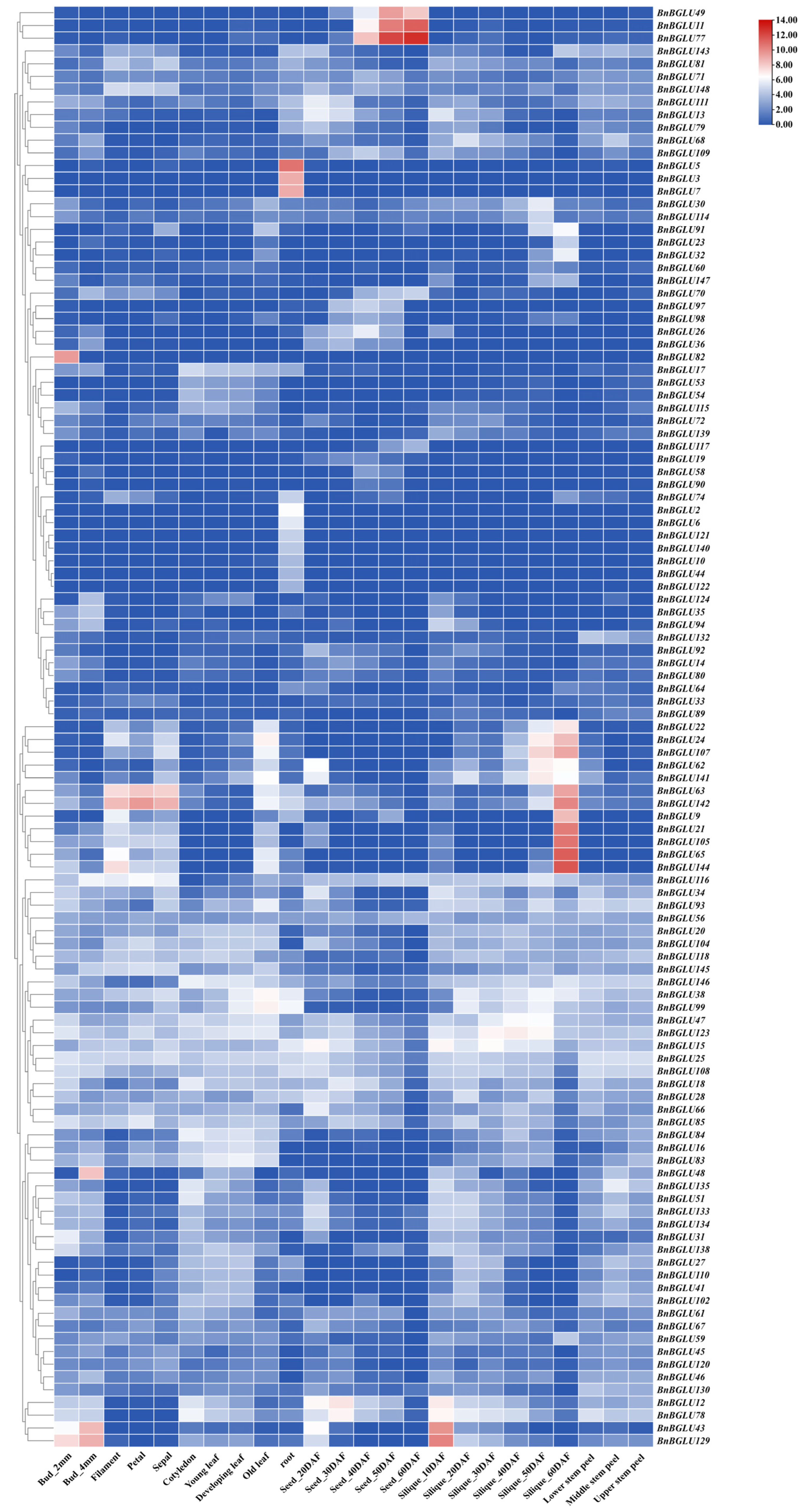
2.8. Tissue Expression Analysis of BnBGLU Genes
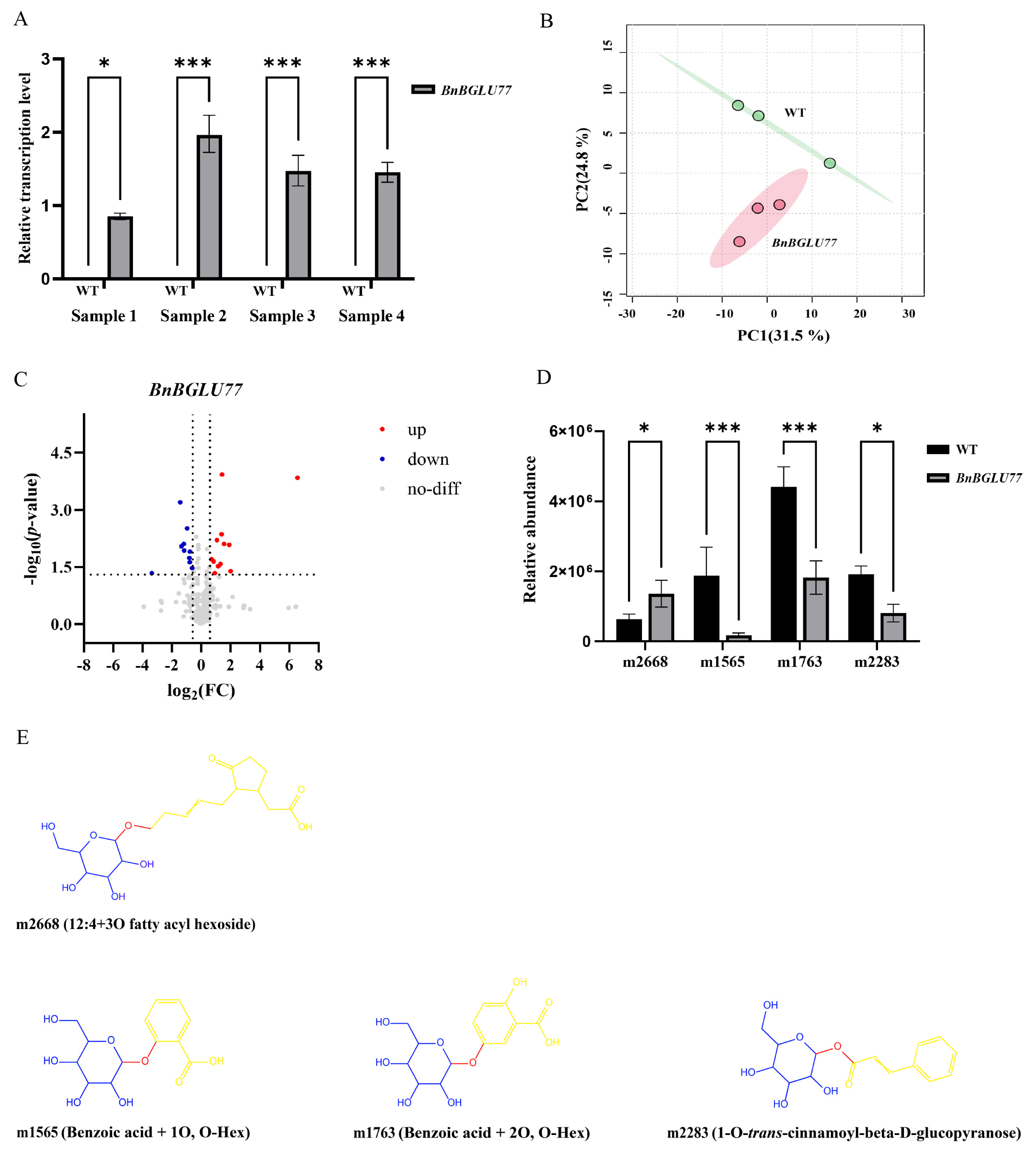
2.9. The Effect of Transient Expression of BnBGLU77 on Metabolites in N. benthamiana (Nicotiana benthamiana)
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of BnBGLU Genes
4.2. Phylogenetic Analysis of the BGLU Family Members
4.3. Conserved Motif Identification and Gene Structure Analysis of the BnBGLU Family
4.4. Analysis of the Chromosomal Localization of BnBGLU Genes
4.5. Analysis of the Colinearity of BnBGLU Genes
4.6. Cis-Regulatory Element Analysis in the Promoter Region of BnBGLU Genes
4.7. Expression Analysis of BnBGLU Genes
4.8. Transient Transformation of BnBGLU77 in N. benthamiana and qRT-PCR Analysis
4.9. Untargeted Metabolomics Analysis of BnBGLU77
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mareri, L.; Parrotta, L.; Cai, G. Environmental Stress and Plants. Int. J. Mol. Sci. 2022, 23, 5416. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Nawaz, M.; Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A Review of Plants Strategies to Resist Biotic and Abiotic Environmental Stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Complex Plant Responses to Drought and Heat Stress under Climate Change. Plant J. Cell Mol. Biol. 2024, 117, 1873–1892. [Google Scholar] [CrossRef]
- Yang, C.; Tian, F.; Ma, J.; Chen, M.; Shi, X.; Chen, D.; Xie, Y.; Zhou, X.; Zhou, Z.; Dai, X.; et al. Glycosylation of Secondary Metabolites: A Multifunctional UDP-Glycosyltransferase, CsUGT74Y1, Promotes the Growth of Plants. J. Agric. Food. Chem. 2023, 71, 18999–19009. [Google Scholar] [CrossRef]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant Secondary Metabolism Linked Glycosyltransferases: An Update on Expanding Knowledge and Scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef]
- Wang, X. Structure, Mechanism and Engineering of Plant Natural Product Glycosyltransferases. FEBS Lett. 2009, 583, 3303–3309. [Google Scholar] [CrossRef]
- Ponnu, J.; Wahl, V.; Schmid, M. Trehalose-6-Phosphate: Connecting Plant Metabolism and Development. Front. Plant Sci. 2011, 2, 70. [Google Scholar] [CrossRef]
- Lu, M.; Zhao, Y.; Feng, Y.; Tang, X.; Zhao, W.; Yu, K.; Pan, Y.; Wang, Q.; Cui, J.; Zhang, M.; et al. 2,4-dihydroxybenzoic Acid, a Novel SA Derivative, Controls Plant Immunity via UGT95B17-mediated Glucosylation: A Case Study in Camellia sinensis. Adv. Sci. 2023, 11, 2307051. [Google Scholar] [CrossRef] [PubMed]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, S.; Ma, X.; Zheng, X.; Liu, Y.; Zhu, Q.; Luo, X.; Cui, J.; Song, C. Glycoside-Specific Metabolomics Reveals the Novel Mechanism of Glycinebetaine-Induced Cold Tolerance by Regulating Apigenin Glycosylation in Tea Plants. New Phytol. 2025, 245, 2616–2631. [Google Scholar] [CrossRef]
- Chandrasekar, B.; Colby, T.; Emran Khan Emon, A.; Jiang, J.; Hong, T.N.; Villamor, J.G.; Harzen, A.; Overkleeft, H.S.; van der Hoorn, R.A.L. Broad-Range Glycosidase Activity Profiling. Mol. Cell. Proteom 2014, 13, 2787–2800. [Google Scholar] [CrossRef]
- Kong, H.; Song, J.; Ma, S.; Yang, J.; Shao, Z.; Li, Q.; Li, Z.; Xie, Z.; Yang, P.; Cao, Y. Genome-Wide Identification and Expression Analysis of the Glycosyl Hydrolase Family 1 Genes in Medicago Sativa Revealed Their Potential Roles in Response to Multiple Abiotic Stresses. BMC Genom. 2024, 25, 20. [Google Scholar] [CrossRef]
- Chuenchor, W.; Pengthaisong, S.; Robinson, R.C.; Yuvaniyama, J.; Oonanant, W.; Bevan, D.R.; Esen, A.; Chen, C.-J.; Opassiri, R.; Svasti, J.; et al. Structural Insights into Rice BGlu1 Beta-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation. J. Mol. Biol. 2008, 377, 1200–1215. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.A.; Vishwakarma, R.A.; Ashraf, N. Functional Characterization of CsBGlu12, a β-Glucosidase from Crocus sativus, Provides Insights into Its Role in Abiotic Stress through Accumulation of Antioxidant Flavonols. J. Biol. Chem. 2017, 292, 4700–4713. [Google Scholar] [CrossRef]
- Ouyang, B.; Wang, G.; Zhang, N.; Zuo, J.; Huang, Y.; Zhao, X. Recent Advances in β-Glucosidase Sequence and Structure Engineering: A Brief Review. Molecules 2023, 28, 4990. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jin, X.; Zhu, L.; Lu, Y.; Ma, Z.; Liu, S.; Chen, X. Glycosyl Hydrolase Catalyzed Glycosylation in Unconventional Media. Appl. Microbiol. Biotechnol. 2020, 104, 9523–9534. [Google Scholar] [CrossRef]
- Godse, R.; Bawane, H.; Tripathi, J.; Kulkarni, R. Unconventional β-Glucosidases: A Promising Biocatalyst for Industrial Biotechnology. Appl. Biochem. Biotechnol. 2021, 193, 2993–3016. [Google Scholar] [CrossRef]
- Bednarek, P.; Pislewska-Bednarek, M.; Svatos, A.; Schneider, B.; Doubsky, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. A Glucosinolate Metabolism Pathway in Living Plant Cells Mediates Broad-Spectrum Antifungal Defense. Science 2009, 323, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Hanson, J.; Pieterse, C.M.J. β-Glucosidase BGLU42 Is a MYB72-Dependent Key Regulator of Rhizobacteria-Induced Systemic Resistance and Modulates Iron Deficiency Responses in Arabidopsis Roots. New Phytol. 2014, 204, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Li, D.; Zhen, C.; Chen, D.; Chen, X. Specific Roles of Os4BGlu10, Os6BGlu24, and Os9BGlu33 in Seed Germination, Root Elongation, and Drought Tolerance in Rice. Planta 2019, 249, 1851–1861. [Google Scholar] [CrossRef]
- Yang, H.; Yao, X.; Wu, W.; He, A.; Ma, C.; Yang, S.; Ruan, J. Genome-Wide Identification and Gene Expression Pattern Analysis of the Glycoside Hydrolase Family 1 in Fagopyrum tataricum. BMC Plant Biol. 2024, 24, 1183. [Google Scholar] [CrossRef]
- Frommann, J.-F.; Pucker, B.; Sielmann, L.M.; Müller, C.; Weisshaar, B.; Stracke, R.; Schweiger, R. Metabolic Fingerprinting Reveals Roles of Arabidopsis thaliana BGLU1, BGLU3, and BGLU4 in Glycosylation of Various Flavonoids. Phytochemistry 2025, 231, 114338. [Google Scholar] [CrossRef]
- Wu, J.; Lv, S.; Zhao, L.; Gao, T.; Yu, C.; Hu, J.; Ma, F. Advances in the Study of the Function and Mechanism of the Action of Flavonoids in Plants under Environmental Stresses. Planta 2023, 257, 108. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D.; Hahn, M.G. Cell Wall Carbohydrates as Signals in Plants. Semin. Cell. Biol. 1993, 4, 93–102. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Lu, H.-Q.; Jiang, K.-X.; Wang, Y.-R.; Wang, Y.-P.; Jiang, J.-J. The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review. Int. J. Mol. Sci. 2022, 24, 357. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Anduro, G.; Ceniceros-Ojeda, E.A.; Casados-Vázquez, L.E.; Bencivenni, C.; Sierra-Beltrán, A.; Murillo-Amador, B.; Tiessen, A. Genome-Wide Analysis of the Beta-Glucosidase Gene Family in Maize (Zea mays L. Var B73). Plant Mol. Biol. 2011, 77, 159–183. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Feng, X.; Peng, F.; Mazoor, M.A.; Zhang, Y.; Zhao, Y.; Han, W.; Lu, J.; Cao, Y.; et al. Analysis of the β-Glucosidase Family Reveals Genes Involved in the Lignification of Stone Cells in Chinese White Pear (Pyrus bretschneideri Rehd.). Front. Plant Sci. 2022, 13, 852001. [Google Scholar] [CrossRef]
- Thorlby, G.; Fourrier, N.; Warren, G. The SENSITIVE TO FREEZING2 Gene, Required for Freezing Tolerance in Arabidopsis thaliana, Encodes a Beta-Glucosidase. Plant Cell 2004, 16, 2192–2203. [Google Scholar] [CrossRef]
- Ishihara, H.; Tohge, T.; Viehöver, P.; Fernie, A.R.; Weisshaar, B.; Stracke, R. Natural Variation in Flavonol Accumulation in Arabidopsis Is Determined by the Flavonol Glucosyltransferase BGLU6. J. Exp. Bot. 2016, 67, 1505–1517. [Google Scholar] [CrossRef]
- Escamilla-Treviño, L.L.; Chen, W.; Card, M.L.; Shih, M.-C.; Cheng, C.-L.; Poulton, J.E. Arabidopsis thaliana Beta-Glucosidases BGLU45 and BGLU46 Hydrolyse Monolignol Glucosides. Phytochemistry 2006, 67, 1651–1660. [Google Scholar] [CrossRef]
- Chapelle, A.; Morreel, K.; Vanholme, R.; Le-Bris, P.; Morin, H.; Lapierre, C.; Boerjan, W.; Jouanin, L.; Demont-Caulet, N. Impact of the Absence of Stem-Specific β-Glucosidases on Lignin and Monolignols. Plant Physiol. 2012, 160, 1204–1217. [Google Scholar] [CrossRef]
- Xu, Z.; Escamilla-Treviño, L.; Zeng, L.; Lalgondar, M.; Bevan, D.; Winkel, B.; Mohamed, A.; Cheng, C.-L.; Shih, M.-C.; Poulton, J.; et al. Functional Genomic Analysis of Arabidopsis thaliana Glycoside Hydrolase Family 1. Plant Mol. Biol. 2004, 55, 343–367. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, W.P.; Cottaz, S.; Driguez, H.; Iori, R.; Palmieri, S.; Henrissat, B. The Crystal Structures of Sinapis alba Myrosinase and a Covalent Glycosyl-Enzyme Intermediate Provide Insights into the Substrate Recognition and Active-Site Machinery of an S-Glycosidase. Structure 1997, 5, 663–675. [Google Scholar] [CrossRef]
- Qiao, X.; Yin, H.; Li, L.; Wang, R.; Wu, J.; Wu, J.; Zhang, S. Different Modes of Gene Duplication Show Divergent Evolutionary Patterns and Contribute Differently to the Expansion of Gene Families Involved in Important Fruit Traits in Pear (Pyrus bretschneideri). Front. Plant Sci. 2018, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants1[OPEN]. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Roth, C.; Liberles, D.A. A Systematic Search for Positive Selection in Higher Plants (Embryophytes). BMC Plant Biol. 2006, 6, 12. [Google Scholar] [CrossRef]
- Brunet, T.D.P.; Doolittle, W.F.; Bielawski, J.P. The Role of Purifying Selection in the Origin and Maintenance of Complex Function. Stud. Hist. Philos. Sci. 2021, 87, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. TIG 2002, 18, 486. [Google Scholar] [CrossRef]
- Halder, K.; Chaudhuri, A.; Abdin, M.Z.; Majee, M.; Datta, A. Chromatin-Based Transcriptional Reprogramming in Plants under Abiotic Stresses. Plants 2022, 11, 1449. [Google Scholar] [CrossRef]
- Richardson, A. Plant Development: Coordinating across Space and Time. Curr. Biol. CB 2020, 30, R1492–R1494. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.F.; Chai, R.S.; Jin, G.L.; Wang, H.; Tang, C.X.; Zhang, Y.S. Responses of Root Architecture Development to Low Phosphorus Availability: A Review. Ann. Bot. 2013, 112, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Malboobi, M.A.; Lefebvre, D.D. A Phosphate-Starvation Inducible Beta-Glucosidase Gene (Psr3.2) Isolated from Arabidopsis thaliana Is a Member of a Distinct Subfamily of the BGA Family. Plant Mol. Biol. 1997, 34, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.P.; Bennett, M.J.; Bowen, H.C.; Broadley, M.R.; Eastwood, D.C.; May, S.T.; Rahn, C.; Swarup, R.; Woolaway, K.E.; White, P.J. Changes in Gene Expression in Arabidopsis Shoots during Phosphate Starvation and the Potential for Developing Smart Plants. Plant Physiol. 2003, 132, 578–596. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, L.; Wang, F.; Deng, M.; Yi, K. Modulating the Root Elongation by Phosphate/Nitrogen Starvation in an OsGLU3 Dependant Way in Rice. Plant Signal. Behav. 2012, 7, 1144–1145. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Romero-Téllez, S.; Lluch, J.M.; González-Lafont, À.; Masgrau, L. Comparing Hydrolysis and Transglycosylation Reactions Catalyzed by Thermus thermophilus β-Glycosidase. A Combined MD and QM/MM Study. Front. Chem. 2019, 7, 200. [Google Scholar] [CrossRef]
- Grellet Bournonville, C.; Filippone, M.P.; Di Peto, P.d.L.Á.; Trejo, M.F.; Couto, A.S.; Mamaní de Marchese, A.; Díaz Ricci, J.C.; Welin, B.; Castagnaro, A.P. Strawberry Fatty Acyl Glycosides Enhance Disease Protection, Have Antibiotic Activity and Stimulate Plant Growth. Sci. Rep. 2020, 10, 8196. [Google Scholar] [CrossRef]
- Mutschler, M.A.; Kennedy, G.G.; Ullman, D.E. Acylsugar-Mediated Resistance as Part of a Multilayered Defense against Thrips, Orthotospoviruses, and Beyond. Curr. Opin. Insect Sci. 2023, 56, 101021. [Google Scholar] [CrossRef]
- Chen, J.; Clinton, M.; Qi, G.; Wang, D.; Liu, F.; Fu, Z.Q. Reprogramming and Remodeling: Transcriptional and Epigenetic Regulation of Salicylic Acid-Mediated Plant Defense. J. Exp. Bot. 2020, 71, 5256–5268. [Google Scholar] [CrossRef]
- Koo, Y.M.; Heo, A.Y.; Choi, H.W. Salicylic Acid as a Safe Plant Protector and Growth Regulator. Plant Pathol. J. 2020, 36, 1–10. [Google Scholar] [CrossRef]
- Bellés, J.M.; Garro, R.; Pallás, V.; Fayos, J.; Rodrigo, I.; Conejero, V. Accumulation of Gentisic Acid as Associated with Systemic Infections but Not with the Hypersensitive Response in Plant-Pathogen Interactions. Planta 2006, 223, 500–511. [Google Scholar] [CrossRef]
- Fayos, J.; Bellés, J.M.; López-Gresa, M.P.; Primo, J.; Conejero, V. Induction of Gentisic Acid 5-O-Beta-D-Xylopyranoside in Tomato and Cucumber Plants Infected by Different Pathogens. Phytochemistry 2006, 67, 142–148. [Google Scholar] [CrossRef]
- López-González, D.; Bruno, L.; Díaz-Tielas, C.; Lupini, A.; Aci, M.M.; Talarico, E.; Madeo, M.L.; Muto, A.; Sánchez-Moreiras, A.M.; Araniti, F. Short-Term Effects of Trans-Cinnamic Acid on the Metabolism of Zea mays L. Roots. Plants 2023, 12, 189. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Robles, A.; Cala Peralta, A.; Zorrilla, J.G.; Soriano, G.; Masi, M.; Vilariño-Rodríguez, S.; Cimmino, A.; Fernández-Aparicio, M. Structure-Activity Relationship (SAR) Study of Trans-Cinnamic Acid and Derivatives on the Parasitic Weed Cuscuta campestris. Plants 2023, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, H.; Ding, Z.; Yan, J.; Yu, H.; Pan, R.; Hu, J.; Guan, Y.; Hua, J. Low Temperature Enhances Plant Immunity via Salicylic Acid Pathway Genes That Are Repressed by Ethylene. Plant Physiol. 2020, 182, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Wang, H.; Yang, C.; Wang, L.; Qi, L.; Guo, Z.; Chen, X. Salicylic Acid Is Required for Broad-Spectrum Disease Resistance in Rice. Int. J. Mol. Sci. 2022, 23, 1354. [Google Scholar] [CrossRef] [PubMed]
- Vlaminck, L.; De Rouck, B.; Desmet, S.; Van Gerrewey, T.; Goeminne, G.; De Smet, L.; Storme, V.; Kyndt, T.; Demeestere, K.; Gheysen, G.; et al. Opposing Effects of Trans- and Cis-Cinnamic Acid during Rice Coleoptile Elongation. Plant Direct 2022, 6, e465. [Google Scholar] [CrossRef]
- Remali, J.; Sahidin, I.; Aizat, W.M. Xanthone Biosynthetic Pathway in Plants: A Review. Front. Plant Sci. 2022, 13, 809497. [Google Scholar] [CrossRef]
- Xiang, L.; Etxeberria, E.; Van den Ende, W. Vacuolar Protein Sorting Mechanisms in Plants. FEBS J. 2013, 280, 979–993. [Google Scholar] [CrossRef]
- Vaschetto, L.M.; Ortiz, N. The Role of Sequence Duplication in Transcriptional Regulation and Genome Evolution. Curr. Genom. 2019, 20, 405–408. [Google Scholar] [CrossRef] [PubMed]
- De la Concepcion, J.C.; Vega Benjumea, J.; Bialas, A.; Terauchi, R.; Kamoun, S.; Banfield, M.J. Functional Diversification Gave Rise to Allelic Specialization in a Rice NLR Immune Receptor Pair. eLife 2021, 10, e71662. [Google Scholar] [CrossRef] [PubMed]
- Gelfman, S.; Burstein, D.; Penn, O.; Savchenko, A.; Amit, M.; Schwartz, S.; Pupko, T.; Ast, G. Changes in Exon-Intron Structure during Vertebrate Evolution Affect the Splicing Pattern of Exons. Genome Res. 2012, 22, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Reiser, L.; Subramaniam, S.; Zhang, P.; Berardini, T. Using the Arabidopsis Information Resource (TAIR) to Find Information about Arabidopsis Genes. Curr. Protocol. 2022, 2, e574. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A Multi-Omics Database with Various Tools for Brassica napus Research and Breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A User-Friendly Online Tool for Drawing Genetic Maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Dong, C.; Qu, G.; Guo, J.; Wei, F.; Gao, S.; Sun, Z.; Jin, L.; Sun, X.; Rochaix, J.-D.; Miao, Y.; et al. Rational Design of Geranylgeranyl Diphosphate Synthase Enhances Carotenoid Production and Improves Photosynthetic Efficiency in Nicotiana Tabacum. Sci. Bull. 2022, 67, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhang, L.; Wu, Y.; Cao, Y.; Lu, C. Comparison of Five Endogenous Reference Genes for Specific PCR Detection and Quantification of Brassica napus. J. Agric. Food Chem. 2010, 58, 2812–2817. [Google Scholar] [CrossRef] [PubMed]
- Mitteer, D.R.; Greer, B.D. Using GraphPad Prism’s Heat Maps for Efficient, Fine-Grained Analyses of Single-Case Data. Behav. Anal. Pract. 2022, 15, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Yin, N.; Chen, S.; Wang, S.; Chen, X.; Zhao, H.; Shen, S.; Fu, F.; Zhou, B.; Xu, X.; et al. Comparative Analysis of the Metabolic Profiles of Yellow- versus Black-Seeded Rapeseed Using UPLC-HESI-MS/MS and Transcriptome Analysis. J. Agric. Food Chem. 2020, 68, 3033–3049. [Google Scholar] [CrossRef]
- Shen, S.; Tang, Y.; Liu, D.; Chen, L.; Zhang, Y.; Ye, K.; Sun, F.; Wei, X.; Du, H.; Zhao, H.; et al. Untargeted Metabolomics Analysis Reveals Differential Accumulation of Flavonoids between Yellow-Seeded and Black-Seeded Rapeseed Varieties. Plants 2025, 14, 753. [Google Scholar] [CrossRef]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A Lipidome Atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a Unified Platform for Metabolomics Data Processing, Analysis and Interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Tang, Y.; Liu, Y.; Shen, S.; Xu, J.; Chen, L.; Li, M.; Zhao, H.; Zhang, T.; Du, H.; et al. Genome-Wide Identification of the Glycosyl Hydrolase Family 1 Genes in Brassica napus L. and Functional Characterization of BnBGLU77. Plants 2025, 14, 2686. https://doi.org/10.3390/plants14172686
Wei X, Tang Y, Liu Y, Shen S, Xu J, Chen L, Li M, Zhao H, Zhang T, Du H, et al. Genome-Wide Identification of the Glycosyl Hydrolase Family 1 Genes in Brassica napus L. and Functional Characterization of BnBGLU77. Plants. 2025; 14(17):2686. https://doi.org/10.3390/plants14172686
Chicago/Turabian StyleWei, Xingzhi, Yunshan Tang, Yuanyuan Liu, Shulin Shen, Jie Xu, Lulu Chen, Meifang Li, Huiyan Zhao, Ti Zhang, Hai Du, and et al. 2025. "Genome-Wide Identification of the Glycosyl Hydrolase Family 1 Genes in Brassica napus L. and Functional Characterization of BnBGLU77" Plants 14, no. 17: 2686. https://doi.org/10.3390/plants14172686
APA StyleWei, X., Tang, Y., Liu, Y., Shen, S., Xu, J., Chen, L., Li, M., Zhao, H., Zhang, T., Du, H., Wan, H., Qu, C., & Yin, N. (2025). Genome-Wide Identification of the Glycosyl Hydrolase Family 1 Genes in Brassica napus L. and Functional Characterization of BnBGLU77. Plants, 14(17), 2686. https://doi.org/10.3390/plants14172686








