Brassica-Specific Orphan Gene CROG1 Confers Clubroot Resistance in Arabidopsis via Phenylpropanoid Pathway Activation
Abstract
1. Introduction
2. Results
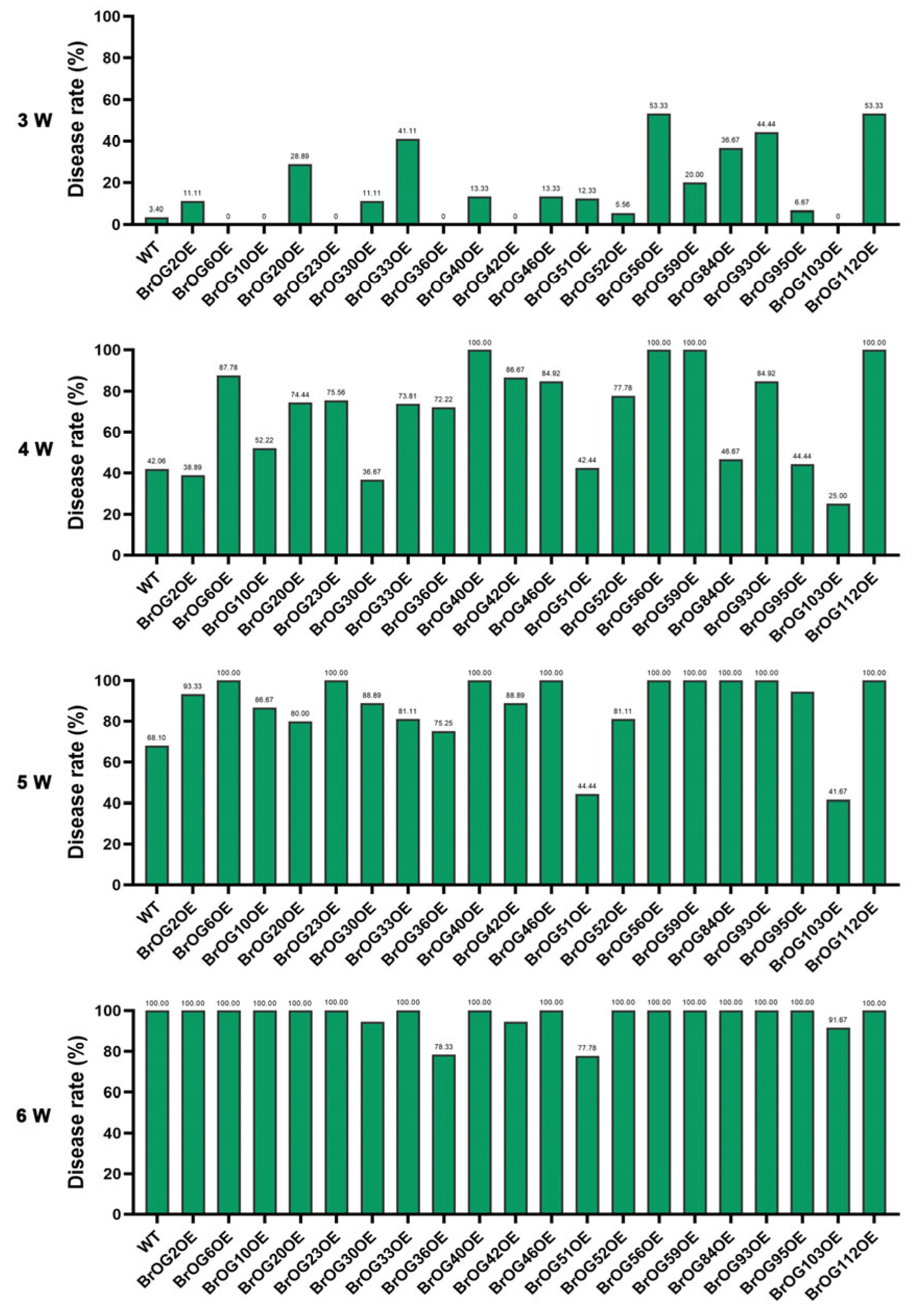
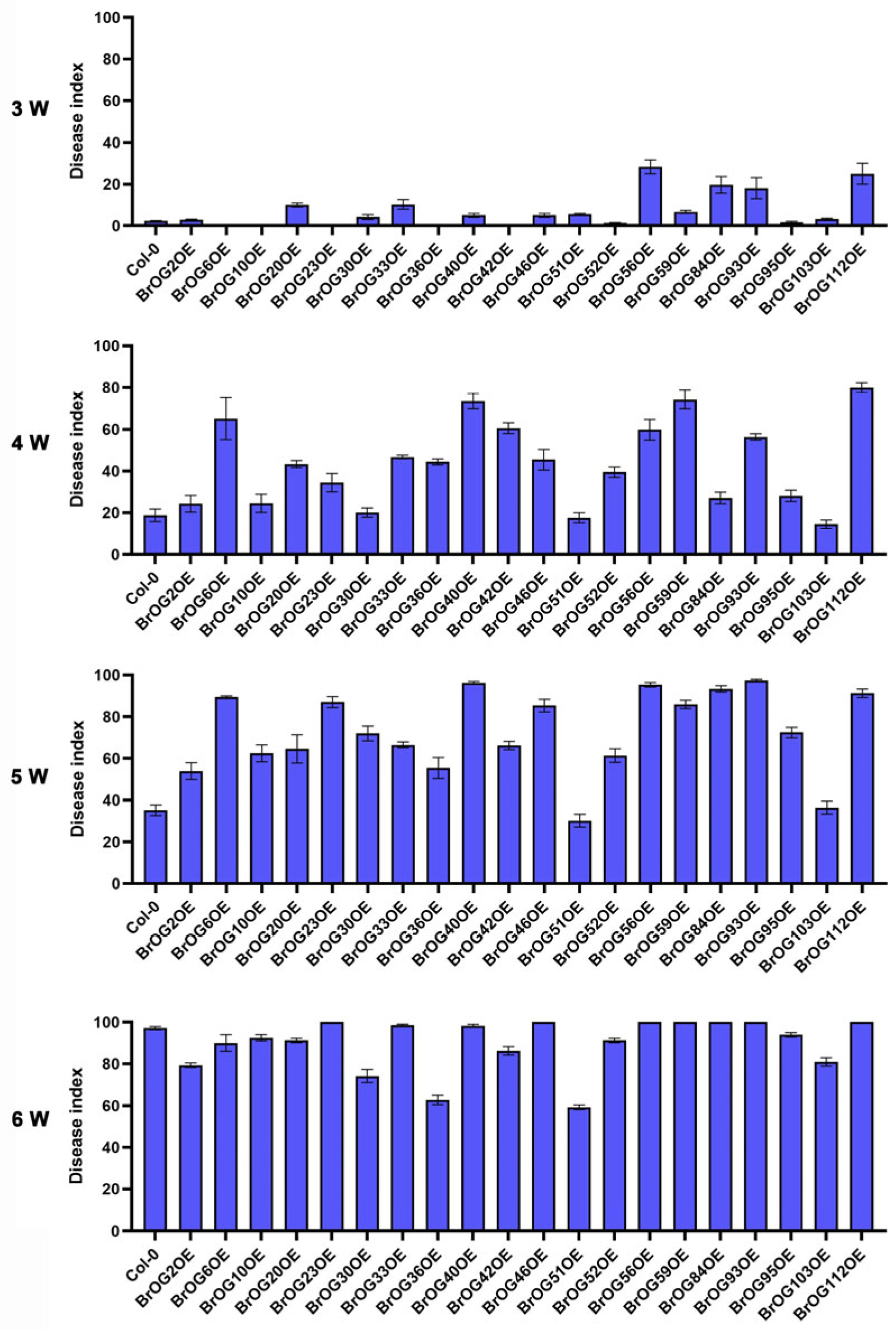
2.1. Characterization of Clubroot Resistance in BrOGs Overexpression Lines
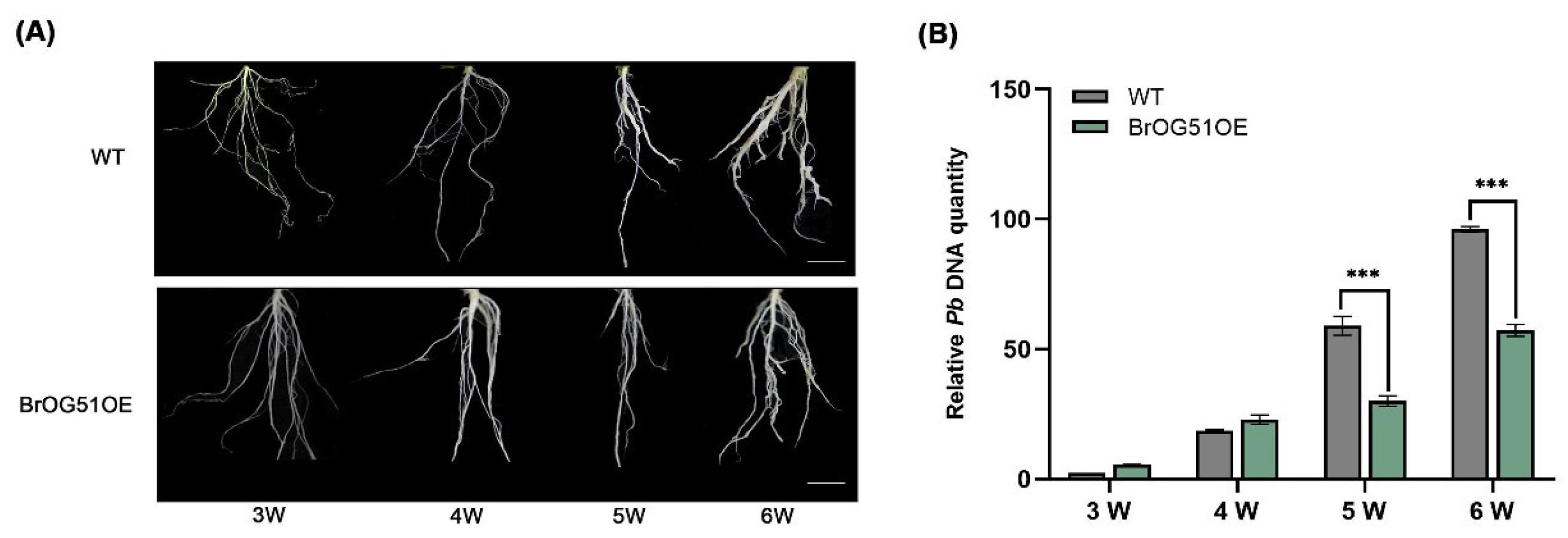
2.2. BrOG51OE Showed Significant Resistance to Both Pseudomonas syringae pv. Tomato DC3000 and P. brassicae Infection
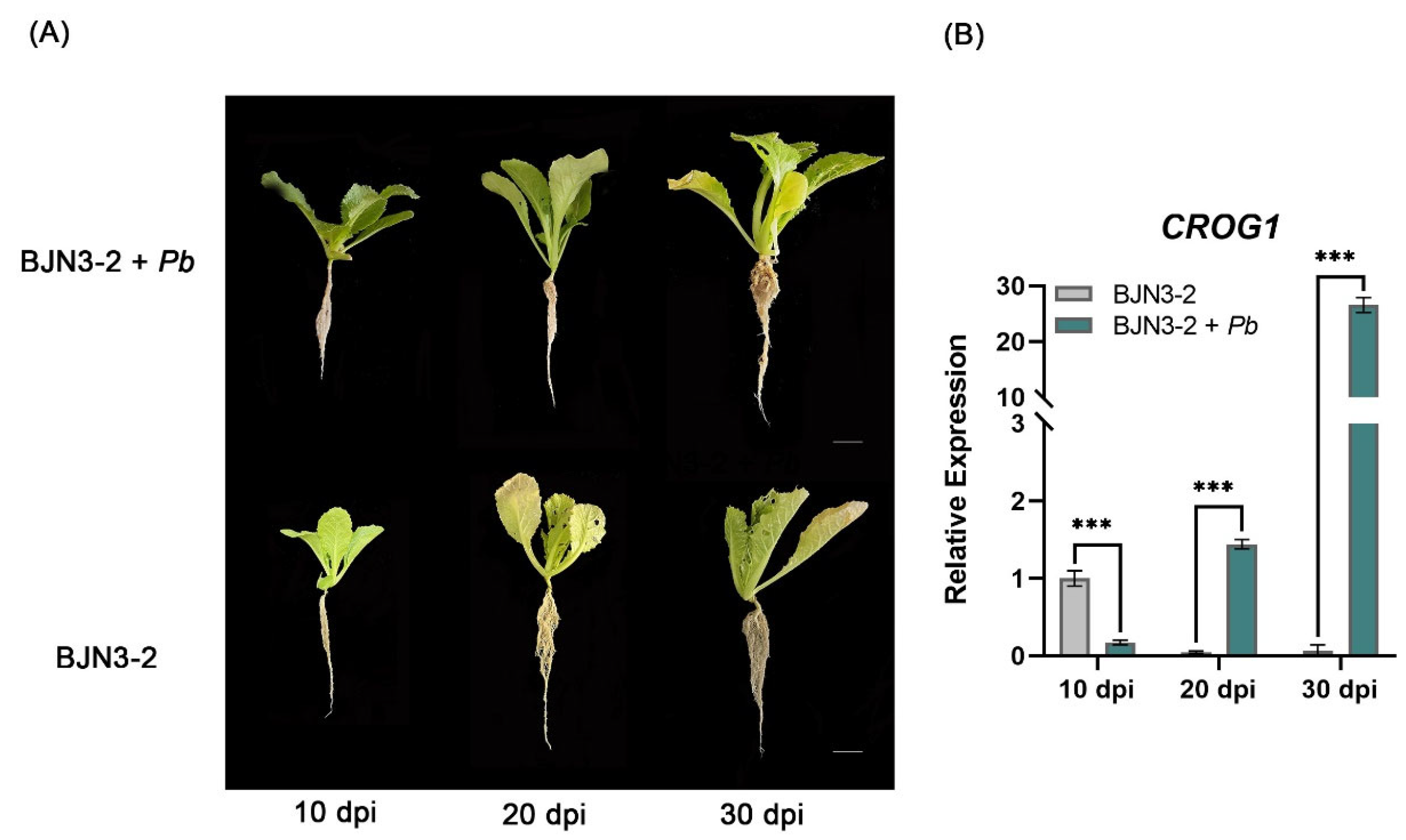
2.3. Analysis of CROG1 Expression in Brassica rapa After Inoculation with P. brassicae
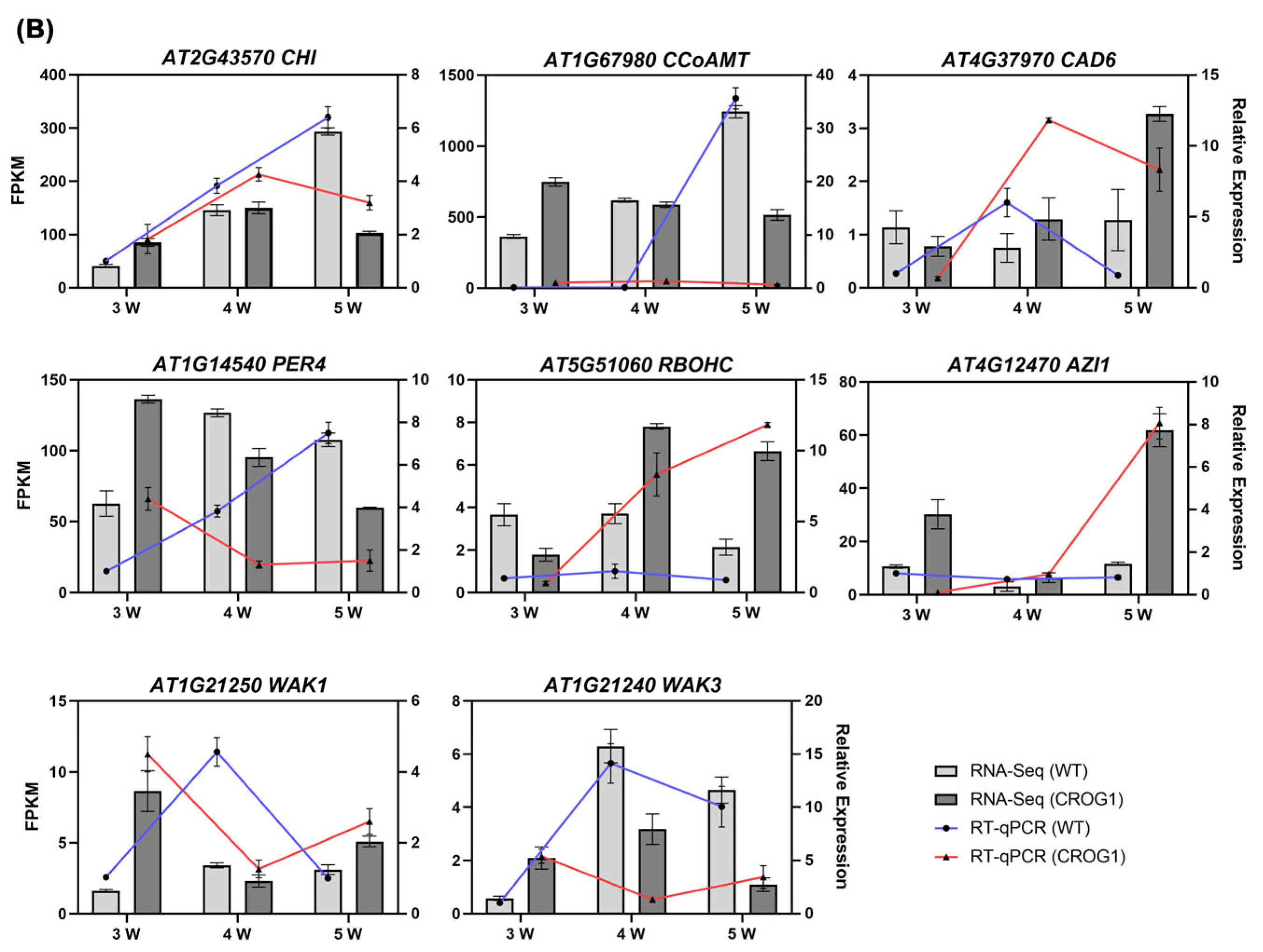
2.4. Transcriptome Analysis of Arabidopsis thaliana CROG1 Transgenic Plants Inoculated with P. brassicae
2.5. CROG1 Is Involved in Clubroot Resistance by Affecting the Phenylpropane Metabolic Pathway
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Plasmodiophora brassicae Inoculation
4.3. Clubroot Disease Assessment
4.4. Inoculation with Pseudomonas syringae pv. Tomato DC3000
4.5. RNA Extraction and RT-qPCR Analysis
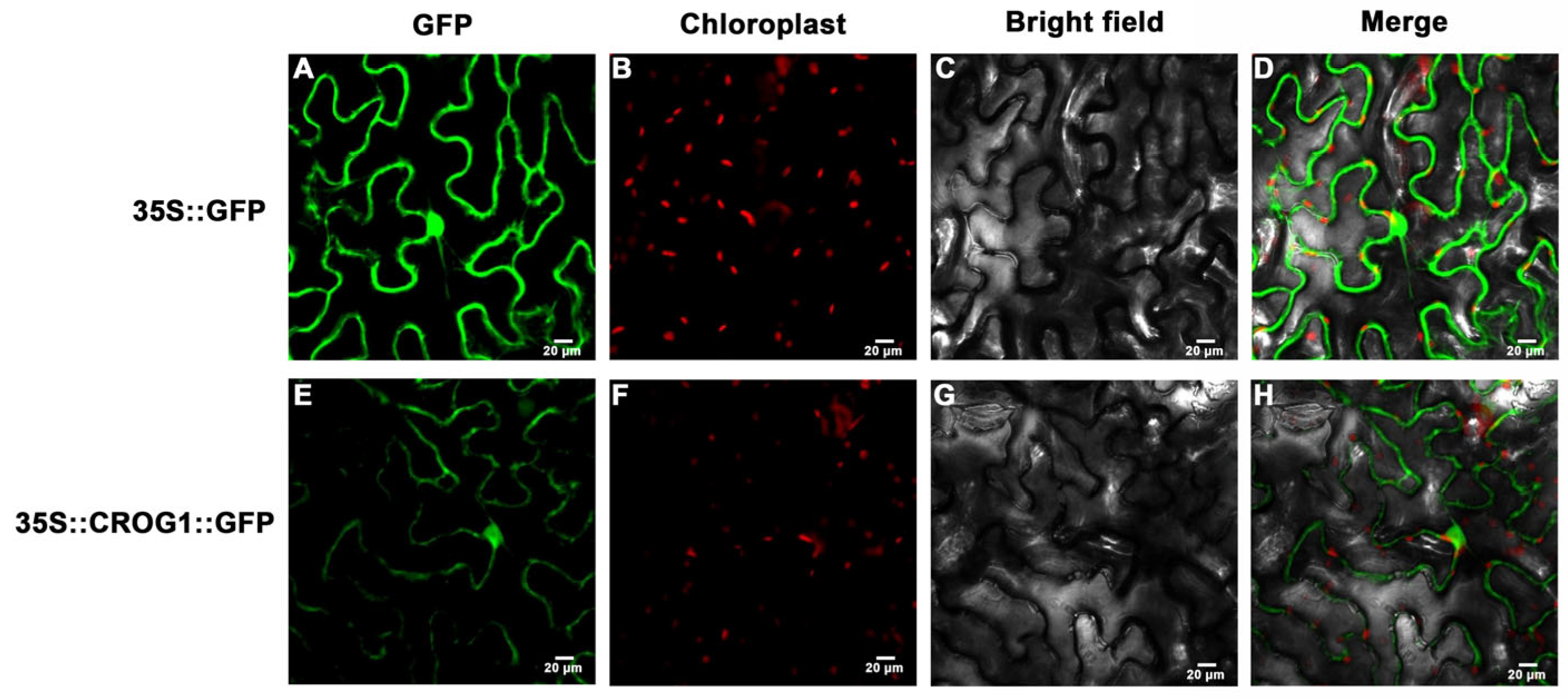
4.6. Subcellular Localization Assays
4.7. Library Construction and RNA-Seq
4.8. RNA-Seq Data Analysis
4.9. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| OGs | Orphan genes |
| CROG1 | Clubroot-related orphan gene 1 |
| CCoAMT | Transcaffeoyl Coenzyme A 3-O-methyltransferase |
| CAD6 | Cinnamyl alcohol dehydrogenase 6 |
| PER4 | PEROXIDASE 4 |
| AZI1 | AZELAIC ACID INDUCED 1 |
| RBOHC | RESPIRATORY BURST OXIDASE HOMOLOG C |
| WAKS | WALL-ASSOCIATED KINASES |
| EGT | Ergothioneine |
| DR | Disease rating |
| DI | Disease index |
| OE | Overexpression |
| dpi | Days post-inoculation |
| wpi | Weeks post-inoculation |
| WT | Wild-type |
| RT-qPCR | Quantitative real-time polymerase chain reaction |
| GO | Gene Ontology |
| DEGs | Differentially expressed genes |
| WGCNA | Weighted correlation network analysis |
| AtQQS | Arabidopsis Qua-Quine Starch |
| FPKM | Fragments per kilobase of transcript per million mapped |
References
- Dixon, G.R. The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Liu, L.J.; Qin, L.; Zhou, Z.Q.; Hendriks, W.G.H.M.; Liu, S.Y.; Wei, Y.D. Refining the life cycle of Plasmodiophora brassicae. Phytopathology 2020, 110, 1704–1712. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Wu, C.Y.; Zhang, F.; Yao, J.; Fan, L.; Liu, Z.R.; Yao, Y.J. Comprehensive review of Plasmodiophora brassicae: Pathogenesis, pathotype diversity, and integrated control methods. Front. Microbiol. 2025, 16, 1531393. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Du, C.Y.; Li, Y.W.; Wang, H.; Zhang, C.T.; Chen, P. Advances in biological control and resistance genes of Brassicaceae clubroot disease-the study case of China. Int. J. Mol. Sci. 2023, 24, 785. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Li, X.; Piao, Z. Multi-omics approaches to improve the studies on the mechanisms of clubroot resistance in Brassica hosts. Acta Hortic. Sin. 2025, 52, 1213–1232. [Google Scholar] [CrossRef]
- Ueno, H.; Matsumoto, E.; Aruga, D.; Kitagawa, S.; Matsumura, H.; Hayashida, N. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 2012, 80, 621–629. [Google Scholar] [CrossRef]
- Hatakeyama, K.; Suwabe, K.; Tomita, R.N.; Kato, T.; Nunome, T.; Fukuoka, H.; Matsumoto, S. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS ONE 2013, 8, e54745. [Google Scholar] [CrossRef]
- Voigt, C.A. Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci. 2014, 5, 168. [Google Scholar] [CrossRef]
- Zhao, Q.; Dixon, R.A. Altering the cell wall and its impact on plant disease: From forage to bioenergy. Annu. Rev. Phytopathol. 2014, 52, 69–91. [Google Scholar] [CrossRef]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef]
- Bradley, D.J.; Kjellbom, P.; Lamb, C.J. Elicitor-induced and wound-induced oxidative cross-linking of a proline-rich plant-cell wall protein—A novel, rapid defense response. Cell 1992, 70, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Schwelm, A.; Fogelqvist, J.; Knaust, A.; Jülke, S.; Lilja, T.; Rosso, G.B.; Karlsson, M.; Shevchenko, A.; Dhandapani, V.; Choi, S.R.; et al. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep. 2015, 5, 11153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q. Lignification: Flexibility, Biosynthesis and Regulation. Trends Plant Sci. 2016, 21, 713–721. [Google Scholar] [CrossRef]
- Nakayama, T.; Takahashi, S.; Waki, T. Formation of flavonoid metabolons: Functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front. Plant Sci. 2019, 10, 821. [Google Scholar] [CrossRef]
- Tu, J.; Qin, L.; Karunakaran, C.; Wei, Y.; Peng, G. Lignin accumulation in cell wall plays a role in clubroot resistance. Front. Plant Sci. 2024, 15, 1401265. [Google Scholar] [CrossRef]
- Zhao, Y.; Bi, K.; Gao, Z.; Chen, T.; Liu, H.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D. Transcriptome Analysis of Arabidopsis thaliana in Response to Plasmodiophora brassicae during Early Infection. Front. Microbiol. 2017, 8, 673. [Google Scholar] [CrossRef]
- Irani, S.; Todd, C.D.; Wei, Y.D.; Bonham-Smith, P.C. Changes in phenylpropanoid pathway gene expression in roots and leaves of susceptible and resistant Brassica napus lines in response to Plasmodiophora brassicae inoculation. Physiol. Mol. Plant P. 2019, 106, 196–203. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, G.; Li, X.; Piao, Z. Effects of Exogenous Ergothioneine on Brassica rapa Clubroot Development Revealed by Transcriptomic Analysis. Int. J. Mol. Sci. 2023, 24, 6380. [Google Scholar] [CrossRef]
- Chu, M.; Song, T.; Falk, K.C.; Zhang, X.; Liu, X.; Chang, A.; Lahlali, R.; McGregor, L.; Gossen, B.D.; Yu, F.; et al. Fine mapping of Rcr1 and analyses ofits effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 2014, 15, 1166. [Google Scholar] [CrossRef]
- Chen, J.J.; Pang, W.X.; Chen, B.; Zhang, C.Y.; Piao, Z.Y. Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and-susceptible alleles in response to Plasmodiophora brassicae during early infection. Front. Plant Sci. 2016, 6, 1183. [Google Scholar] [CrossRef]
- Irani, S.; Trost, B.; Waldner, M.; Nayidu, N.; Tu, J.; Kusalik, A.J.; Todd, C.D.; Wei, Y.; Bonham-Smith, P.C. Transcriptome analysis of response to Plasmodiophora brassicae infection in the Arabidopsis shoot and root. BMC Genom. 2018, 19, 23. [Google Scholar] [CrossRef]
- Ciaghi, S.; Schwelm, A.; Neuhauser, S. Transcriptomic response in symptomless roots of clubroot infected kohlrabi (Brassica oleracea var. gongylodes) mirrors resistant plants. BMC Plant Biol. 2019, 19, 288. [Google Scholar] [CrossRef]
- Wen, R.; Song, T.; Gossen, B.D.; Peng, G. Comparative transcriptome analysis of canola carrying a single vs stacked resistance genes against clubroot. Front. Plant Sci. 2024, 15, 1358605. [Google Scholar] [CrossRef]
- Li, L.; Long, Y.; Li, H.; Wu, X. Comparative transcriptome analysis reveals key pathways and hub genes in rapeseed during the early stage of Plasmodiophora brassicae infection. Front. Genet. 2020, 10, 1275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, M.; Ma, J.; Chen, J.; Kong, L.; Zhan, Z.; Li, X.; Piao, Z. Metabolomics and transcriptomics reveal the function of trigonelline and its synthesis gene BrNANMT in clubroot susceptibility of Brassica rapa. Plant Cell Environ. 2025. [Google Scholar] [CrossRef] [PubMed]
- Bhandary, P.; Seetharam, A.S.; Arendsee, Z.W.; Hur, M.; Wurtele, E.S. Raising orphans from a metadata morass: A researcher’s guide to reuse of public’ omics data. Plant Sci. 2018, 267, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Guo, B.; He, S. The roles and evolutionary patterns of intronless genes in deuterostomes. Comp. Funct. Genomics 2011, 2011, 680673. [Google Scholar] [CrossRef]
- Jiang, M.L.; Dong, X.S.; Lang, H.; Pang, W.X.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Mining of Brassica-Specific Genes (BSGs) and Their Induction in Different Developmental Stages and under Plasmodiophora brassicae Stress in Brassica rapa. Int. J. Mol. Sci. 2018, 19, 2064. [Google Scholar] [CrossRef]
- Jiang, M.L.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Construction and evaluation of Brassica rapa orphan genes overexpression library. Front. Plant Sci. 2025, 16, 1532449. [Google Scholar] [CrossRef]
- Barros, J.; Escamilla-Trevino, L.; Song, L.; Rao, X.; Serrani-Yarce, J.C.; Palacios, M.D.; Engle, N.; Choudhury, F.K.; Tschaplinski, T.J.; Venables, B.J.; et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019, 10, 1994. [Google Scholar] [CrossRef]
- Gao, H.; Wang, X.-Y.; Han, Y.-Y.; Du, G.-L.; Feng, H.; Xu, Z.-Q. Accumulation of the azelaic acid-induced protein AZI1 affects lignin synthesis and deposition in Arabidopsis thaliana. Plant Growth Regul. 2015, 75, 317–330. [Google Scholar] [CrossRef]
- Jiang, M.L.; Li, X.N.; Dong, X.S.; Zu, Y.; Zhan, Z.X.; Piao, Z.Y.; Lang, H. Research Advances and Prospects of Orphan Genes in Plants. Front. Plant Sci. 2022, 13, 947129. [Google Scholar] [CrossRef] [PubMed]
- Tanvir, R.; Ping, W.; Sun, J.; Cain, M.; Li, X.; Li, L. AtQQS orphan gene and NtNF-YC4 boost protein accumulation and pest resistance in tobacco (Nicotiana tabacum). Plant Sci. 2022, 317, 111198. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, Y.; Yang, X.; Li, X.; Lang, H. Brassica rapa orphan gene BR1 delays flowering time in Arabidopsis. Front. Plant Sci. 2023, 14, 1135684. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Jiang, M.; Zhan, Z.; Li, X.; Piao, Z. Orphan gene BR2 positively regulates bolting resistance through the vernalization pathway in Chinese cabbage. Hortic. Res. 2024, 11, uhae216. [Google Scholar] [CrossRef]
- Liao, R.; Zhang, R.; Li, X.; Jiang, M. Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay. Plants 2025, 14, 1947. [Google Scholar] [CrossRef]
- Lang, H.; Zhang, Y.; Zhao, S.; Li, K.; Li, X.; Jiang, M. Temporal transcriptome analysis reveals core pathways and orphan gene EARLY FLOWERING 1 regulating floral transition in Chinese cabbage. Plants 2025, 14, 2236. [Google Scholar] [CrossRef]
- Yadeta, K.A.; Valkenburg, D.J.; Hanemian, M.; Marco, Y.; Thomma, B.P. The brassicaceae-specific EWR1 gene provides resistance to vascular wilt pathogens. PLoS ONE 2014, 9, e88230. [Google Scholar] [CrossRef]
- Lahlali, R.; Song, T.; Chu, M.; Yu, F.; Kumar, S.; Karunakaran, C.; Peng, G. Evaluating changes in cell-wall components associated with clubroot resistance using fourier transform infrared spectroscopy and RT-PCR. Int. J. Mol. Sci. 2017, 18, 2058. [Google Scholar] [CrossRef]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef]
- Wan, J.; He, M.; Hou, Q.; Zou, L.; Yang, Y.; Wei, Y.; Chen, X. Cell wall associated immunity in plants. Stress Biol. 2021, 1, 3. [Google Scholar] [CrossRef]
- Cai, J.J.; Woo, P.C.; Lau, S.K.; Smith, D.K.; Yuen, K.Y. Accelerated evolutionary rate may be responsible for the emergence oflineage-specific genes in ascomycota. J. Mol. Evol. 2006, 63, 1–11. [Google Scholar] [CrossRef]
- Mahalak, K.K.; Chamberlin, H.M. Orphan genes find a home: Interspecific competition and gene network evolution. PLoS Genet. 2015, 11, e1005254. [Google Scholar] [CrossRef]
- Schlötterer, C. Genes from scratch-the evolutionary fate of de novo genes. Trends Genet. 2015, 31, 215–219. [Google Scholar] [CrossRef]
- Weisman, C.M.; Murray, A.W.; Eddy, S.R. Many, but not all, lineage-specific genes can be explained by homology detection failure. PLoS Biol. 2020, 18, e3000862. [Google Scholar] [CrossRef]
- Arendsee, Z.W.; Li, L.; Wurtele, E.S. Coming of age: Orphan genes in plants. Trends Plant Sci. 2014, 19, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Liang, Y.; Zhan, Z.; Li, X.; Piao, Z. Development of a sinitic clubroot differential set for the pathotype classification of Plasmodiophora brassicae. Front. Plant Sci. 2020, 11, 568771. [Google Scholar] [CrossRef]
- Kong, L.; Liu, J.; Zhang, W.; Li, X.; Zhang, Y.; Chen, X.; Zhan, Z.; Piao, Z. Genome-wide identification and characterization of the trehalose-6-phosphate synthetase gene family in Chinese cabbage (Brassica rapa) and Plasmodiophora brassicae during their interaction. Int. J. Mol. Sci. 2023, 24, 929. [Google Scholar] [CrossRef] [PubMed]
- Siemens, J.; Nagel, M.; Ludwig-Müller, J.; Sacristán, M.D. The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: Parameters for disease quantification and screening of mutant lines. J. Phytopathol. 2002, 150, 592–605. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Zhou, Y.; Sun, Y.; Li, X. Brassica-Specific Orphan Gene CROG1 Confers Clubroot Resistance in Arabidopsis via Phenylpropanoid Pathway Activation. Plants 2025, 14, 2683. https://doi.org/10.3390/plants14172683
Zheng J, Zhou Y, Sun Y, Li X. Brassica-Specific Orphan Gene CROG1 Confers Clubroot Resistance in Arabidopsis via Phenylpropanoid Pathway Activation. Plants. 2025; 14(17):2683. https://doi.org/10.3390/plants14172683
Chicago/Turabian StyleZheng, Jingyi, Yana Zhou, Yan Sun, and Xiaonan Li. 2025. "Brassica-Specific Orphan Gene CROG1 Confers Clubroot Resistance in Arabidopsis via Phenylpropanoid Pathway Activation" Plants 14, no. 17: 2683. https://doi.org/10.3390/plants14172683
APA StyleZheng, J., Zhou, Y., Sun, Y., & Li, X. (2025). Brassica-Specific Orphan Gene CROG1 Confers Clubroot Resistance in Arabidopsis via Phenylpropanoid Pathway Activation. Plants, 14(17), 2683. https://doi.org/10.3390/plants14172683





